Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid DNA
2.2. Cells
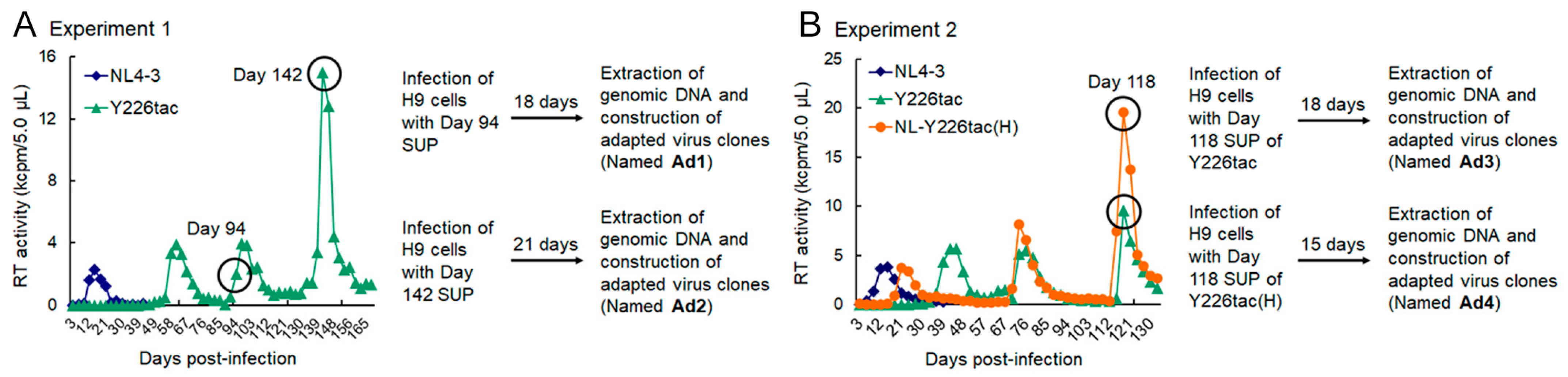
2.3. Adaptation Experiments
2.4. Replication Assays
2.5. Western Blotting Analysis
2.6. Semiquantitative RT-PCR Analysis of Splicing Products
3. Results
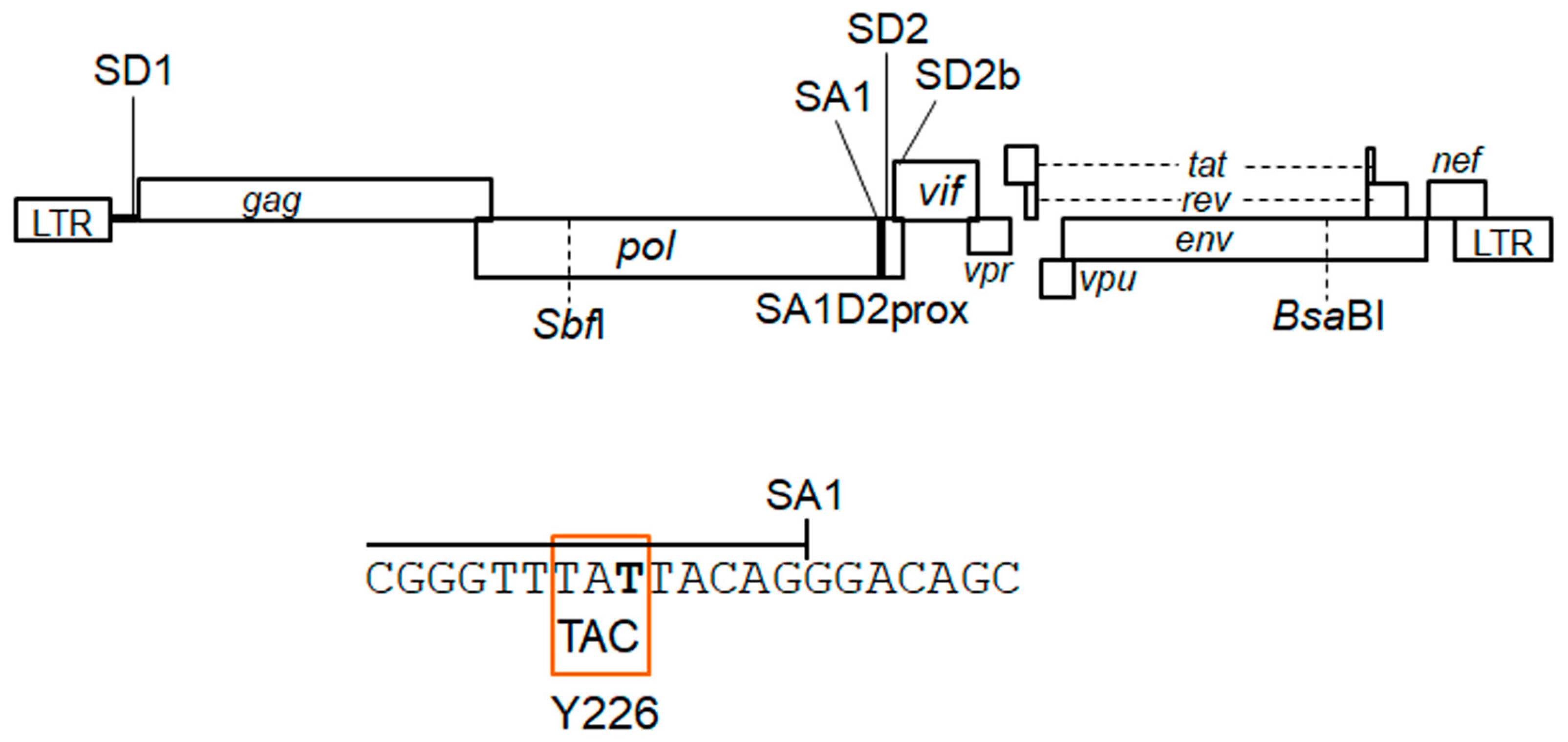
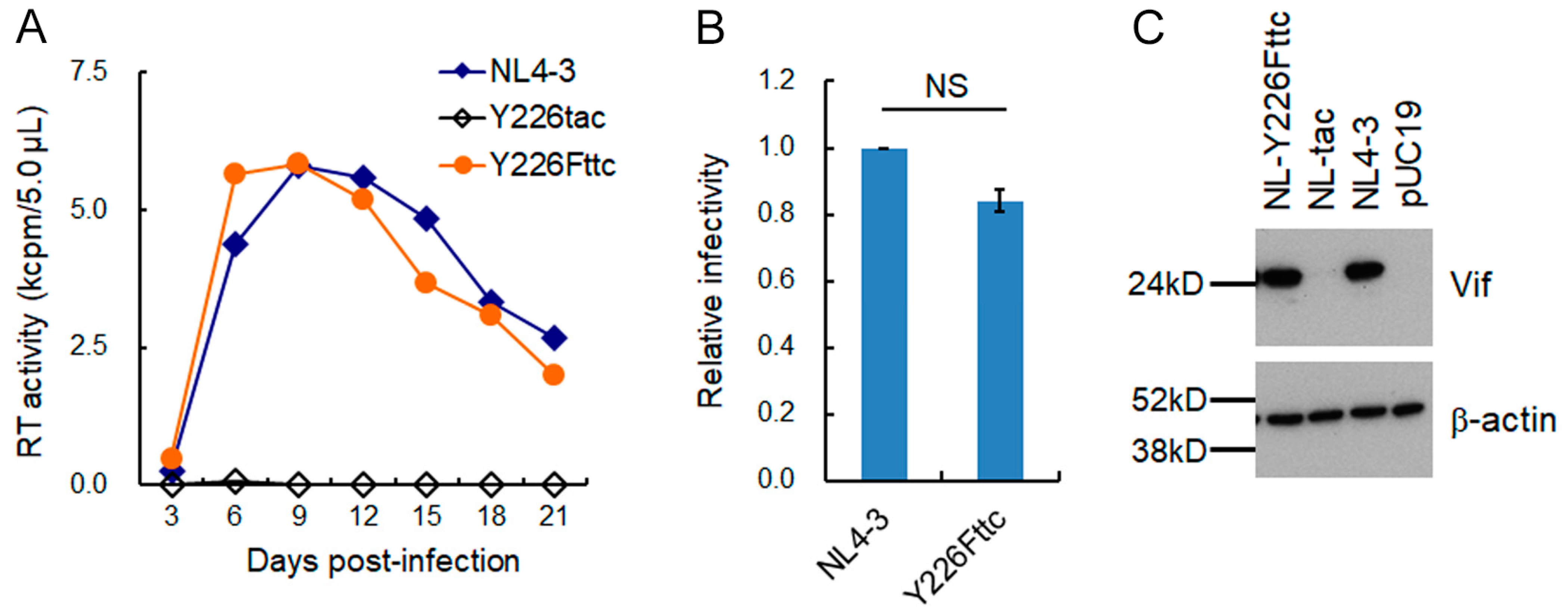
3.1. Extremely Low Vif-Type NL-Y226tac Acquired an Adaptive Mutation That Increases Vif Expression and Viral Replication Potential during a Long-Term Infection in High A3G-Expression Cells H9
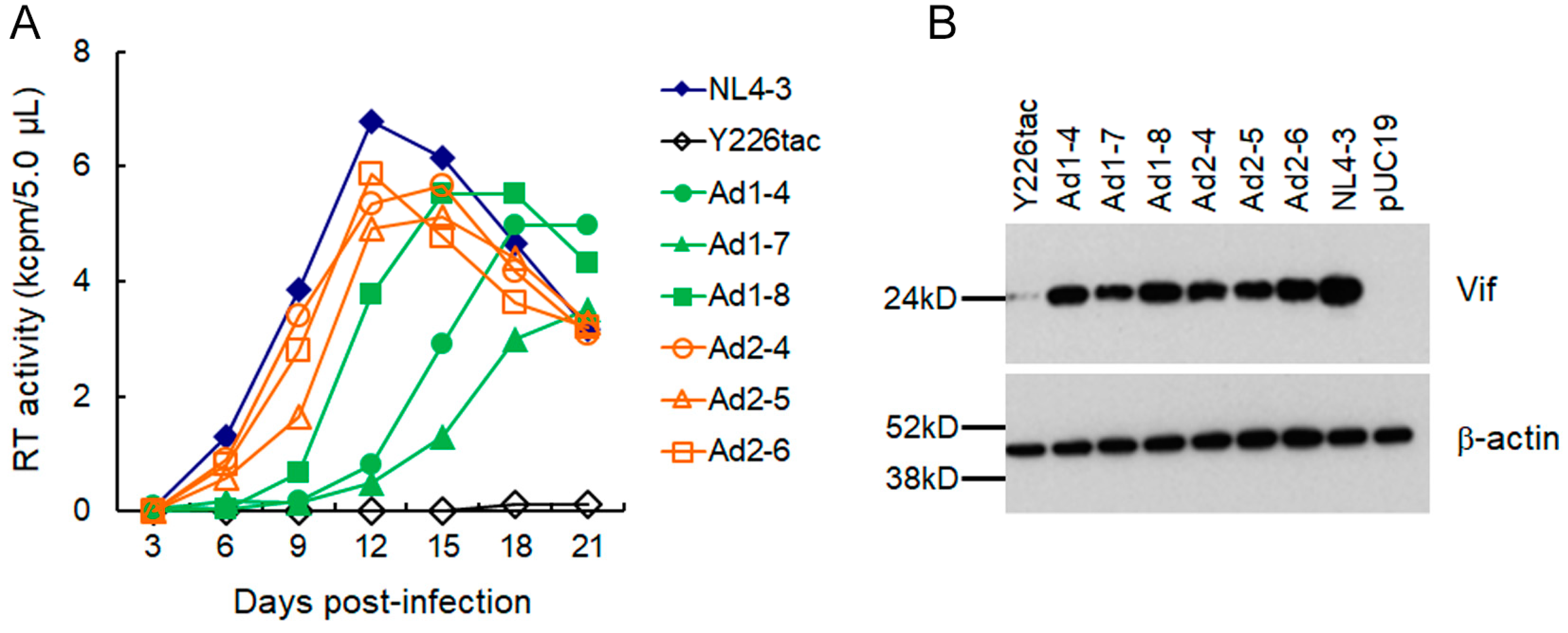
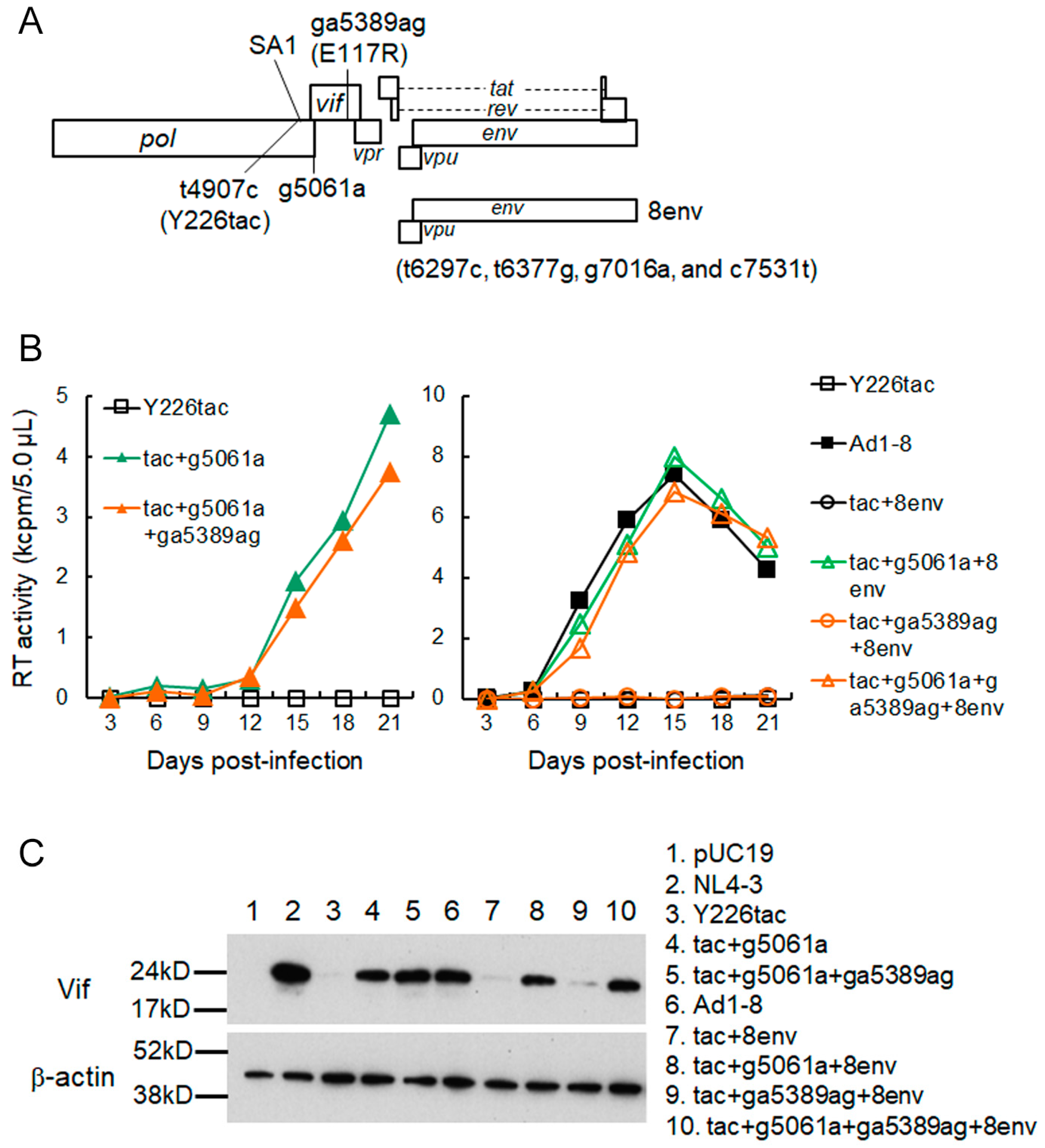
3.2. A Mutation (g5061a) within the SD2b Site Is an Adaptive Mutation Responsible for Increasing Vif Expression and Viral Replication Ability
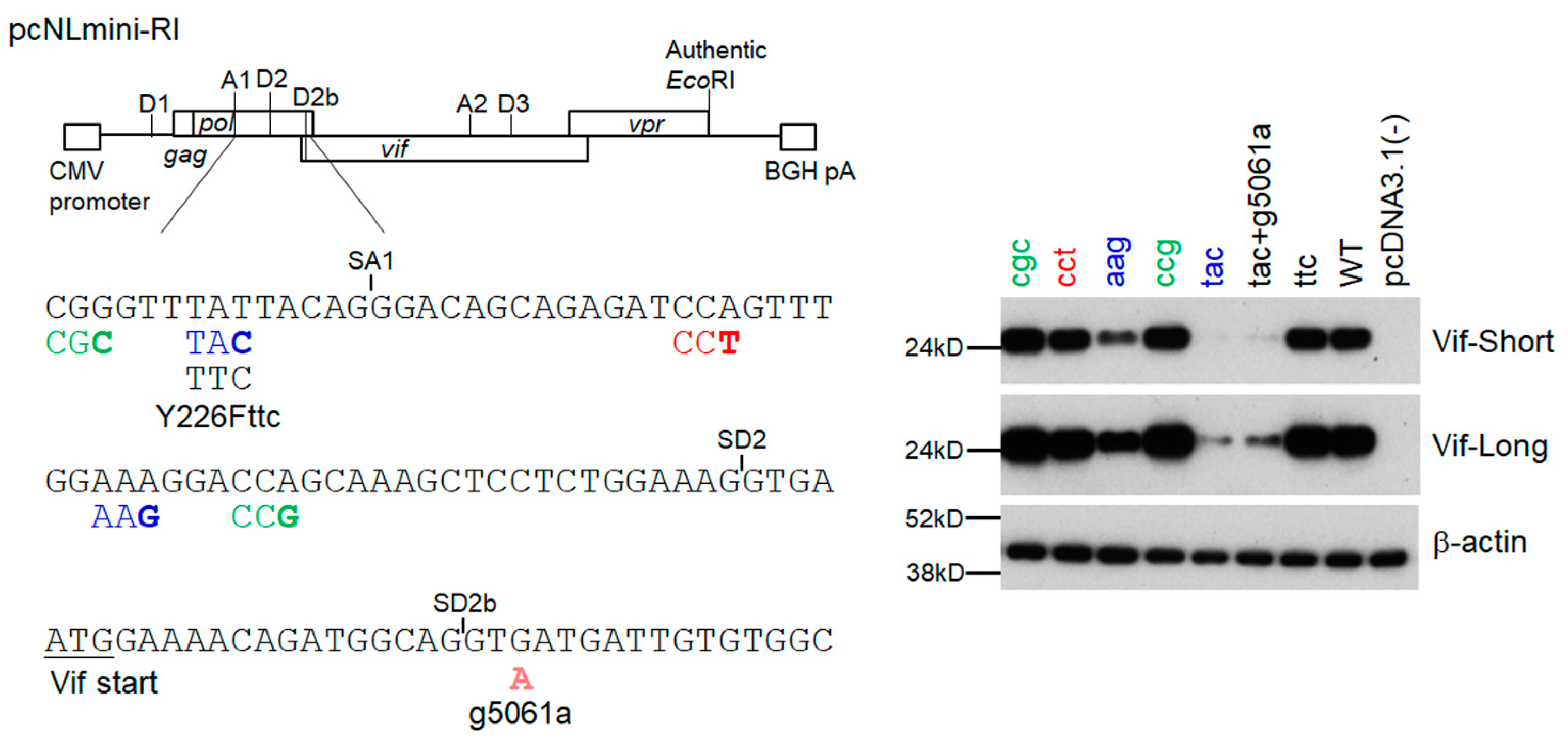
3.3. Another Adaptation Experiment Demonstrated the Importance of a Mutation within SA1 or SD2b for Enhancement of Vif Expression Level
3.4. Adaptive Mutations g5061a and Y226Fttc Enhance the vif mRNA Production by Increasing the Splicing Site Usage of SD2b and SA1, Respectively
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grifoni, A.; Sidney, J.; Vita, R.; Peters, B.; Crotty, S.; Weiskopf, D.; Sette, A. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe 2021, 29, 1076–1092. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Rochman, N.D.; Wolf, Y.I.; Koonin, E.V. Molecular adaptations during viral epidemics. EMBO Rep. 2022, 23, e55393. [Google Scholar] [CrossRef]
- Gilbertson, B.; Duncan, M.; Subbarao, K. Role of the viral polymerase during adaptation of influenza A viruses to new hosts. Curr. Opin. Virol. 2023, 62, 101363. [Google Scholar] [CrossRef]
- Johnson, M.M.; Jones, C.E.; Clark, D.N. The Effect of treatment-associated mutations on HIV replication and transmission cycles. Viruses 2023, 15, 107. [Google Scholar] [CrossRef]
- Martínez, M.A.; Jordan-Paiz, A.; Franco, S.; Nevot, M. Synonymous virus genome recoding as a tool to impact viral fitness. Trends Microbiol. 2016, 24, 134–147. [Google Scholar] [CrossRef]
- Gonçalves-Carneiro, D.; Bieniasz, P.D. Mechanisms of attenuation by genetic recoding of viruses. mBio 2021, 12, e02238-20. [Google Scholar] [CrossRef]
- Jordan-Paiz, A.; Franco, S.; Martínez, M.A. Impact of synonymous genome recoding on the HIV life cycle. Front. Microbiol. 2021, 12, 606087. [Google Scholar] [CrossRef]
- Takata, M.A.; Soll, S.J.; Emery, A.; Blanco-Melo, D.; Swanstrom, R.; Bieniasz, P.D. Global synonymous mutagenesis identifies cis-acting RNA elements that regulate HIV-1 splicing and replication. PLoS Pathog. 2018, 14, e1006824. [Google Scholar] [CrossRef]
- Schwartz, S.; Felber, B.K.; Benko, D.M.; Fenyö, E.M.; Pavlakis, G.N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 1990, 64, 2519–2529. [Google Scholar] [CrossRef]
- Schwartz, S.; Felber, B.K.; Fenyö, E.M.; Pavlakis, G.N. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 1990, 64, 5448–5456. [Google Scholar] [CrossRef]
- Purcell, D.F.J.; Martin, M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar] [CrossRef]
- Amendt, B.A.; Si, Z.H.; Stoltzfus, C.M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: Evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 1995, 15, 4606–4615. [Google Scholar] [CrossRef]
- Emery, A.; Swanstrom, R. HIV-1: To splice or not to splice, that is the question. Viruses 2021, 13, 181. [Google Scholar] [CrossRef]
- Caputi, M. The regulation of HIV-1 mRNA biogenesis. In RNA Processing; Grabowski, P., Ed.; InTech: Rijeka, Croatia, 2011; pp. 79–100. [Google Scholar]
- Karn, J.; Stoltzfus, C.M. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb. Perspect. Med. 2012, 2, a006916. [Google Scholar] [CrossRef]
- Holmes, R.K.; Malim, M.H.; Bishop, K.N. APOBEC-mediated viral restriction: Not simply editing? Trends Biochem. Sci. 2007, 32, 118–128. [Google Scholar] [CrossRef]
- Kirchhoff, F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 2010, 8, 55–67. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Venkatesh, S.; Bieniasz, P.D. Intrinsic cellular defenses against human immunodeficiency viruses. Immunity 2012, 37, 399–411. [Google Scholar] [CrossRef]
- Harris, R.S.; Hultquist, J.F.; Evans, D.T. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 2012, 287, 40875–40883. [Google Scholar] [CrossRef]
- Malim, M.H.; Bieniasz, P.D. HIV restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med. 2012, 2, a006940. [Google Scholar] [CrossRef]
- Desimmie, B.A.; Delviks-Frankenberrry, K.A.; Burdick, R.C.; Qi, D.; Izumi, T.; Pathak, V.K. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J. Mol. Biol. 2014, 426, 1220–1245. [Google Scholar] [CrossRef]
- Stupfler, B.; Verriez, C.; Gallois-Montbrun, S.; Marquet, R.; Paillart, J.C. Degradation-independent inhibition of APOBEC3G by the HIV-1 Vif protein. Viruses 2021, 13, 617. [Google Scholar] [CrossRef]
- Brillen, A.L.; Walotka, L.; Hillebrand, F.; Müller, L.; Widera, M.; Theiss, S.; Schaal, H. Analysis of competing HIV-1 splice donor sites uncovers a tight cluster of splicing regulatory elements within exon 2/2b. J. Virol. 2017, 91, e00389-17. [Google Scholar] [CrossRef]
- Kammler, S.; Otte, M.; Hauber, I.; Kjems, J.; Hauber, J.; Schaal, H. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology 2006, 3, 89. [Google Scholar] [CrossRef]
- Exline, C.M.; Feng, Z.; Stoltzfus, C.M. Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression. J. Virol. 2008, 82, 3921–3931. [Google Scholar] [CrossRef]
- Widera, M.; Erkelenz, S.; Hillebrand, F.; Krikoni, A.; Widera, D.; Kaisers, W.; Deenen, R.; Gombert, M.; Dellen, R.; Pfeiffer, T.; et al. An intronic G run within HIV-1 intron 2 is critical for splicing regulation of vif mRNA. J. Virol. 2013, 87, 2707–2720. [Google Scholar] [CrossRef]
- Nomaguchi, M.; Miyake, A.; Doi, N.; Fujiwara, S.; Miyazaki, Y.; Tsunetsugu-Yokota, Y.; Yokoyama, M.; Sato, H.; Masuda, T.; Adachi, A. Natural single-nucleotide polymorphisms in the 3′ region of the HIV-1 pol gene modulate viral replication ability. J. Virol. 2014, 88, 4145–4160. [Google Scholar] [CrossRef]
- Nomaguchi, M.; Doi, N.; Sakai, Y.; Ode, H.; Iwatani, Y.; Ueno, T.; Matsumoto, Y.; Miyazaki, Y.; Masuda, T.; Adachi, A. Natural single-nucleotide variations in the HIV-1 genomic SA1prox region can alter viral replication ability by regulating Vif expression levels. J. Virol. 2016, 90, 4563–4578. [Google Scholar] [CrossRef]
- Nomaguchi, M.; Doi, N.; Yoshida, T.; Koma, T.; Adachi, S.; Ode, H.; Iwatani, Y.; Yokoyama, M.; Sato, H.; Adachi, A. Production of HIV-1 vif mRNA is modulated by natural nucleotide variations and SLSA1 RNA structure in SA1D2prox genomic region. Front. Microbiol. 2017, 8, 2542. [Google Scholar] [CrossRef]
- Adachi, A.; Gendelman, H.E.; Koenig, S.; Folks, T.; Willey, R.; Rabson, A.; Martin, M.A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986, 59, 284–291. [Google Scholar] [CrossRef]
- Koma, T.; Doi, N.; Takemoto, M.; Watanabe, K.; Yamamoto, H.; Nakashima, S.; Adachi, A.; Nomaguchi, M. The expression level of HIV-1 Vif is optimized by nucleotide changes in the genomic SA1D2prox region during the viral adaptation process. Viruses 2021, 13, 2079. [Google Scholar] [CrossRef]
- Lebkowski, J.S.; Clancy, S.; Calos, M.P. Simian virus 40 replication in adenovirus-transformed human cells antagonizes gene expression. Nature 1985, 317, 169–171. [Google Scholar] [CrossRef]
- Platt, E.J.; Wehrly, K.; Kuhmann, S.E.; Chesebro, B.; Kabat, D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998, 72, 2855–2864. [Google Scholar] [CrossRef]
- Platt, E.J.; Bilska, M.; Kozak, S.L.; Kabat, D.; Montefiori, D.C. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J. Virol. 2009, 83, 8289–8292. [Google Scholar] [CrossRef]
- Kamada, K.; Igarashi, T.; Martin, M.A.; Khamsri, B.; Hatcho, K.; Yamashita, T.; Fujita, M.; Uchiyama, T.; Adachi, A. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. USA 2006, 103, 16959–16964. [Google Scholar] [CrossRef]
- Willey, R.L.; Smith, D.H.; Lasky, L.A.; Theodore, T.S.; Earl, P.L.; Moss, B.; Capon, D.J.; Martin, M.A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 1988, 62, 139–147. [Google Scholar] [CrossRef]
- Nomaguchi, M.; Yokoyama, M.; Kono, K.; Nakayama, E.E.; Shioda, T.; Doi, N.; Fujiwara, S.; Saito, A.; Akari, H.; Miyakawa, K.; et al. Generation of rhesus macaque-tropic HIV-1 clones that are resistant to major anti-HIV-1 restriction factors. J. Virol. 2013, 87, 11447–11461. [Google Scholar] [CrossRef]
- Widera, M.; Hillebrand, F.; Erkelenz, S.; Vasudevan, A.A.; Münk, C.; Schaal, H. A functional conserved intronic G run in HIV-1 intron 3 is critical to counteract APOBEC3G-mediated host restriction. Retrovirology 2014, 11, 72. [Google Scholar] [CrossRef]
- Doi, N.; Yokoyama, M.; Koma, T.; Kotani, O.; Sato, H.; Adachi, A.; Nomaguchi, M. Concomitant enhancement of HIV-1 replication potential and neutralization-resistance in concert with three adaptive mutations in Env V1/C2/C4 domains. Front. Microbiol. 2019, 10, 2. [Google Scholar] [CrossRef]
- Mulder, L.C.; Harari, A.; Simon, V. Cytidine deamination induced HIV-1 drug resistance. Proc. Natl. Acad. Sci. USA 2008, 105, 5501–5506. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Russell, R.A.; Pathak, V.K.; Coffin, J.M. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009, 5, e1000367. [Google Scholar] [CrossRef] [PubMed]
- Wood, N.; Bhattacharya, T.; Keele, B.F.; Giorgi, E.; Liu, M.; Gaschen, B.; Daniels, M.; Ferrari, G.; Haynes, B.F.; McMichael, A.; et al. HIV evolution in early infection: Selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009, 5, e1000414. [Google Scholar] [CrossRef] [PubMed]
- Sadler, H.A.; Stenglein, M.D.; Harris, R.S.; Mansky, L.M. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 2010, 84, 7396–7404. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, D.; Anwar, F.; Davenport, M.P. APOBEC3 has not left an evolutionary footprint on the HIV-1 genome. J. Virol. 2011, 85, 9139–9146. [Google Scholar] [CrossRef] [PubMed]
- Armitage, A.E.; Deforche, K.; Chang, C.H.; Wee, E.; Kramer, B.; Welch, J.J.; Gerstoft, J.; Fugger, L.; McMichael, A.; Rambaut, A.; et al. APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete “all or nothing” phenomenon. PLoS Genet. 2012, 8, e1002550. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Lorenzo-Redondo, R.; Little, S.J.; Chung, Y.S.; Phalora, P.K.; Maljkovic Berry, I.; Archer, J.; Penugonda, S.; Fischer, W.; Richman, D.D.; et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 2014, 10, e1004281. [Google Scholar] [CrossRef]
- Emery, A.; Zhou, S.; Pollom, E.; Swanstrom, R. Characterizing HIV-1 splicing by using next-generation sequencing. J. Virol. 2017, 91, e02515-16. [Google Scholar] [CrossRef]
- Whelan, S. Viral replication strategies. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lipponcott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 105–126. [Google Scholar]
- Freed, E.O.; Martin, M.A. Human immunodeficiency viruses: Replication. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lipponcott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2, pp. 1502–1560. [Google Scholar]
- Rein, A. RNA packaging in HIV. Trends Microbiol. 2019, 27, 715–723. [Google Scholar] [CrossRef]
- Pasternak, A.O.; Berkhout, B. The splice of life: Does RNA processing have a role in HIV-1 persistence? Viruses 2021, 13, 1751. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Gao, X.; Sandig, H.; Limmer, K.; Pavon-Eternod, M.; Jones, T.E.; Landry, S.; Pan, T.; Weitzman, M.D.; et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 2012, 491, 125–128. [Google Scholar] [CrossRef]
- Martrus, G.; Nevot, M.; Andres, C.; Clotet, B.; Martinez, M.A. Changes in codon-pair bias of human immunodeficiency virus type 1 have profound effects on virus replication in cell culture. Retrovirology 2013, 10, 78. [Google Scholar] [CrossRef] [PubMed]
- Jordan-Paiz, A.; Nevot, M.; Lamkiewicz, K.; Lataretu, M.; Franco, S.; Marz, M.; Martinez, M.A. HIV-1 lethality and loss of Env protein expression induced by single synonymous substitutions in the virus genome intronic-splicing silencer. J. Virol. 2020, 94, e01108-20. [Google Scholar] [CrossRef] [PubMed]
- Pollom, E.; Dang, K.K.; Potter, E.L.; Gorelick, R.J.; Burch, C.L.; Weeks, K.M.; Swanstrom, R. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 2013, 9, e1003294. [Google Scholar] [CrossRef] [PubMed]
- Assis, R. Strong epistatic selection on the RNA secondary structure of HIV. PLoS Pathog. 2014, 10, e1004363. [Google Scholar] [CrossRef]
- Riquelme-Barrios, S.; Pereira-Montecinos, C.; Valiente-Echeverría, F.; Soto-Rifo, R. Emerging roles of N6-methyladenosine on HIV-1 RNA metabolism and viral replication. Front. Microbiol. 2018, 9, 576. [Google Scholar] [CrossRef]



| NL-Ad1-4 | NL-Ad1-7 | NL-Ad1-8 | ||||||
|---|---|---|---|---|---|---|---|---|
| Nt Change | Region | NS/S Change in the Region | Nt Change | Region | NS/S Change in the Region | Nt Change | Region | NS/S Change in the Region |
| t3707c | Pol-RT | T386acc | ||||||
| t4907c | Pol-IN | Y226tac | t4907c | Pol-IN | Y226tac | t4907c | Pol-IN | Y226tac |
| g5061a | Vif | V7 | g5061a | Vif | V7 | g5061a | Vif | V7 |
| g5061a | Pol-IN | D278N | g5061a | Pol-IN | D278N | g5061a | Pol-IN | D278N |
| g5224a | Vif | A62T | ||||||
| ga5389ag | Vif | E117R | ga5389ag | Vif | E117R | ga5389ag | Vif | E117R |
| a5627g | Vpr | L23ttg | a5627g | Vpr | L23ttg | |||
| t6297c | Vpu | D79 | ||||||
| t6297c | Env(SP) | M26T | ||||||
| g6308a | Env(SP) | A30T | ||||||
| t6377g | Env(C1) | F53V | ||||||
| g6443a | Env(C1) | V75I | ||||||
| g6654a | Env(V1) | G145E | ||||||
| g7016a | Env(C2) | E266K | ||||||
| g7505a | Env(C4) | G429R | ||||||
| c7531t | Env(C4) | I437att | ||||||
| NL-Ad2-4 | NL-Ad2-5 | NL-Ad2-6 | ||||||
|---|---|---|---|---|---|---|---|---|
| Nt Change | Region | NS/S Change in the Region | Nt Change | Region | NS/S Change in the Region | Nt Change | Region | NS/S Change in the Region |
| c3058t | Pol-RT | P170L | c3058t | Pol-RT | P170L | c3058t | Pol-RT | P170L |
| t4907c | Pol-IN | Y226tac | t4907c | Pol-IN | Y226tac | t4907c | Pol-IN | Y226tac |
| g5061a | Vif | V7 | g5061a | Vif | V7 | g5061a | Vif | V7 |
| g5061a | Pol-IN | D278N | g5061a | Pol-IN | D278N | g5061a | Pol-IN | D278N |
| ga5389ag | Vif | E117R | ga5389ag | Vif | E117R | ga5389ag | Vif | E117R |
| g6273a | Vpu | G71gga | ||||||
| g6273a | Env(SP) | G18D | ||||||
| g6443a | Env(C1) | V75I | g6443a | Env(C1) | V75I | g6443a | Env(C1) | V75I |
| g7160a | Env(V3) | A314T | ||||||
| t7282a | Env(C3) | N354K | ||||||
| t7408a | Env(V4) | S396R | ||||||
| g7499a | Env(C4) | E427K | ||||||
| NL-Ad3-2 | NL-Ad3-4 | NL-Ad3-8 | ||||||
|---|---|---|---|---|---|---|---|---|
| Nt Change | Region | NS/S Change in the Region | Nt Change | Region | NS/S Change in the Region | Nt Change | Region | NS/S Change in the Region |
| g3839a | E430gaa | g3839a | E430gaa | g3839a | Pol-RT | E430gaa | ||
| g4080a | Pol-RT | D511N | ||||||
| t4907c | Pol-IN | Y226tac | t4907c | Pol-IN | Y226tac | t4907c | Pol-IN | Y226tac |
| g5061a | Vif | V7 | g5061a | Vif | V7 | g5061a | Vif | V7 |
| g5061a | Pol-IN | D278N | g5061a | Pol-IN | D278N | g5061a | Pol-IN | D278N |
| a5447t | Q136L | |||||||
| g5484a | Vif | L148tta | g5484a | Vif | L148tta | |||
| t6069c | Vpu | P3ccc | ||||||
| g6150a | Vpu | R30aga | ||||||
| g6225a | Vpu | E55gaa | ||||||
| g6225a | Env(SP) | R2K | ||||||
| g6229a | Vpu | E57K | ||||||
| g6229a | Env(SP) | V3gta | ||||||
| a6513t | Env(C1) | N98I | ||||||
| g6845a | Env(C2) | E209K | g6845a | Env(C2) | E209K | |||
| g7499a | Env(C4) | E427K | ||||||
| a7507g | Env(C4) | G429ggg | a7507g | Env(C4) | G429ggg | |||
| NL-Ad4-2 | NL-Ad4-4 | ||||
|---|---|---|---|---|---|
| Nt Change | Region | NS/S Change in the Region | Nt Change | Region | NS/S Change in the Region |
| a4906t | Pol-IN | Y226Fttc | a4906t | Pol-IN | Y226Fttc |
| g5138t | Vif | R33M | |||
| c5512a | Vif | Q158K | |||
| c5622t | Vpr | L22F | |||
| g5672a | Vpr | W38stop | |||
| g5788a | Vpr | R77Q | |||
| t6584c | Env(C1) | L122cta | |||
| c6830t | Env(C2) | P204S | c6830g(C2) | Env | P204S |
| g6907a | Env(C2) | K229aag | |||
| MaxEnt Score | |||||||
|---|---|---|---|---|---|---|---|
| Mutation | Sequence around the SD2b Site | Hbond Score | Mutation | Sequence around the SA1 Site | ME | MM | WMM |
| WT | CAGGTGATGAT | 12.4 | WT | AATTTTCGGGTTTATTACAGGGA | 6.41 | 7.07 | 7.00 |
| g5061a | CAGGTAATGAT | 15.8 | Y226tac | AATTTTCGGGTTTACTACAGGGA | 5.92 | 5.72 | 6.90 |
| Y226Fttc | AATTTTCGGGTTTTCTACAGGGA | 8.07 | 7.48 | 9.19 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koma, T.; Doi, N.; Le, B.Q.; Kondo, T.; Ishizue, M.; Tokaji, C.; Tsukada, C.; Adachi, A.; Nomaguchi, M. Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation. Viruses 2023, 15, 2424. https://doi.org/10.3390/v15122424
Koma T, Doi N, Le BQ, Kondo T, Ishizue M, Tokaji C, Tsukada C, Adachi A, Nomaguchi M. Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation. Viruses. 2023; 15(12):2424. https://doi.org/10.3390/v15122424
Chicago/Turabian StyleKoma, Takaaki, Naoya Doi, Bao Quoc Le, Tomoyuki Kondo, Mitsuki Ishizue, Chiaki Tokaji, Chizuko Tsukada, Akio Adachi, and Masako Nomaguchi. 2023. "Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation" Viruses 15, no. 12: 2424. https://doi.org/10.3390/v15122424
APA StyleKoma, T., Doi, N., Le, B. Q., Kondo, T., Ishizue, M., Tokaji, C., Tsukada, C., Adachi, A., & Nomaguchi, M. (2023). Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation. Viruses, 15(12), 2424. https://doi.org/10.3390/v15122424





