The Role of Late Presenters in HIV-1 Transmission Clusters in Europe
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Group
2.2. Drug Resistance Analysis and Subtyping
2.3. Transmission Cluster (TC) Identification
2.4. Study Variables
- Migration status—Based on country of origin and country of follow-up (if country of origin and country of follow-up are the same, then the patient was classified as native; otherwise, as a migrant);
- Age at resistance test—Based on the difference between year of birth and date of the first drug resistance test;
- Region of origin—Based on country of origin;
- Recentness of infection—Based on ambiguity rate of genomic sequences. We defined chronic infection as the ambiguity rate being higher than 0.45%; otherwise, recent infection was defined, as previously described [21];
- LP vs. NLP at HIV diagnosis—Based on CD4 count, LPs were defined as individuals with a baseline CD4 count ≤ 350 cells/mm3 and NLPs were defined as individuals with a baseline CD4 count > 350 cells/mm3 [7].
2.5. Statistical Analysis
3. Results
3.1. Characteristics of European Population of Individuals Living with HIV-1
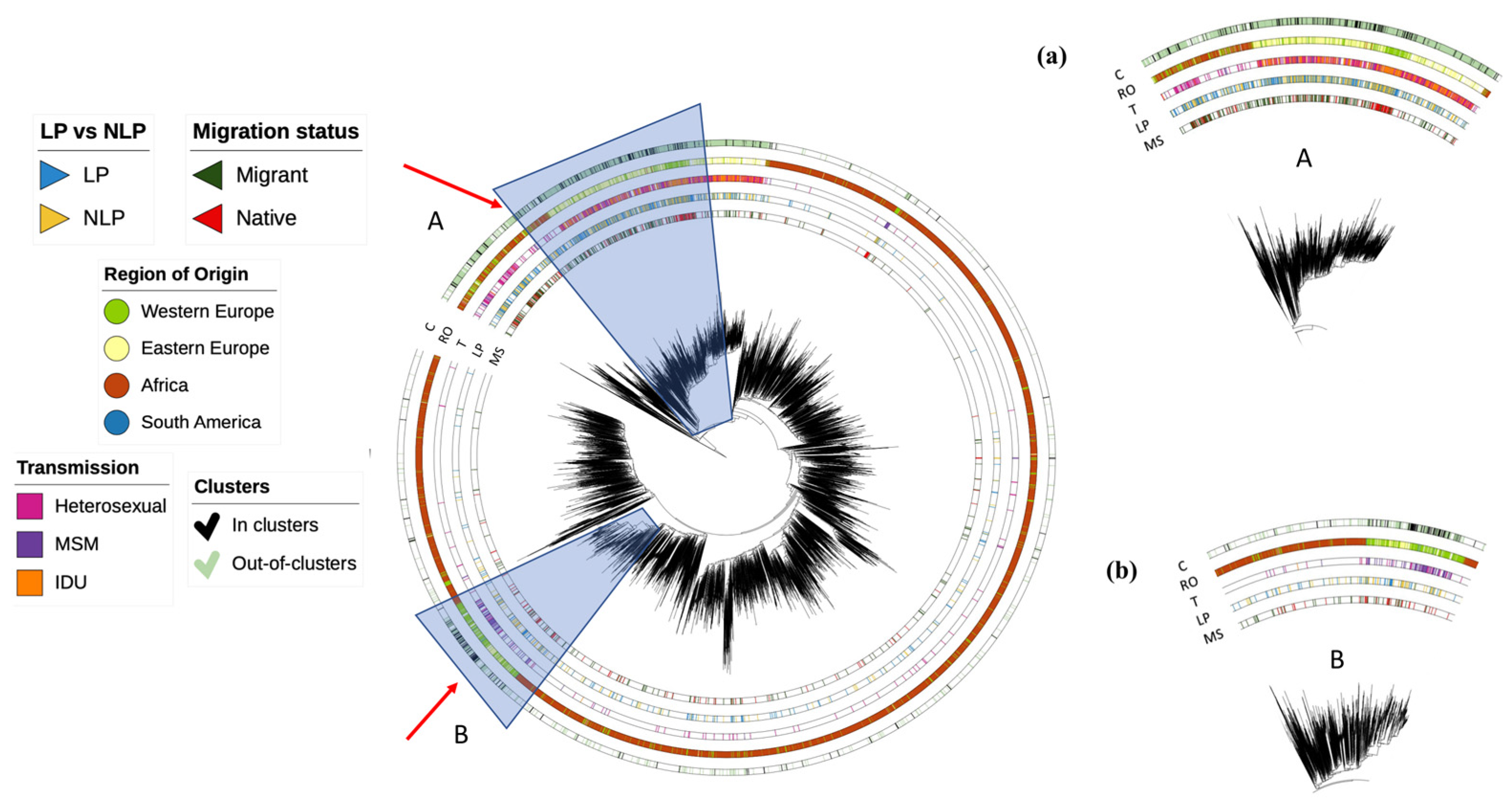
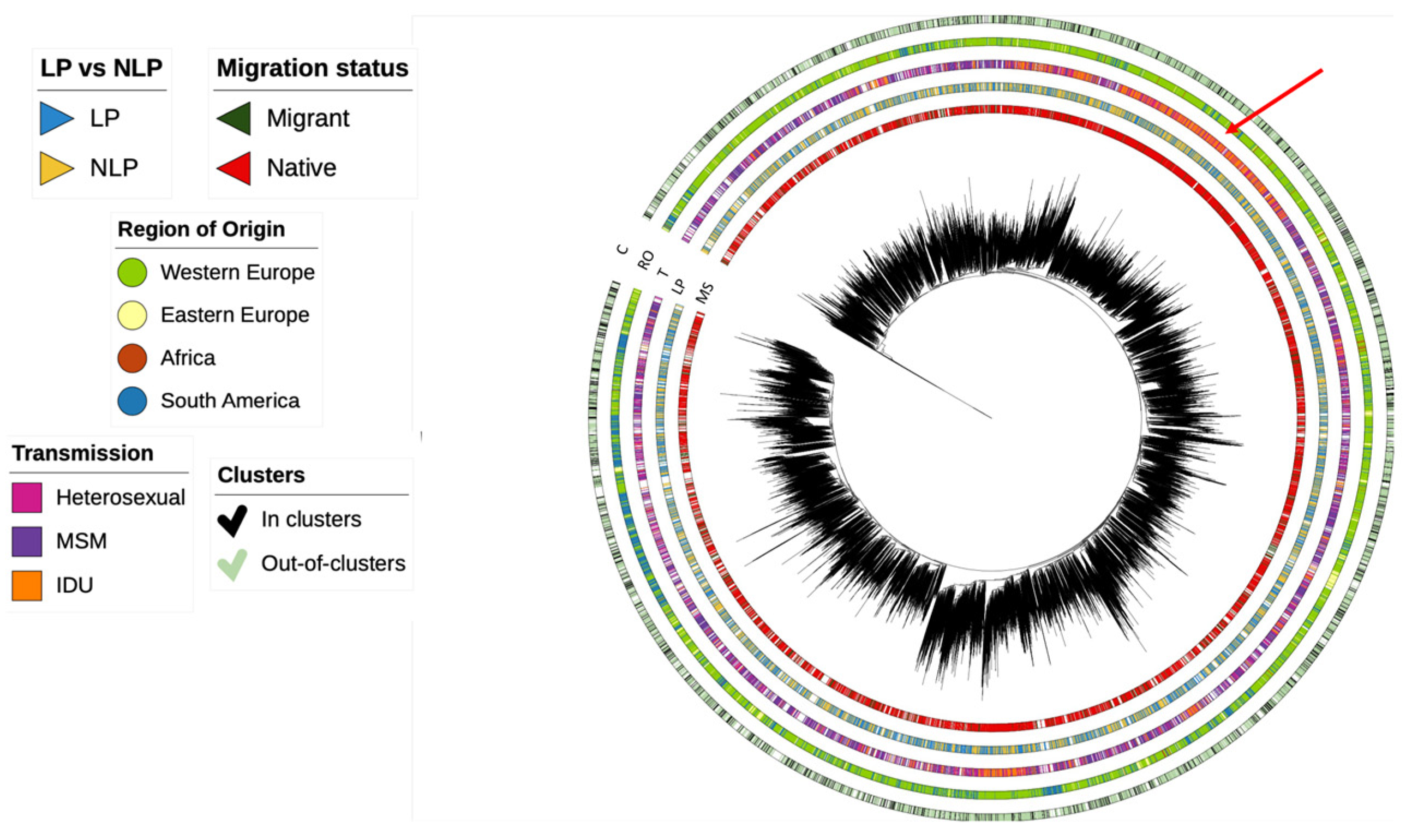
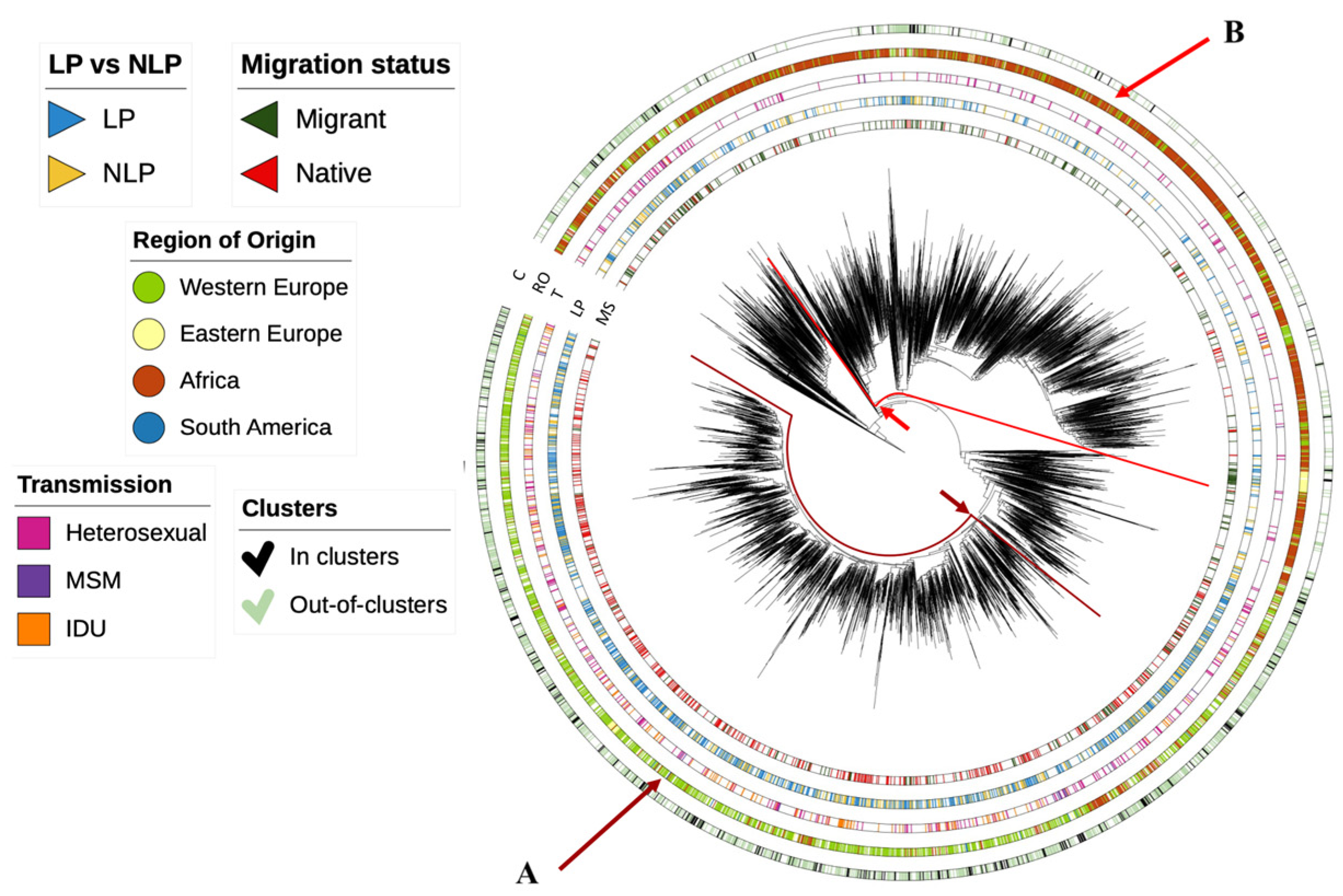
3.2. Dynamics of Subtype A HIV-1 Epidemic in Europe
3.3. Dynamics of Subtype B HIV-1 Epidemic in Europe
3.4. Dynamics of Subtype G HIV-1 Epidemic in Europe
3.5. Determinants Associated with Transmission Clusters of HIV-1 in Europe for Different Subtypes
3.6. Size of Transmission Clusters
3.7. Transmission Clusters in LP vs. NLP
4. Discussion
5. Conclusions
Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global HIV & AIDS Statistics—Fact Sheet|UNAIDS. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on 25 January 2022).
- Grabowski, M.K.; Herbeck, J.T.; Poon, A.F.Y. Genetic Cluster Analysis for HIV Prevention. Curr. HIV/AIDS Rep. 2018, 15, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Abecasis, A.B.; Wensing, A.M.; Paraskevis, D.; Vercauteren, J.; Theys, K.; Van de Vijver, D.A.; Albert, J.; Asjö, B.; Balotta, C.; Beshkov, D.; et al. HIV-1 Subtype Distribution and Its Demographic Determinants in Newly Diagnosed Patients in Europe Suggest Highly Compartmentalized Epidemics. Retrovirology 2013, 10, 7. Available online: https://retrovirology.biomedcentral.com/articles/10.1186/1742-4690-10-7 (accessed on 14 January 2013). [CrossRef] [PubMed]
- Brenner, B.G.; Roger, M.; Routy, J.; Moisi, D.; Ntemgwa, M.; Matte, C.; Baril, J.; Thomas, R.; Rouleau, D.; Bruneau, J.; et al. High Rates of Forward Transmission Events after Acute/Early HIV-1 Infection. J. Infect. Dis. 2007, 195, 951–959. Available online: https://academic.oup.com/jid/article-lookup/doi/10.1086/512088 (accessed on 1 April 2007). [CrossRef]
- Factors That Increase HIV Risk|HIV Transmission|HIV Basics|HIV/AIDS|CDC. Available online: https://www.cdc.gov/hiv/basics/hiv-transmission/increase-hiv-risk.html (accessed on 5 May 2023).
- Miranda, M.N.S.; Pingarilho, M.; Pimentel, V.; Martins, M.D.R.O.; Vandamme, A.-M.; Bobkova, M.; Böhm, M.; Seguin-Devaux, C.; Paredes, R.; Rubio, R.; et al. Determinants of HIV-1 Late Presentation in Patients Followed in Europe. Pathogens 2021, 10, 835. Available online: https://www.mdpi.com/2076-0817/10/7/835 (accessed on 2 July 2021). [CrossRef]
- Antinori, A.; Coenen, T.; Costagiola, D.; Dedes, N.; Ellefson, M.; Gatell, J.; Girardi, E.; Johnson, M.; Kirk, O.; Lundgren, J.; et al. Late Presentation of HIV Infection: A Consensus Definition. HIV Med. 2011, 12, 61–64. Available online: https://onlinelibrary.wiley.com/doi/10.1111/j.1468-1293.2010.00857.x (accessed on 3 December 2010). [CrossRef] [PubMed]
- Late Presentation Working Groups in EuroSIDA and COHERE. Estimating the Burden of HIV Late Presentation and Its Attributable Morbidity and Mortality Across Europe 2010–2016. BMC Infect. Dis. 2020, 20, 728. Available online: https://bmcinfectdis.biomedcentral.com/articles/10.1186/s12879-020-05261-7 (accessed on 7 October 2020).
- Rava, M.; Domínguez-Domínguez, L.; Bisbal, O.; López-Cortés, L.F.; Busca, C.; Antela, A.; González-Ruano, P.; Hernández, C.; Iribarren, J.-A.; Rubio, R.; et al. Late Presentation for HIV Remains a Major Health Issue in Spain: Results from a Multicenter Cohort Study, 2004–2018. PLoS ONE 2021, 16, e0249864. [Google Scholar] [CrossRef]
- Pineda-Peña, A.-C.; Pingarilho, M.; Li, G.; Vrancken, B.; Libin, P.; Gomes, P.; Camacho, R.J.; Theys, K.; Abecasis, A.B.; on behalf of the Portuguese HIV-1 Resistance Study Group. Drivers of HIV-1 Transmission: The Portuguese Case. PLoS ONE 2019, 14, e0218226. [Google Scholar] [CrossRef]
- Euresist Data Analysis—Database. Available online: http://engine.euresist.org/database/ (accessed on 4 January 2021).
- Bennett, D.E.; Camacho, R.J.; Otelea, D.; Kuritzkes, D.R.; Fleury, H.; Kiuchi, M.; Heneine, W.; Kantor, R.; Jordan, M.R.; Schapiro, J.M.; et al. Drug Resistance Mutations for Surveillance of Transmitted HIV-1 Drug-Resistance: 2009 Update. PLoS ONE 2009, 4, e4724. [Google Scholar] [CrossRef]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.-M.; et al. Genome Detective: An automated system for virus identification from high-throughput sequencing data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef]
- Struck, D.; Lawyer, G.; Ternes, A.M.; Schmit, J.C.; Bercoff, D.P. COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014, 42, e144. [Google Scholar] [CrossRef]
- Pond, S.L.K.; Posada, D.; Stawiski, E.; Chappey, C.; Poon, A.F.; Hughes, G.; Fearnhill, E.; Gravenor, M.B.; Brown, A.J.L.; Frost, S.D. An Evolutionary Model-Based Algorithm for Accurate Phylogenetic Breakpoint Mapping and Subtype Prediction in HIV-1. PLoS Comput. Biol. 2009, 5, e1000581. [Google Scholar] [CrossRef]
- Stoesser, G.; Griffith, M.; Griffith, O.L. HIV Sequence Database. In Dictionary of Bioinformatics and Computational Biology; John Wiley & Sons, Ltd.: Chichester, UK, 2004; pp. 52–61. Available online: https://onlinelibrary.wiley.com/doi/10.1002/9780471650126.dob0322.pub2 (accessed on 7 April 2014).
- Libin, P.J.K.; Deforche, K.; Abecasis, A.B.; Theys, K. VIRULIGN: Fast Codon-Correct Alignment and Annotation of Viral Genomes. Hancock, J., Editor. Bioinformatics 2019, 35, 1763–1765. Available online: https://academic.oup.com/bioinformatics/article/35/10/1763/5123354 (accessed on 8 October 2018). [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Ragonnet-Cronin, M.; Hodcroft, E.; Hué, S.; Fearnhill, E.; Delpech, V.; Brown, A.J.L.; Lycett, S. Automated Analysis of Phylogenetic Clusters. BMC Bioinform. 2013, 14, 317. Available online: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-14-317 (accessed on 6 November 2013). [CrossRef]
- Andersson, E.; Shao, W.; Bontell, I.; Cham, F.; Cuong, D.D.; Wondwossen, A.; Morris, L.; Hunt, G.; Sönnerborg, A.; Bertagnolio, S.; et al. Evaluation of Sequence Ambiguities of the HIV-1 Pol Gene as a Method to Identify Recent HIV-1 Infection in Transmitted Drug Resistance Surveys. Infect. Genet Evol. 2013, 18, 125–131. Available online: https://linkinghub.elsevier.com/retrieve/pii/S156713481300141X (accessed on 11 April 2013). [CrossRef]
- Paraskevis, D.; Beloukas, A.; Stasinos, K.; Pantazis, N.; de Mendoza, C.; Bannert, N.; Meyer, L.; Zangerle, R.; Gill, J.; Prins, M.; et al. HIV-1 Molecular Transmission Clusters in Nine European Countries and Canada: Association with Demographic and Clinical Factors. BMC Med. 2019, 17, 4. Available online: https://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-018-1241-1 (accessed on 8 January 2019). [CrossRef]
- Petersen, A.; Cowan, S.A.; Nielsen, J.; Fischer, T.K.; Fonager, J. Characterisation of HIV-1 Transmission Clusters and Drug-resistant Mutations in Denmark, 2004 to 2016. Eurosurveillance 2018, 23, 1–9. Available online: https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2018.23.44.1700633 (accessed on 1 November 2018). [CrossRef]
- Delgado, E.; Benito, S.; Montero, V.; Cuevas, M.T.; Fernández-García, A.; Sánchez-Martínez, M.; García-Bodas, E.; Díez-Fuertes, F.; Gil, H.; Cañada, J.; et al. Diverse Large HIV-1 Non-subtype B Clusters Are Spreading Among Men Who Have Sex With Men in Spain. Front. Microbiol. 2019, 10, 1–19. Available online: https://www.frontiersin.org/article/10.3389/fmicb.2019.00655/full (accessed on 3 April 2019). [CrossRef]
- Hanke, K.; Faria, N.R.; Kühnert, D.; Yousef, K.P.; Hauser, A.; Meixenberger, K.; Hofmann, A.; Bremer, V.; Bartmeyer, B.; Pybus, O.; et al. Reconstruction of the Genetic History and the Current Spread of HIV-1 Subtype A in Germany. J. Virol. 2019, 93, e02238-18. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Bozzi, G.; Franzetti, M.; Binda, F.; Simonetti, F.R.; De Luca, A.; Micheli, V.; Meraviglia, P.; Bagnarelli, P.; Di Biagio, A.; et al. HIV-1 A1 Subtype Epidemic in Italy Originated from Africa and Eastern Europe and Shows a High Frequency of Transmission Chains Involving Intravenous Drug Users. PLoS ONE 2016, 11, e0146097. [Google Scholar] [CrossRef] [PubMed]
- Pingarilho, M.; Pimentel, V.; Miranda, M.N.S.; Silva, A.R.; Diniz, A.; Ascenção, B.B.; Piñeiro, C.; Koch, C.; Rodrigues, C.; Caldas, C.; et al. HIV-1-Transmitted Drug Resistance and Transmission Clusters in Newly Diagnosed Patients in Portugal Between 2014 and 2019. Front. Microbiol. 2022, 13, 1220. Available online: https://www.frontiersin.org/article/10.3389/fmicb.2022.823208 (accessed on 25 April 2022). [CrossRef] [PubMed]
- Kostaki, E.G.; Gova, M.; Adamis, G.; Xylomenos, G.; Chini, M.; Mangafas, N.; Lazanas, M.; Metallidis, S.; Tsachouridou, O.; Papastamopoulos, V.; et al. A Nationwide Study about the Dispersal Pat. Terns of the Predominant HIV-1 Subtypes A1 and B in Greece: Inference of the Molecular Transmission Clusters. Viruses 2020, 12, 1183. Available online: https://www.mdpi.com/1999-4915/12/10/1183 (accessed on 19 October 2020). [CrossRef]
- MacCarthy, S.; Brignol, S.; Reddy, M.; Nunn, A.; Dourado, I. Late Presentation to HIV/AIDS Care in Brazil Among Men Who Self-Identify as Heterosexual. Rev. Saude Publica 2016, 50, 54. Available online: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0034-89102016000100233&lng=en&tlng=en (accessed on 16 August 2016). [CrossRef]
- Araújo, P.M.M.; Carvalho, A.; Pingarilho, M.; Faria, D.; Pinho, R.; Ferreira, J.; Proença, P.; Nunes, S.; Mouro, M.; Teófilo, E.; et al. Characterization of a large cluster of HIV-1 A1 infections detected in Portugal and connected to several Western European countries. Sci. Rep. 2019, 9, 7223. [Google Scholar] [CrossRef]
- Załęski, A.; Lembas, A.; Dyda, T.; Siwak, E.; Osińska, J.; Suchacz, M.; Stempkowska-Rejek, J.; Strycharz, M.; Orzechowska, J.; Wiercińska-Drapało, A. Changes in Primary HIV-1 Drug Resistance Due to War Migration from Eastern Europe. J. Immigr. Minor. Health 2023, 1–8. [Google Scholar] [CrossRef]

| Subtype A | Subtype B | Subtype G | |||||
|---|---|---|---|---|---|---|---|
| In Clusters/Out of Clusters | Final Model (Stepwise) | Final Model (Stepwise) | Final Model (Stepwise) | ||||
| aOR (95%CI) | p-Value | aOR (95%CI) | p-Value | aOR (95%CI) | p-Value | ||
| Sex | Female | Ref | Ref | Ref | Ref | Ref | Ref |
| Male | 4.72 (1.78–12.51) | 0.002 | 1.03 (0.72–1.47) | 0.877 | 1.74 (0.55–5.47) | 0.345 | |
| Transmission Route | Heterosexual | Ref | Ref | ||||
| MSM | 1.50 (1.17–1.92) | 0.001 | |||||
| IDU | 1.50 (0.92–2.46) | 0.106 | |||||
| Other | 1.54 (0.60–3.94) | 0.365 | |||||
| Migration Status | Migrant | Ref | Ref | ||||
| Native | 2.43 (1.02–5.77) | 0.044 | |||||
| Recentness of Infection | Chronic | Ref | Ref | Ref | Ref | Ref | Ref |
| Recent | 3.31 (1.37–8.01) | 0.008 | 2.29 (1.89–2.78) | <0.001 | 4.38 (1.38–13.65) | 0.012 | |
| LP vs. NLP | LP | Ref | Ref | ||||
| NLP | 1.99 (0.84–4.70) | 0.117 | |||||
| In Clusters | ||||
|---|---|---|---|---|
| LP | NLP | p-Value | p-Value Compared | |
| Subtype A | ||||
| Age at resistance test | 0.226 | |||
| <18 (n = 1) | 1 (100%) | 0 (0%) | - | |
| 19–30 (n = 12) | 3 (25.0%) | 9 (75.0%) | 0.014 | |
| 31–55 (n = 19) | 9 (47.4%) | 10 (52.6%) | 0.749 | |
| >56 (n = 6) | 4 (66.7%) | 2 (33.3%) | 0.247 | |
| Recentness of infection | 0.203 | |||
| Chronic (n = 18) | 10 (55.6%) | 8 (44.4%) | 0.501 | |
| Recent (n = 20) | 7 (35.6%) | 13 (64.4%) | 0.011 | |
| Viral load at diagnosis (log10 copies/mL) | 0.025 | |||
| ≤4.0 (n = 7) | 0 (0%) | 7 (100%) | - | |
| 4.1–5.0 (n = 13) | 7 (53.8%) | 6 (46.2%) | 0.698 | |
| ≥5.1 (n = 17) | 10 (58.8%) | 7 (41.2%) | 0.305 | |
| Subtype B | ||||
| Sex | 0.109 | |||
| Male (n = 913) | 451 (49.4%) | 462 (50.6%) | 0.608 | |
| Female (n = 104) | 60 (57.7%) | 44 (42.3%) | 0.026 | |
| Age at resistance test | <0.001 | |||
| <18 (n = 4) | 0 (0%) | 4 (100%) | - | |
| 19–30 (n = 245) | 89 (36.3%) | 156 (63.7%) | <0.001 | |
| 31–55 (n = 685) | 363 (53.0%) | 322 (47.0%) | 0.026 | |
| >56 (n = 88) | 61 (69.3%) | 27 (30.7%) | <0.001 | |
| Region of origin | 0.109 | |||
| Western Europe (n = 770) | 387 (50.3%) | 383 (49.7%) | 0.200 | |
| Eastern Europe (n = 31) | 17 (54.8%) | 14 (45.2%) | 0.450 | |
| Africa (n = 10) | 7 (70.0%) | 3 (30.0%) | 0.074 | |
| South America (n = 25) | 17 (68.0%) | 8 (32.0%) | 0.011 | |
| Other (n = 15) | 11 (73.3%) | 4 (26.7%) | 0.011 | |
| Recentness of infection | <0.001 | |||
| Chronic (n = 454) | 310 (68.3%) | 144 (31.7%) | <0.001 | |
| Recent (n = 578) | 206 (35.6%) | 372 (64.4%) | <0.001 | |
| Viral load at diagnosis (log10 copies/mL) | <0.001 | |||
| ≤4.0 (n = 228) | 69 (30.3%) | 159 (69.7%) | <0.001 | |
| 4.1–5.0 (n = 390) | 167 (42.8%) | 223 (57.2%) | <0.001 | |
| ≥5.1 (n = 397) | 270 (68.0%) | 127 (32.0%) | <0.001 | |
| Migrant status | 0.025 | |||
| Yes (n = 107) | 66 (61.7%) | 41 (38.3%) | 0.001 | |
| No (n = 744) | 373 (50.1%) | 371 (49.9%) | 0.939 | |
| Subtype G | ||||
| Sex | 0.046 | |||
| Male (n = 8) | 2 (25.0%) | 6 (75.0%) | 0.046 | |
| Female (n = 8) | 6 (75.0%) | 2 (25.0%) | 0.046 | |
| Region of origin | 0.054 | |||
| Western Europe (n = 6) | 2 (33.3%) | 4 (66.7%) | 0.247 | |
| Eastern Europe (n = 1) | 0 (0%) | 1 (100%) | - | |
| Africa (n = 5) | 5 (100%) | 0 (0%) | - | |
| South America (n = 25) | 0 (0%) | 1 (100%) | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miranda, M.N.S.; Pimentel, V.; Gomes, P.; Martins, M.d.R.O.; Seabra, S.G.; Kaiser, R.; Böhm, M.; Seguin-Devaux, C.; Paredes, R.; Bobkova, M.; et al. The Role of Late Presenters in HIV-1 Transmission Clusters in Europe. Viruses 2023, 15, 2418. https://doi.org/10.3390/v15122418
Miranda MNS, Pimentel V, Gomes P, Martins MdRO, Seabra SG, Kaiser R, Böhm M, Seguin-Devaux C, Paredes R, Bobkova M, et al. The Role of Late Presenters in HIV-1 Transmission Clusters in Europe. Viruses. 2023; 15(12):2418. https://doi.org/10.3390/v15122418
Chicago/Turabian StyleMiranda, Mafalda N. S., Victor Pimentel, Perpétua Gomes, Maria do Rosário O. Martins, Sofia G. Seabra, Rolf Kaiser, Michael Böhm, Carole Seguin-Devaux, Roger Paredes, Marina Bobkova, and et al. 2023. "The Role of Late Presenters in HIV-1 Transmission Clusters in Europe" Viruses 15, no. 12: 2418. https://doi.org/10.3390/v15122418





