Rousettus aegyptiacus Fruit Bats Do Not Support Productive Replication of Cedar Virus upon Experimental Challenge
Abstract
1. Introduction
2. Materials and Methods
2.1. CedV Acquisition and Propagation
2.2. R. aegyptiacus Challenge Experiment
2.3. Total RNA Extraction and Detection of Viral RNA by Real-Time RTq-PCR
2.4. Monitoring of Body Core Temperature and Locomotor Activity Using Data Loggers
2.5. Serological Analysis
3. Results
3.1. Clinical Signs and Virological Analysis
3.2. Serology
3.3. Body Temperature and Locomotor Activity of Selected Individuals
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yob, J.M.; Field, H.; Rashdi, A.M.; Morrissy, C.; van der Heide, B.; Rota, P.; bin Adzhar, A.; White, J.; Daniels, P.; Jamaluddin, A.; et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001, 7, 439–441. [Google Scholar] [CrossRef]
- Selvey, L.A.; Wells, R.M.; McCormack, J.G.; Ansford, A.J.; Murray, K.; Rogers, R.J.; Lavercombe, P.S.; Selleck, P.; Sheridan, J.W. Infection of humans and horses by a newly described morbillivirus. Med. J. Aust. 1995, 162, 642–645. [Google Scholar] [CrossRef]
- Taylor, J.; Thompson, K.; Annand, E.J.; Massey, P.D.; Bennett, J.; Eden, J.S.; Horsburgh, B.A.; Hodgson, E.; Wood, K.; Kerr, J.; et al. Novel variant Hendra virus genotype 2 infection in a horse in the greater Newcastle region, New South Wales, Australia. One Health 2022, 15, 100423. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.B.; Goh, K.J.; Wong, K.T.; Kamarulzaman, A.; Tan, P.S.; Ksiazek, T.G.; Zaki, S.R.; Paul, G.; Lam, S.K.; Tan, C.T. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 1999, 354, 1257–1259. [Google Scholar] [CrossRef] [PubMed]
- Satter, S.M.; Aquib, W.R.; Sultana, S.; Sharif, A.R.; Nazneen, A.; Alam, M.R.; Siddika, A.; Akther Ema, F.; Chowdhury, K.I.A.; Alam, A.N.; et al. Tackling a global epidemic threat: Nipah surveillance in Bangladesh, 2006–2021. PLoS Negl. Trop. Dis. 2023, 17, e0011617. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Deb, N.; Roy, P.; Jaiswal, V.; Sah, S.; Pandey, Y.; Reddy Edara, R.S.; Mohanty, A.; Henao-Martínez, A.F.; Sah, R. Recent Nipah virus outbreak in India: Lessons and imperatives. Ther. Adv. Infect. Dis. 2023, 10, 20499361231208535. [Google Scholar] [CrossRef]
- Ching, P.K.; de los Reyes, V.C.; Sucaldito, M.N.; Tayag, E.; Columna-Vingno, A.B.; Malbas, F.F., Jr.; Bolo, G.C., Jr.; Sejvar, J.J.; Eagles, D.; Playford, G.; et al. Outbreak of henipavirus infection, Philippines, 2014. Emerg. Infect. Dis. 2015, 21, 328–331. [Google Scholar] [CrossRef]
- Wong, K.T.; Shieh, W.J.; Kumar, S.; Norain, K.; Abdullah, W.; Guarner, J.; Goldsmith, C.S.; Chua, K.B.; Lam, S.K.; Tan, C.T.; et al. Nipah virus infection: Pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am. J. Pathol. 2002, 161, 2153–2167. [Google Scholar] [CrossRef]
- Enserink, M. Emerging diseases. Malaysian researchers trace Nipah virus outbreak to bats. Science 2000, 289, 518–519. [Google Scholar] [CrossRef]
- Clayton, B.A.; Wang, L.F.; Marsh, G.A. Henipaviruses: An updated review focusing on the pteropid reservoir and features of transmission. Zoonoses Public Health 2013, 60, 69–83. [Google Scholar] [CrossRef]
- Halpin, K.; Hyatt, A.D.; Fogarty, R.; Middleton, D.; Bingham, J.; Epstein, J.H.; Rahman, S.A.; Hughes, T.; Smith, C.; Field, H.E.; et al. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: A comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 2011, 85, 946–951. [Google Scholar] [CrossRef]
- Li, H.; Kim, J.V.; Pickering, B.S. Henipavirus zoonosis: Outbreaks, animal hosts and potential new emergence. Front. Microbiol. 2023, 14, 1167085. [Google Scholar] [CrossRef] [PubMed]
- Marsh, G.A.; de Jong, C.; Barr, J.A.; Tachedjian, M.; Smith, C.; Middleton, D.; Yu, M.; Todd, S.; Foord, A.J.; Haring, V.; et al. Cedar virus: A novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012, 8, e1002836. [Google Scholar] [CrossRef] [PubMed]
- Pryce, R.; Azarm, K.; Rissanen, I.; Harlos, K.; Bowden, T.A.; Lee, B. A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus. Life Sci. Alliance 2020, 3, e201900578. [Google Scholar] [CrossRef] [PubMed]
- Lieu, K.G.; Marsh, G.A.; Wang, L.F.; Netter, H.J. The non-pathogenic Henipavirus Cedar paramyxovirus phosphoprotein has a compromised ability to target STAT1 and STAT2. Antivir. Res. 2015, 124, 69–76. [Google Scholar] [CrossRef]
- Schountz, T.; Campbell, C.; Wagner, K.; Rovnak, J.; Martellaro, C.; DeBuysscher, B.L.; Feldmann, H.; Prescott, J. Differential Innate Immune Responses Elicited by Nipah Virus and Cedar Virus Correlate with Disparate In Vivo Pathogenesis in Hamsters. Viruses 2019, 11, 291. [Google Scholar] [CrossRef]
- Huaman, C.; Clouse, C.; Rader, M.; Yan, L.; Bai, S.; Gunn, B.M.; Amaya, M.; Laing, E.D.; Broder, C.C.; Schaefer, B.C. An in vivo BSL-2 model for henipavirus infection based on bioluminescence imaging of recombinant Cedar virus replication in mice. Front. Chem. Biol. 2024, 3, 1363498. [Google Scholar] [CrossRef]
- Laing, E.D.; Amaya, M.; Navaratnarajah, C.K.; Feng, Y.R.; Cattaneo, R.; Wang, L.F.; Broder, C.C. Rescue and characterization of recombinant cedar virus, a non-pathogenic Henipavirus species. Virol. J. 2018, 15, 56. [Google Scholar] [CrossRef]
- Laing, E.D.; Navaratnarajah, C.K.; Cheliout Da Silva, S.; Petzing, S.R.; Xu, Y.; Sterling, S.L.; Marsh, G.A.; Wang, L.F.; Amaya, M.; Nikolov, D.B.; et al. Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus. Proc. Natl. Acad. Sci. USA 2019, 116, 20707–20715. [Google Scholar] [CrossRef]
- Amman, B.R.; Jones, M.E.; Sealy, T.K.; Uebelhoer, L.S.; Schuh, A.J.; Bird, B.H.; Coleman-McCray, J.D.; Martin, B.E.; Nichol, S.T.; Towner, J.S. Oral shedding of Marburg virus in experimentally infected Egyptian fruit bats (Rousettus aegyptiacus). J. Wildl. Dis. 2015, 51, 113–124. [Google Scholar] [CrossRef]
- Schlottau, K.; Eggerbauer, E.; Freuling, C.M.; Beer, M.; Muller, T.; Hoffmann, B. Rapid molecular species identification of indigenous bats from Germany for surveillance purposes. Infect. Genet. Evol. 2020, 78, 104140. [Google Scholar] [CrossRef]
- Schuh, A.J.; Amman, B.R.; Guito, J.C.; Graziano, J.C.; Sealy, T.K.; Kirejczyk, S.G.M.; Towner, J.S. Natural reservoir Rousettus aegyptiacus bat host model of orthonairovirus infection identifies potential zoonotic spillover mechanisms. Sci. Rep. 2022, 12, 20936. [Google Scholar] [CrossRef] [PubMed]
- Halwe, N.J.; Gorka, M.; Hoffmann, B.; Rissmann, M.; Breithaupt, A.; Schwemmle, M.; Beer, M.; Kandeil, A.; Ali, M.A.; Kayali, G.; et al. Egyptian Fruit Bats (Rousettus aegyptiacus) Were Resistant to Experimental Inoculation with Avian-Origin Influenza A Virus of Subtype H9N2, But Are Susceptible to Experimental Infection with Bat-Borne H9N2 Virus. Viruses 2021, 13, 672. [Google Scholar] [CrossRef] [PubMed]
- Seifert, S.N.; Letko, M.C.; Bushmaker, T.; Laing, E.D.; Saturday, G.; Meade-White, K.; van Doremalen, N.; Broder, C.C.; Munster, V.J. Rousettus aegyptiacus Bats Do Not Support Productive Nipah Virus Replication. J. Infect. Dis. 2020, 221, S407–S413. [Google Scholar] [CrossRef]
- Markotter, W.; Geldenhuys, M.; Jansen van Vuren, P.; Kemp, A.; Mortlock, M.; Mudakikwa, A.; Nel, L.; Nziza, J.; Paweska, J.; Weyer, J. Paramyxo- and Coronaviruses in Rwandan Bats. Trop. Med. Infect. Dis. 2019, 4, 99. [Google Scholar] [CrossRef]
- Mortlock, M.; Geldenhuys, M.; Dietrich, M.; Epstein, J.H.; Weyer, J.; Pawęska, J.T.; Markotter, W. Seasonal shedding patterns of diverse henipavirus-related paramyxoviruses in Egyptian rousette bats. Sci. Rep. 2021, 11, 24262. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Duengkae, P.; Chaiyes, A.; Kaewpom, T.; Rodpan, A.; Yingsakmongkon, S.; Petcharat, S.; Phengsakul, P.; Maneeorn, P.; Hemachudha, T. Longitudinal study of age-specific pattern of coronavirus infection in Lyle’s flying fox (Pteropus lylei) in Thailand. Virol. J. 2018, 15, 38. [Google Scholar] [CrossRef]
- Dietrich, M.; Kearney, T.; Seamark, E.C.J.; Paweska, J.T.; Markotter, W. Synchronized shift of oral, faecal and urinary microbiotas in bats and natural infection dynamics during seasonal reproduction. R Soc. Open Sci. 2018, 5, 180041. [Google Scholar] [CrossRef] [PubMed]
- Pickering, B.S.; Collignon, B.; Smith, G.; Marszal, P.; Kobinger, G.; Weingartl, H.M. Detection of Zaire ebolavirus in swine: Assay development and optimization. Transbound Emerg. Dis. 2018, 65, 77–84. [Google Scholar] [CrossRef]
- Fischer, K.; Diederich, S.; Smith, G.; Reiche, S.; Pinho Dos Reis, V.; Stroh, E.; Groschup, M.H.; Weingartl, H.M.; Balkema-Buschmann, A. Indirect ELISA based on Hendra and Nipah virus proteins for the detection of henipavirus specific antibodies in pigs. PLoS ONE 2018, 13, e0194385. [Google Scholar] [CrossRef]
- Fischer, K.; dos Reis, V.P.; Finke, S.; Sauerhering, L.; Stroh, E.; Karger, A.; Maisner, A.; Groschup, M.H.; Diederich, S.; Balkema-Buschmann, A. Expression, characterisation and antigenicity of a truncated Hendra virus attachment protein expressed in the protozoan host Leishmania tarentolae. J. Virol. Methods 2016, 228, 48–54. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Balkema-Buschmann, A.; Fischer, K.; McNabb, L.; Diederich, S.; Singanallur, N.B.; Ziegler, U.; Keil, G.M.; Kirkland, P.D.; Penning, M.; Sadeghi, B.; et al. Serological Hendra Virus Diagnostics Using an Indirect ELISA-Based DIVA Approach with Recombinant Hendra G and N Proteins. Microorganisms 2022, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
- Rissmann, M.; Friedrichs, V.; Kley, N.; Straube, M.; Sadeghi, B.; Balkema-Buschmann, A. Baseline of Physiological Body Temperature and Hematological Parameters in Captive Rousettus aegyptiacus and Eidolon helvum Fruit Bats. Front. Physiol. 2022, 13, 910157. [Google Scholar] [CrossRef] [PubMed]
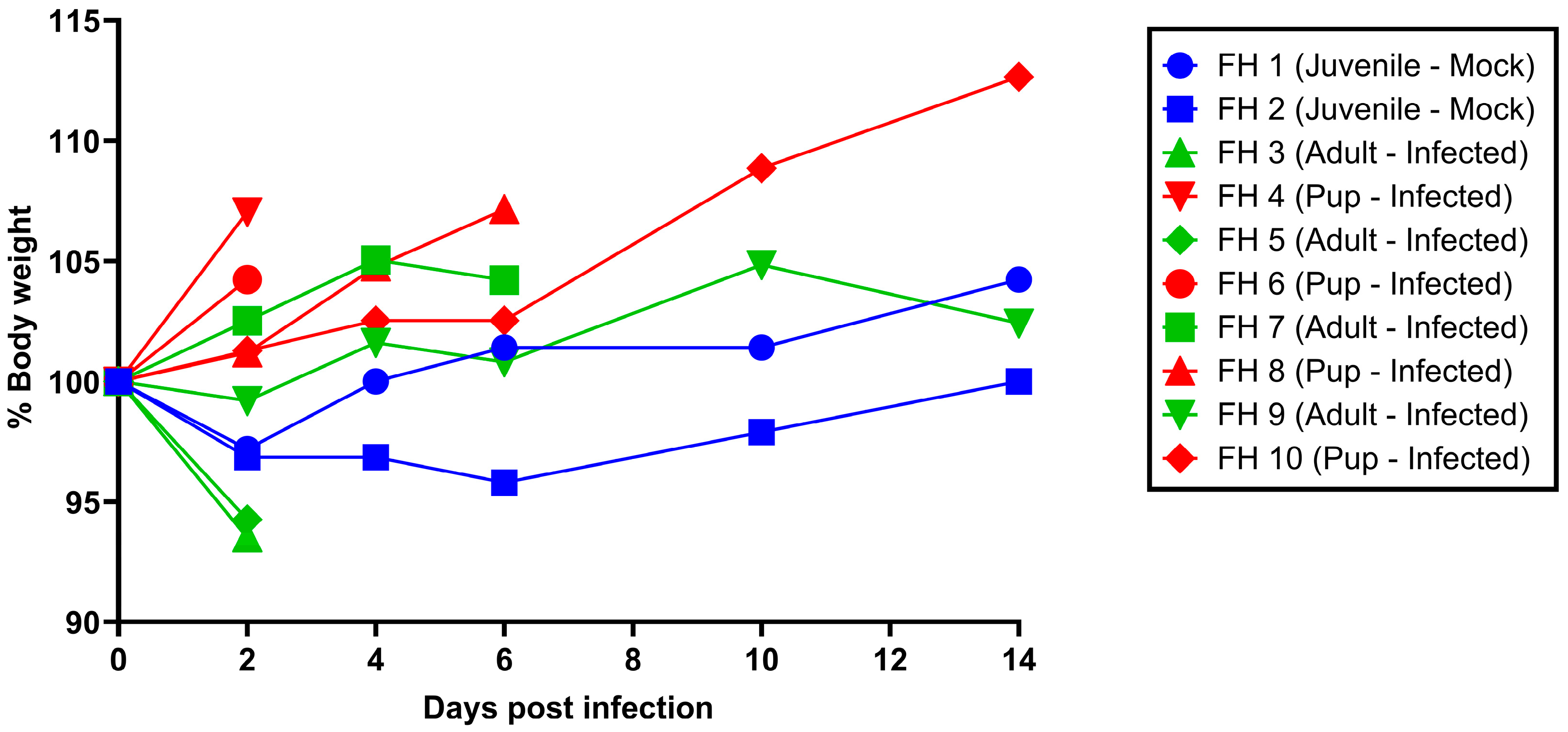
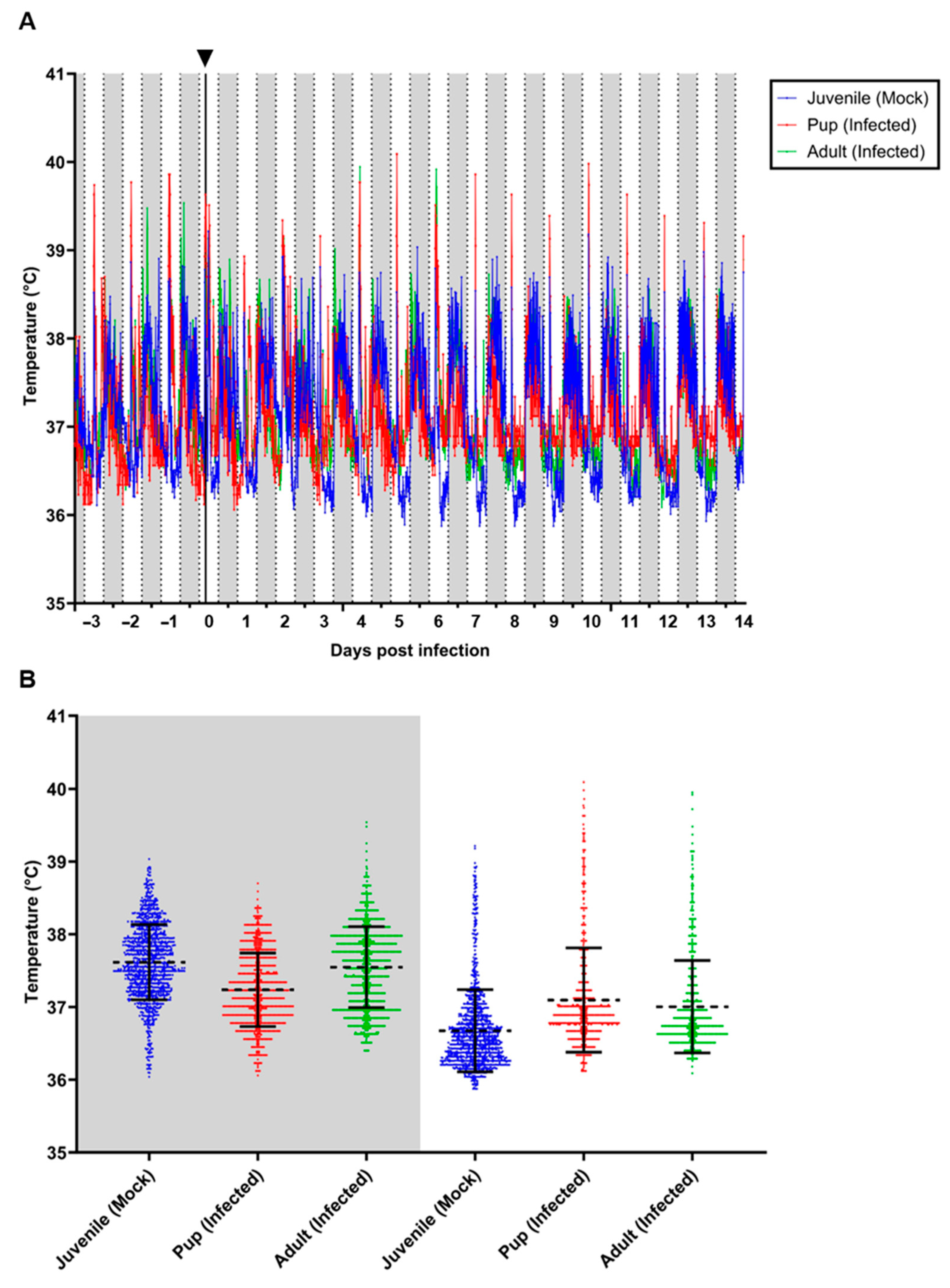
| Animal | Age Group | dpi | Sample | Ct-Value RT-qPCR |
|---|---|---|---|---|
| FH 3 | adult | 2 | Nasal lavage Anal swab Organ: nasal conchae | 36.97 35.11 39.2 |
| FH 4 | pup of FH 3 | 2 | Organ: kidney | 37.6 |
| FH 5 | adult | 2 | Organ: nasal conchae | 36.9 |
| FH 6 | pup of FH 5 | 2 | Oral swab Nasal lavage | 38.72 39.13 |
| Animal ID and Status | PP CedV G Protein | PP CedV N Protein |
|---|---|---|
| FH 1 (mock-inoculated juvenile) | 4.0 | 3.5 |
| FH 2 (mock-inoculated juvenile) | 2.9 | 3.4 |
| FH 3 (inoculated adult; necropsy 2 dpi) | 3.1 | 3.3 |
| FH 4 (inoculated pup of FH 3; necropsy 2 dpi) | 3.6 | 3.7 |
| FH 5 (inoculated adult; necropsy 2 dpi) | 4.6 | 4.1 |
| FH 6 (inoculated pup of FH 5; necropsy 2 dpi) | 4.3 | 6.2 |
| FH 7 (inoculated adult; necropsy 6 dpi) | 3.1 | 3.7 |
| FH 8 (inoculated pup of FH 7; necropsy 6 dpi) | 3.3 | 3.6 |
| FH 9 (inoculated adult; necropsy 14 dpi) | 4.2 | 3.5 |
| FH 10 (inoculated pup of FH 9; necropsy 14 dpi) | 5.1 | 2.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohl, B.-P.; Diederich, S.; Fischer, K.; Balkema-Buschmann, A. Rousettus aegyptiacus Fruit Bats Do Not Support Productive Replication of Cedar Virus upon Experimental Challenge. Viruses 2024, 16, 1359. https://doi.org/10.3390/v16091359
Mohl B-P, Diederich S, Fischer K, Balkema-Buschmann A. Rousettus aegyptiacus Fruit Bats Do Not Support Productive Replication of Cedar Virus upon Experimental Challenge. Viruses. 2024; 16(9):1359. https://doi.org/10.3390/v16091359
Chicago/Turabian StyleMohl, Björn-Patrick, Sandra Diederich, Kerstin Fischer, and Anne Balkema-Buschmann. 2024. "Rousettus aegyptiacus Fruit Bats Do Not Support Productive Replication of Cedar Virus upon Experimental Challenge" Viruses 16, no. 9: 1359. https://doi.org/10.3390/v16091359
APA StyleMohl, B.-P., Diederich, S., Fischer, K., & Balkema-Buschmann, A. (2024). Rousettus aegyptiacus Fruit Bats Do Not Support Productive Replication of Cedar Virus upon Experimental Challenge. Viruses, 16(9), 1359. https://doi.org/10.3390/v16091359







