Repetitive Exposure to Bacteriophage Cocktails against Pseudomonas aeruginosa or Escherichia coli Provokes Marginal Humoral Immunity in Naïve Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.1.1. Differential Blood Count and Plasma Preparation
2.1.2. Peritoneal Lavage, Bronchoalveolar Lavage and Organ Removal
2.1.3. Isolation of Spleen and Lymph Node Leukocytes and Flow Cytometry Analysis
2.1.4. Histopathology Analysis
2.1.5. Anti-Phage Antibody Measurement via ELISA
2.1.6. Cytokine Analysis via LEGENdplexTM
2.2. Bacteriophages and Bacterial Strains
2.2.1. Plaque Assays
2.2.2. Transmission Electron Microscopy Analysis of Phage Suspensions
2.3. Data Analysis
3. Results
3.1. Intraperitoneally Injected Phages Reach the Lungs and Are Harmless to Naïve Mice
3.2. Innate Immune Response toward Phage Cocktails
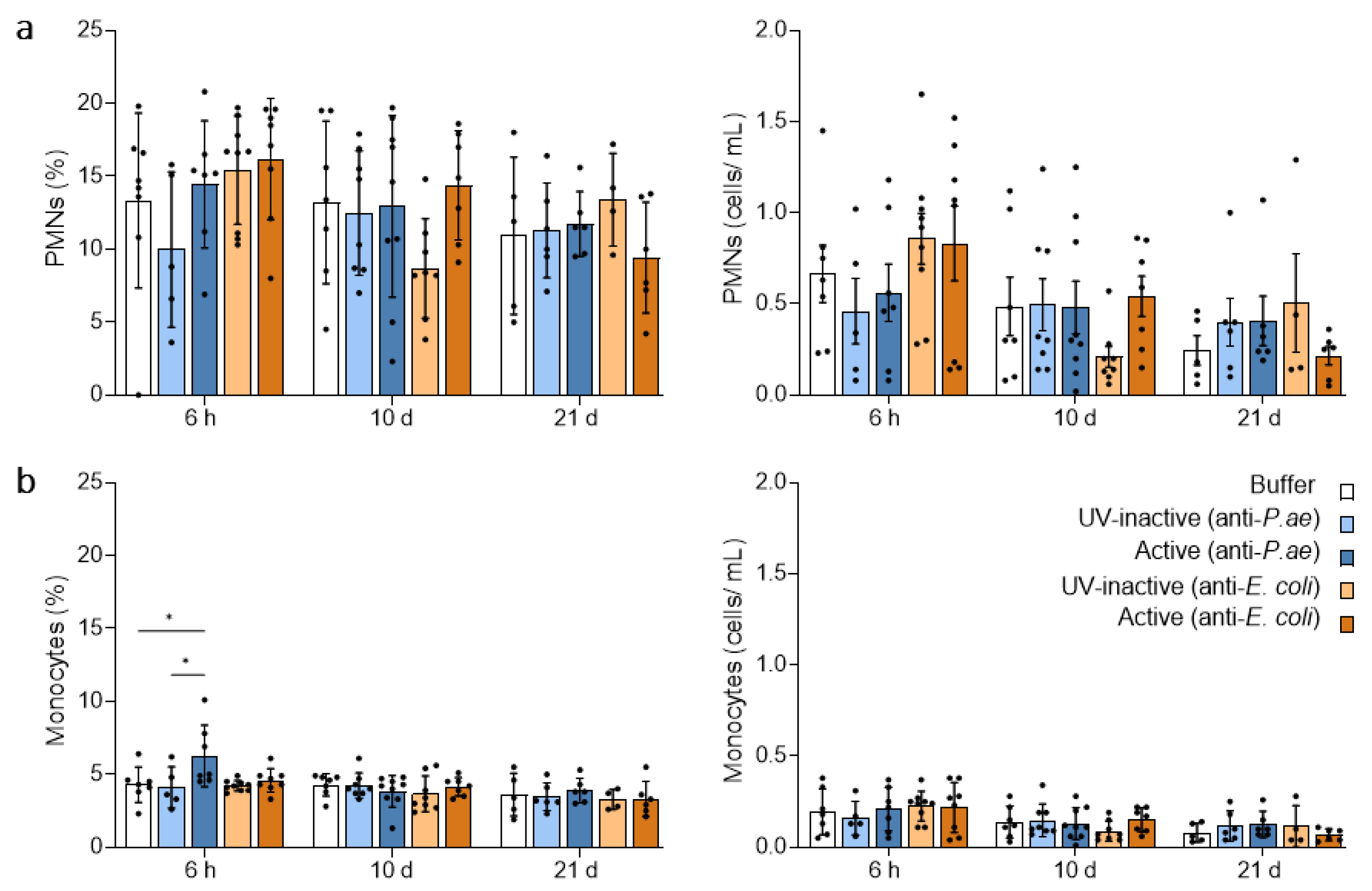
3.2.1. Blood Neutrophils and Monocytes Remain Unaffected by Phage Treatment
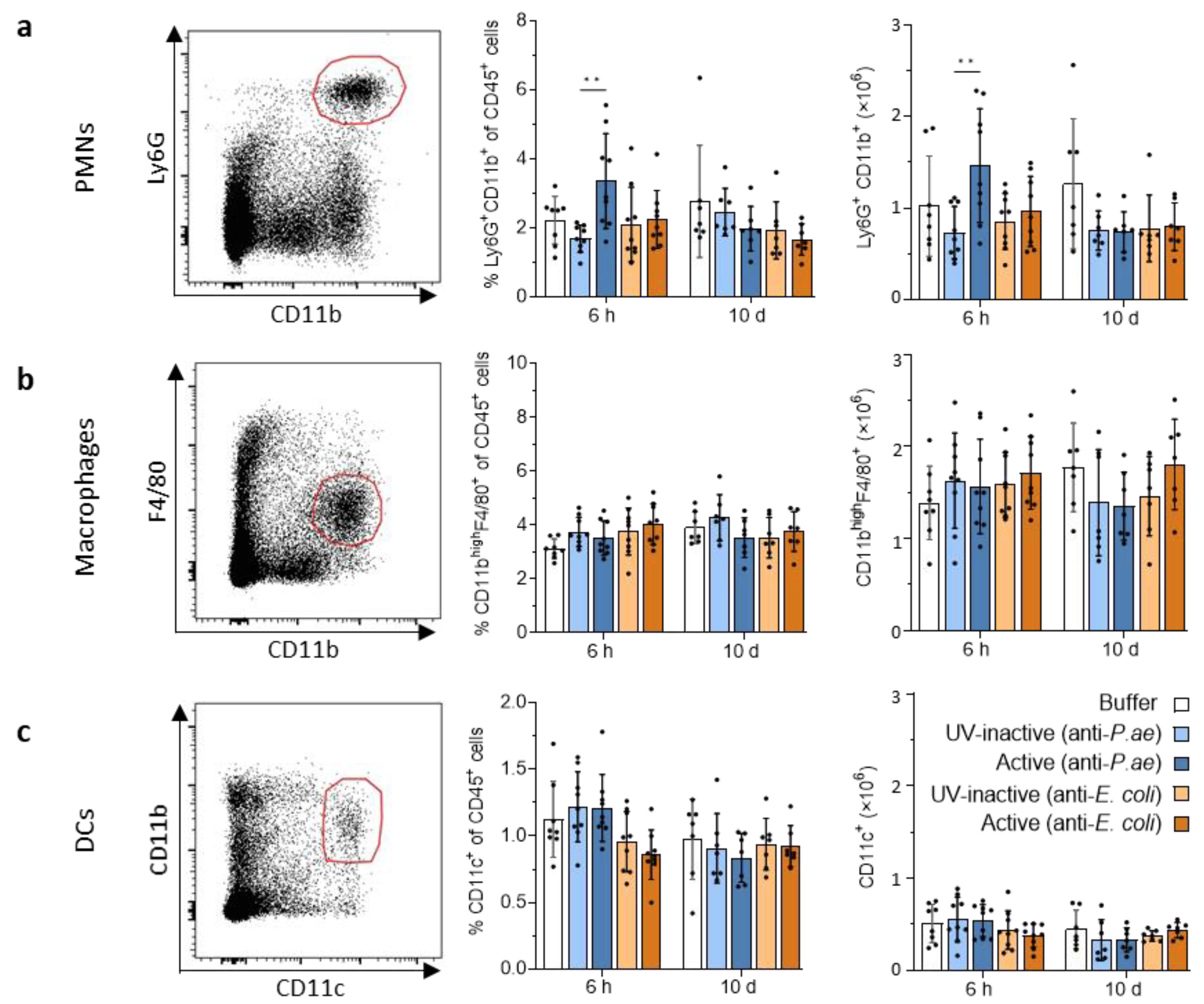
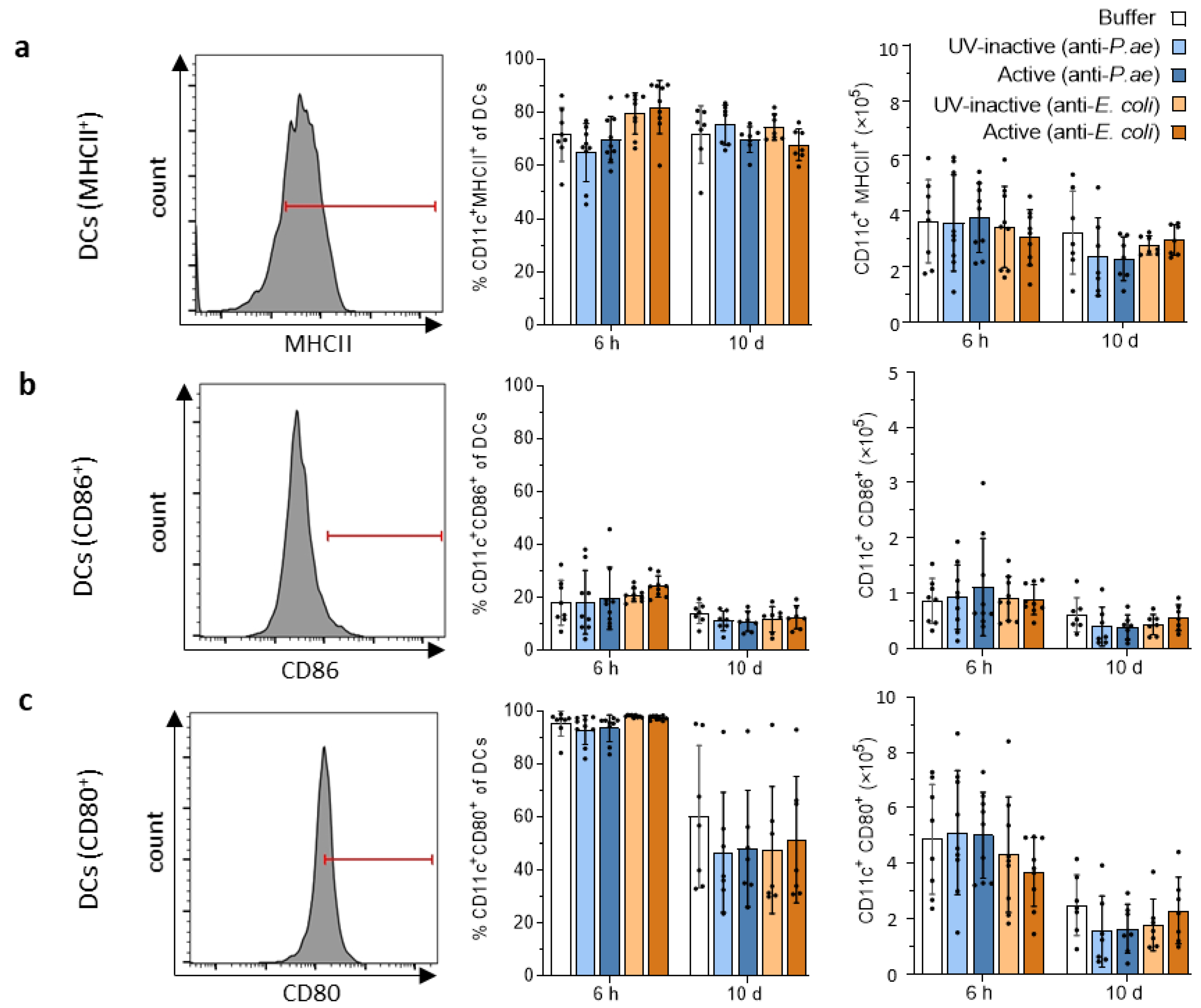
3.2.2. Lymphatic Innate Immune Cells in the Spleen and Draining Lymph Nodes Do Not Display Signs of Activation after Phage Treatment
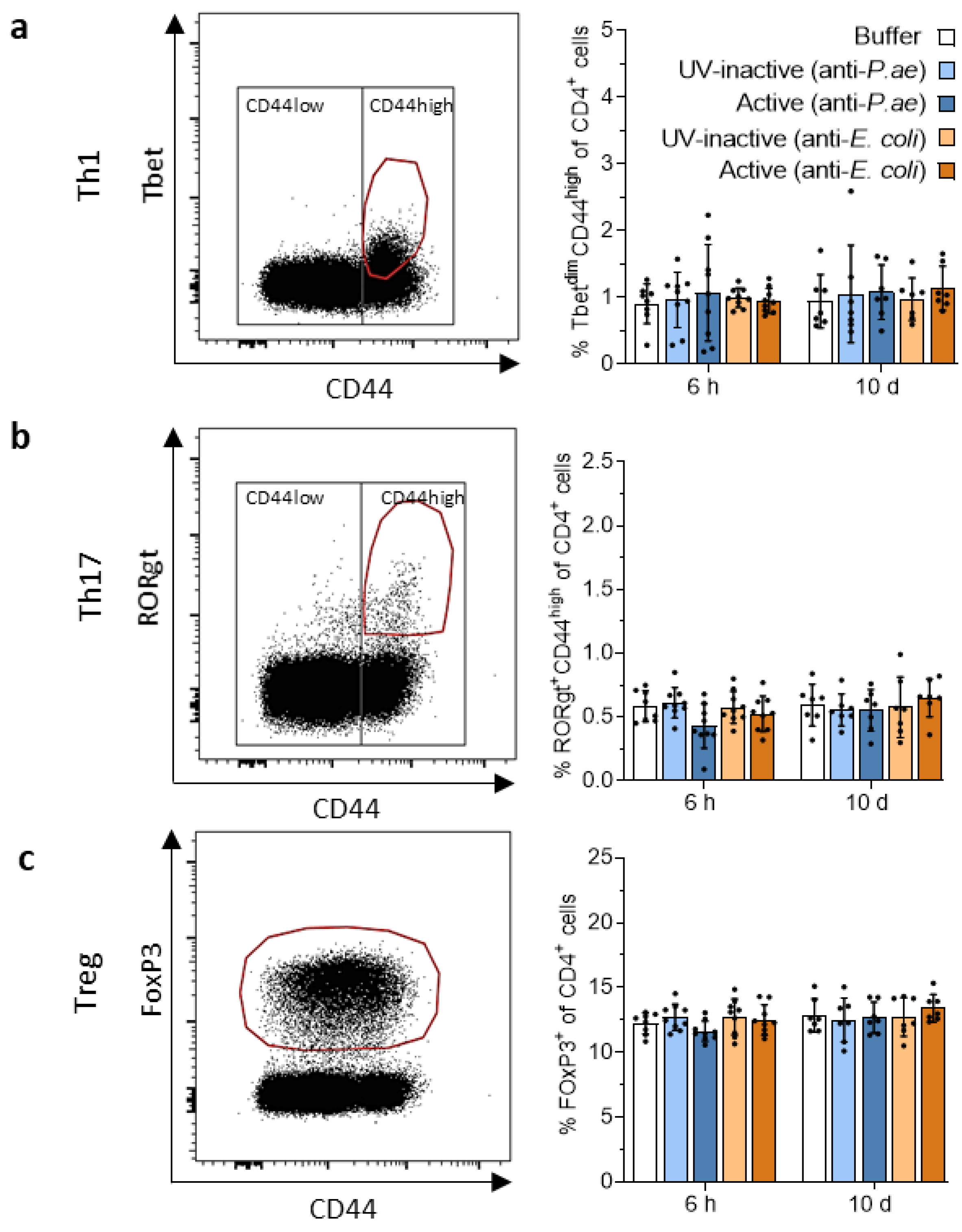
3.3. Adaptive Immune Response against Phage Cocktails—Cellular Immunity
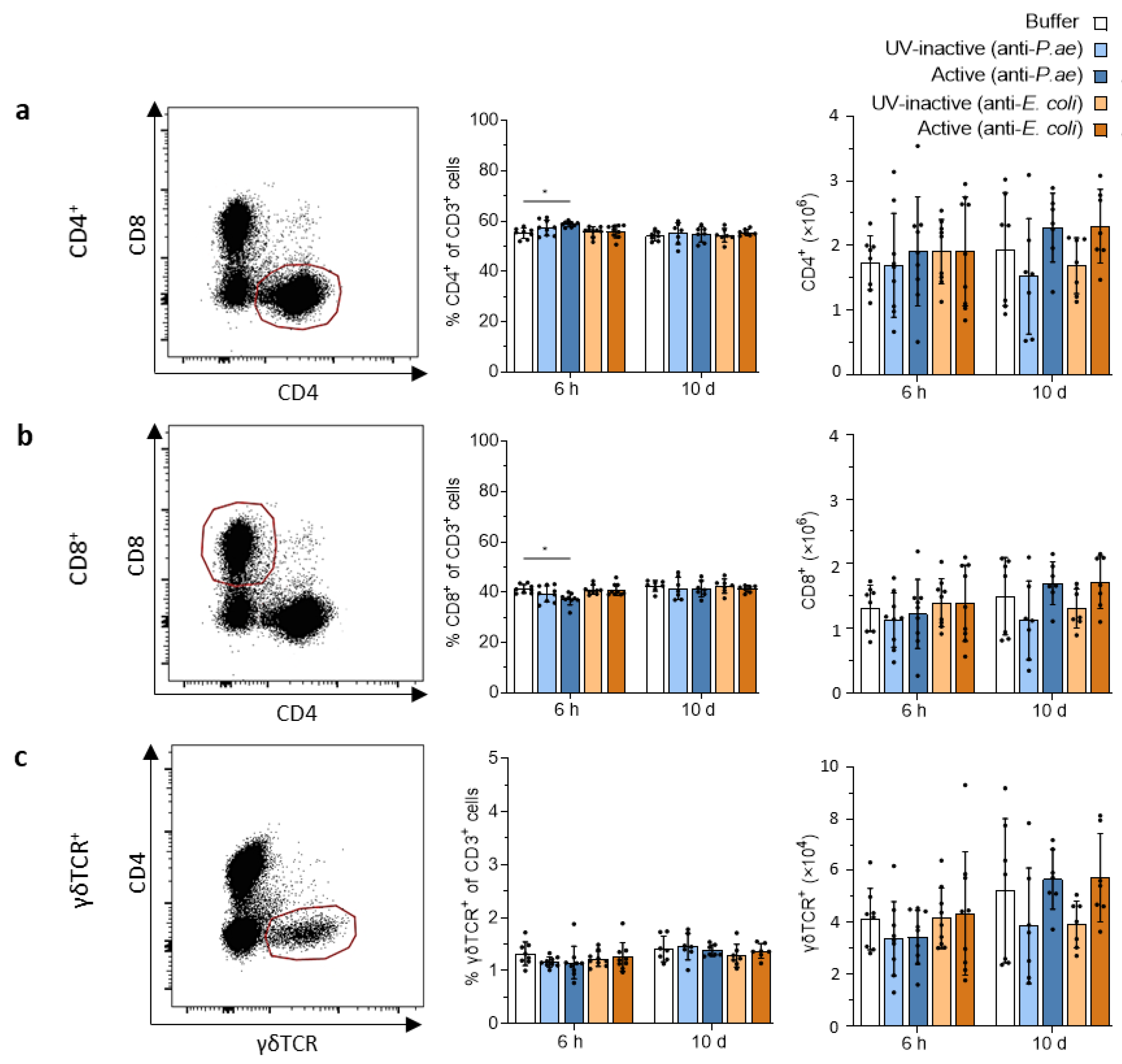
3.3.1. Phage Treatment Does Not Trigger Adaptive T-Cell Responses in Draining Lymph Nodes or Spleen
3.3.2. Minimal Pro-Inflammatory Cytokine and Chemokine Secretion after Phage Treatment
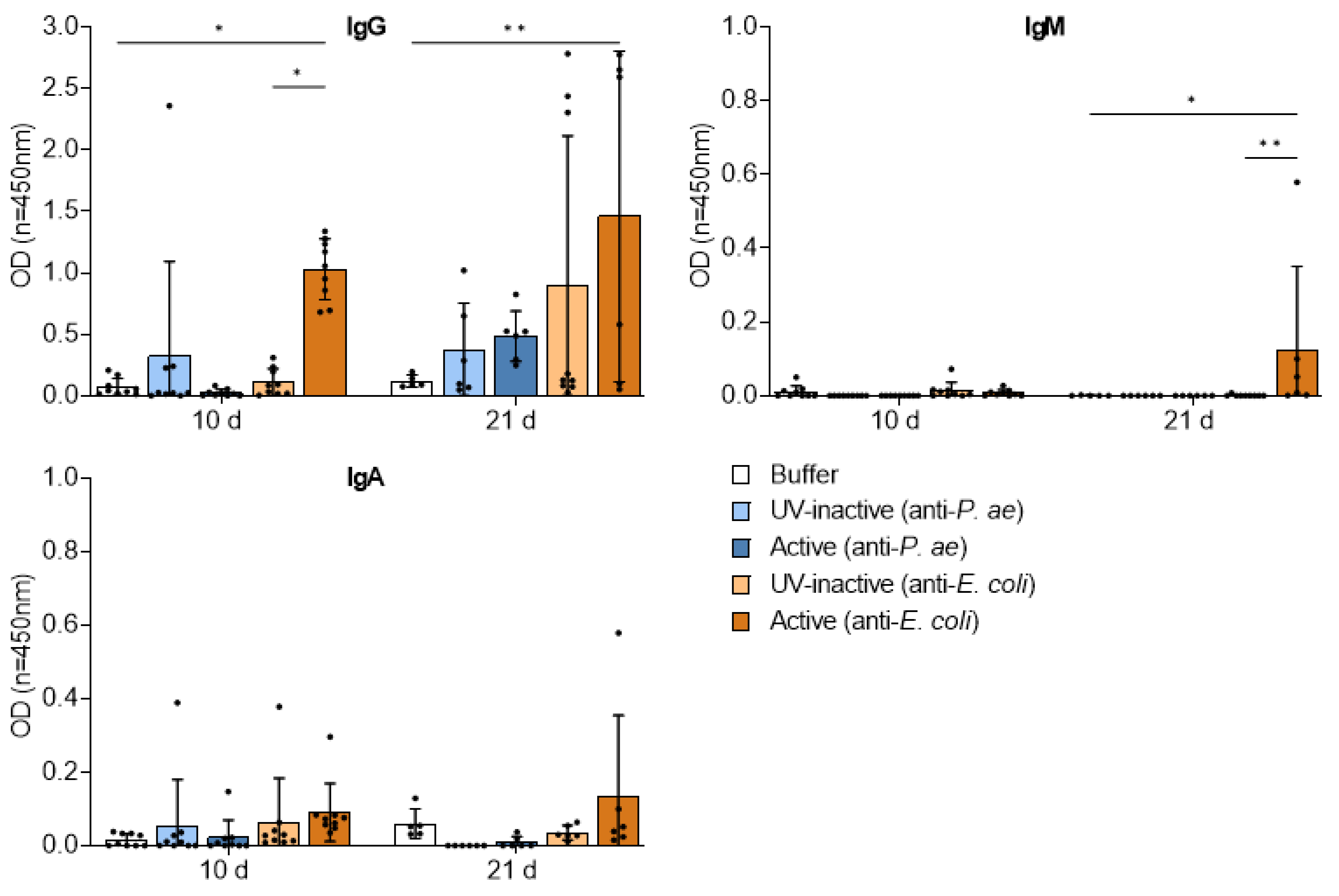
3.4. Adaptive Immune Response against Phage Cocktails—Humoral Immunity
3.4.1. Phage Treatment Induces Minimal to Low-Grade Germinal Center Formation
3.4.2. Repetitive Phage Treatment Evokes Phage-Specific Antibody Response
3.4.3. Destruction of Phage Structures by UV-Inactivation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anyaegbunam, N.J.; Anekpo, C.C.; Anyaegbunam, Z.K.G.; Doowuese, Y.; Chinaka, C.B.; Odo, O.J.; Sharndama, H.C.; Okeke, O.P.; Mba, I.E. The resurgence of phage-based therapy in the era of increasing antibiotic resistance: From research progress to challenges and prospects. Microbiol. Res. 2022, 264, 127155. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Kalanuria, A.A.; Ziai, W.; Zai, W.; Mirski, M. Ventilator-associated pneumonia in the ICU. Crit. Care 2014, 18, 208. [Google Scholar] [CrossRef] [PubMed]
- Ben Lakhal, H.; M’Rad, A.; Naas, T.; Brahmi, N. Antimicrobial Susceptibility among Pathogens Isolated in Early- versus Late-Onset Ventilator-Associated Pneumonia. Infect. Dis. Rep. 2021, 13, 401–410. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Wienhold, S.-M.; Brack, M.C.; Nouailles, G.; Krishnamoorthy, G.; Korf, I.H.E.; Seitz, C.; Wienecke, S.; Dietert, K.; Gurtner, C.; Kershaw, O.; et al. Preclinical Assessment of Bacteriophage Therapy against Experimental Acinetobacter baumannii Lung Infection. Viruses 2021, 14, 33. [Google Scholar] [CrossRef]
- Forti, F.; Roach, D.R.; Cafora, M.; Pasini, M.E.; Horner, D.S.; Fiscarelli, E.V.; Rossitto, M.; Cariani, L.; Briani, F.; Debarbieux, L.; et al. Design of a Broad-Range Bacteriophage Cocktail That Reduces Pseudomonas aeruginosa Biofilms and Treats Acute Infections in Two Animal Models. Antimicrob. Agents Chemother. 2018, 62, e02573-17. [Google Scholar] [CrossRef]
- Prazak, J.; Iten, M.; Cameron, D.R.; Save, J.; Grandgirard, D.; Resch, G.; Goepfert, C.; Leib, S.L.; Takala, J.; Jakob, S.M.; et al. Bacteriophages Improve Outcomes in Experimental Staphylococcus aureus Ventilator-associated Pneumonia. Am. J. Respir. Crit. Care Med. 2019, 200, 1126–1133. [Google Scholar] [CrossRef]
- Lebeaux, D.; Merabishvili, M.; Caudron, E.; Lannoy, D.; van Simaey, L.; Duyvejonck, H.; Guillemain, R.; Thumerelle, C.; Podglajen, I.; Compain, F.; et al. A Case of Phage Therapy against Pandrug-Resistant Achromobacter xylosoxidans in a 12-Year-Old Lung-Transplanted Cystic Fibrosis Patient. Viruses 2021, 13, 60. [Google Scholar] [CrossRef]
- Paul, K.; Merabishvili, M.; Hazan, R.; Christner, M.; Herden, U.; Gelman, D.; Khalifa, L.; Yerushalmy, O.; Coppenhagen-Glazer, S.; Harbauer, T.; et al. Bacteriophage Rescue Therapy of a Vancomycin-Resistant Enterococcus faecium Infection in a One-Year-Old Child following a Third Liver Transplantation. Viruses 2021, 13, 1785. [Google Scholar] [CrossRef] [PubMed]
- Law, N.; Logan, C.; Yung, G.; Furr, C.-L.L.; Lehman, S.M.; Morales, S.; Rosas, F.; Gaidamaka, A.; Bilinsky, I.; Grint, P.; et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 2019, 47, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Jennes, S.; Merabishvili, M.; Soentjens, P.; Pang, K.W.; Rose, T.; Keersebilck, E.; Soete, O.; François, P.-M.; Teodorescu, S.; Verween, G.; et al. Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury-a case report. Crit. Care 2017, 21, 129. [Google Scholar] [CrossRef] [PubMed]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Sarker, S.A.; Sultana, S.; Reuteler, G.; Moine, D.; Descombes, P.; Charton, F.; Bourdin, G.; McCallin, S.; Ngom-Bru, C.; Neville, T.; et al. Oral Phage Therapy of Acute Bacterial Diarrhea with Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. eBioMedicine 2016, 4, 124–137. [Google Scholar] [CrossRef]
- Leitner, L.; Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Chkonia, I.; Rigvava, S.; Chkhotua, A.; Changashvili, G.; McCallin, S.; Schneider, M.P.; et al. Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: A randomised, placebo-controlled, double-blind clinical trial. Lancet Infect. Dis. 2021, 21, 427–436. [Google Scholar] [CrossRef]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.-A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.H.; Anggård, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef]
- Jamal, M.; Bukhari, S.M.A.U.S.; Andleeb, S.; Ali, M.; Raza, S.; Nawaz, M.A.; Hussain, T.; Rahman, S.U.; Shah, S.S.A. Bacteriophages: An overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 2019, 59, 123–133. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Febvre, H.P.; Rao, S.; Gindin, M.; Goodwin, N.D.M.; Finer, E.; Vivanco, J.S.; Lu, S.; Manter, D.K.; Wallace, T.C.; Weir, T.L. PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Roach, D.R.; Debarbieux, L. Phage therapy: Awakening a sleeping giant. Emerg. Top. Life Sci. 2017, 1, 93–103. [Google Scholar] [CrossRef]
- Górski, A.; Borysowski, J.; Międzybrodzki, R. Phage Therapy: Towards a Successful Clinical Trial. Antibiotics 2020, 9, 827. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Weber-Dabrowska, B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell. Mol. Life Sci. 2005, 62, 511–519. [Google Scholar] [CrossRef]
- Townsend, E.M.; Kelly, L.; Muscatt, G.; Box, J.D.; Hargraves, N.; Lilley, D.; Jameson, E. The Human Gut Phageome: Origins and Roles in the Human Gut Microbiome. Front. Cell. Infect. Microbiol. 2021, 11, 643214. [Google Scholar] [CrossRef]
- Carroll-Portillo, A.; Lin, H.C. Bacteriophage and the Innate Immune System: Access and Signaling. Microorganisms 2019, 7, 625. [Google Scholar] [CrossRef]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef]
- Gembara, K.; Dąbrowska, K. Phage-specific antibodies. Curr. Opin. Biotechnol. 2021, 68, 186–192. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Borysowski, J.; Dąbrowska, K.; Wierzbicki, P.; Ohams, M.; Korczak-Kowalska, G.; Olszowska-Zaremba, N.; Łusiak-Szelachowska, M.; Kłak, M.; et al. Phage as a modulator of immune responses: Practical implications for phage therapy. Adv. Virus Res. 2012, 83, 41–71. [Google Scholar] [CrossRef] [PubMed]
- Forthal, D.N.; Moog, C. Fc receptor-mediated antiviral antibodies. Curr. Opin. HIV AIDS 2009, 4, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Archana, A.; Patel, P.S.; Kumar, R.; Nath, G. Neutralizing antibody response against subcutaneously injected bacteriophages in rabbit model. Virusdisease 2021, 32, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Majewska, J.; Kaźmierczak, Z.; Lahutta, K.; Lecion, D.; Szymczak, A.; Miernikiewicz, P.; Drapała, J.; Harhala, M.; Marek-Bukowiec, K.; Jędruchniewicz, N.; et al. Induction of Phage-Specific Antibodies by Two Therapeutic Staphylococcal Bacteriophages Administered per os. Front. Immunol. 2019, 10, 2607. [Google Scholar] [CrossRef] [PubMed]
- Naghizadeh, M.; Karimi Torshizi, M.A.; Rahimi, S.; Engberg, R.M.; Sørensen Dalgaard, T. Effect of serum anti-phage activity on colibacillosis control by repeated phage therapy in broilers. Vet. Microbiol. 2019, 234, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Łusiak-Szelachowska, M.; Międzybrodzki, R.; Fortuna, W.; Borysowski, J.; Górski, A. Anti-phage serum antibody responses and the outcome of phage therapy. Folia Microbiol. (Praha) 2021, 66, 127–131. [Google Scholar] [CrossRef]
- Żaczek, M.; Łusiak-Szelachowska, M.; Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Międzybrodzki, R.; Owczarek, B.; Kopciuch, A.; Fortuna, W.; Rogóż, P.; Górski, A. Antibody Production in Response to Staphylococcal MS-1 Phage Cocktail in Patients Undergoing Phage Therapy. Front. Microbiol. 2016, 7, 1681. [Google Scholar] [CrossRef]
- Majewska, J.; Beta, W.; Lecion, D.; Hodyra-Stefaniak, K.; Kłopot, A.; Kaźmierczak, Z.; Miernikiewicz, P.; Piotrowicz, A.; Ciekot, J.; Owczarek, B.; et al. Oral Application of T4 Phage Induces Weak Antibody Production in the Gut and in the Blood. Viruses 2015, 7, 4783–4799. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Zaczek, M.; Weber-Dąbrowska, B.; Międzybrodzki, R.; Kłak, M.; Fortuna, W.; Letkiewicz, S.; Rogóż, P.; Szufnarowski, K.; Jończyk-Matysiak, E.; et al. Phage neutralization by sera of patients receiving phage therapy. Viral Immunol. 2014, 27, 295–304. [Google Scholar] [CrossRef]
- Dufour, N.; Delattre, R.; Chevallereau, A.; Ricard, J.-D.; Debarbieux, L. Phage Therapy of Pneumonia Is Not Associated with an Overstimulation of the Inflammatory Response Compared to Antibiotic Treatment in Mice. Antimicrob. Agents Chemother. 2019, 63, e00379-19. [Google Scholar] [CrossRef]
- Rohde, C.; Resch, G.; Pirnay, J.-P.; Blasdel, B.G.; Debarbieux, L.; Gelman, D.; Górski, A.; Hazan, R.; Huys, I.; Kakabadze, E.; et al. Expert Opinion on Three Phage Therapy Related Topics: Bacterial Phage Resistance, Phage Training and Prophages in Bacterial Production Strains. Viruses 2018, 10, 178. [Google Scholar] [CrossRef]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage Cocktail Development for Bacteriophage Therapy: Toward Improving Spectrum of Activity Breadth and Depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef]
- Kaur, G.; Agarwal, R.; Sharma, R.K. Bacteriophage Therapy for Critical and High-Priority Antibiotic-Resistant Bacteria and Phage Cocktail-Antibiotic Formulation Perspective. Food Environ. Virol. 2021, 13, 433–446. [Google Scholar] [CrossRef]
- Vaitekenas, A.; Tai, A.S.; Ramsay, J.P.; Stick, S.M.; Kicic, A. Pseudomonas aeruginosa Resistance to Bacteriophages and Its Prevention by Strategic Therapeutic Cocktail Formulation. Antibiotics 2021, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell Host Microbe 2017, 22, 38–47.e4. [Google Scholar] [CrossRef] [PubMed]
- Selezska, K.; Kazmierczak, M.; Müsken, M.; Garbe, J.; Schobert, M.; Häussler, S.; Wiehlmann, L.; Rohde, C.; Sikorski, J. Pseudomonas aeruginosa population structure revisited under environmental focus: Impact of water quality and phage pressure. Environ. Microbiol. 2012, 14, 1952–1967. [Google Scholar] [CrossRef] [PubMed]
- Garbe, J.; Wesche, A.; Bunk, B.; Kazmierczak, M.; Selezska, K.; Rohde, C.; Sikorski, J.; Rohde, M.; Jahn, D.; Schobert, M. Characterization of JG024, a pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol. 2010, 10, 301. [Google Scholar] [CrossRef]
- Maura, D.; Morello, E.; Du Merle, L.; Bomme, P.; Le Bouguénec, C.; Debarbieux, L. Intestinal colonization by enteroaggregative Escherichia coli supports long-term bacteriophage replication in mice. Environ. Microbiol. 2012, 14, 1844–1854. [Google Scholar] [CrossRef] [PubMed]
- Hornischer, K.; Khaledi, A.; Pohl, S.; Schniederjans, M.; Pezoldt, L.; Casilag, F.; Muthukumarasamy, U.; Bruchmann, S.; Thöming, J.; Kordes, A.; et al. BACTOME-a reference database to explore the sequence- and gene expression-variation landscape of Pseudomonas aeruginosa clinical isolates. Nucleic Acids Res. 2019, 47, D716–D720. [Google Scholar] [CrossRef]
- Summers, C.; Rankin, S.M.; Condliffe, A.M.; Singh, N.; Peters, A.M.; Chilvers, E.R. Neutrophil kinetics in health and disease. Trends Immunol. 2010, 31, 318–324. [Google Scholar] [CrossRef] [PubMed]
- den Haan, J.M.M.; Arens, R.; van Zelm, M.C. The activation of the adaptive immune system: Cross-talk between antigen-presenting cells, T cells and B cells. Immunol. Lett. 2014, 162, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Worbs, T.; Hammerschmidt, S.I.; Förster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2017, 17, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Steinman, R.M.; Inaba, K. The binding of antigen presenting cells to T lymphocytes. Adv. Exp. Med. Biol. 1988, 237, 31–41. [Google Scholar] [CrossRef]
- Lemos, M.P.; Fan, L.; Lo, D.; Laufer, T.M. CD8alpha+ and CD11b+ dendritic cell-restricted MHC class II controls Th1 CD4+ T cell immunity. J. Immunol. 2003, 171, 5077–5084. [Google Scholar] [CrossRef] [PubMed]
- Richards, D.M.; Delacher, M.; Goldfarb, Y.; Kägebein, D.; Hofer, A.-C.; Abramson, J.; Feuerer, M. Treg Cell Differentiation: From Thymus to Peripheral Tissue. Prog. Mol. Biol. Transl. Sci. 2015, 136, 175–205. [Google Scholar] [CrossRef] [PubMed]
- de Salvo, C.; Buela, K.-A.; Pizarro, T.T. Cytokine-Mediated Regulation of Innate Lymphoid Cell Plasticity in Gut Mucosal Immunity. Front. Immunol. 2020, 11, 585319. [Google Scholar] [CrossRef] [PubMed]
- Soni, B.; Singh, S. Cytokine Milieu in Infectious Disease: A Sword or a Boon? J. Interferon Cytokine Res. 2020, 40, 24–32. [Google Scholar] [CrossRef]
- Gómez-López, V.M.; Jubinville, E.; Rodríguez-López, M.I.; Trudel-Ferland, M.; Bouchard, S.; Jean, J. Inactivation of Foodborne Viruses by UV Light: A Review. Foods 2021, 10, 3141. [Google Scholar] [CrossRef]
- Podlacha, M.; Grabowski, Ł.; Kosznik-Kawśnicka, K.; Zdrojewska, K.; Stasiłojć, M.; Węgrzyn, G.; Węgrzyn, A. Interactions of Bacteriophages with Animal and Human Organisms-Safety Issues in the Light of Phage Therapy. Int. J. Mol. Sci. 2021, 22, 8937. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Jończyk-Matysiak, E.; Żaczek, M.; Borysowski, J. Phage-specific diverse effects of bacterial viruses on the immune system. Future Microbiol. 2019, 14, 1171–1174. [Google Scholar] [CrossRef]
- Rouse, M.D.; Stanbro, J.; Roman, J.A.; Lipinski, M.A.; Jacobs, A.; Biswas, B.; Regeimbal, J.; Henry, M.; Stockelman, M.G.; Simons, M.P. Impact of Frequent Administration of Bacteriophage on Therapeutic Efficacy in an A. baumannii Mouse Wound Infection Model. Front. Microbiol. 2020, 11, 414. [Google Scholar] [CrossRef]
- Park, K.; Cha, K.E.; Myung, H. Observation of inflammatory responses in mice orally fed with bacteriophage T7. J. Appl. Microbiol. 2014, 117, 627–633. [Google Scholar] [CrossRef]
- Cha, K.; Oh, H.K.; Jang, J.Y.; Jo, Y.; Kim, W.K.; Ha, G.U.; Ko, K.S.; Myung, H. Characterization of Two Novel Bacteriophages Infecting Multidrug-Resistant (MDR) Acinetobacter baumannii and Evaluation of Their Therapeutic Efficacy in Vivo. Front. Microbiol. 2018, 9, 696. [Google Scholar] [CrossRef] [PubMed]
- de Sousa-Pereira, P.; Woof, J.M. IgA: Structure, Function, and Developability. Antibodies 2019, 8, 57. [Google Scholar] [CrossRef]
- Ladjemi, M.Z.; Martin, C.; Lecocq, M.; Detry, B.; Nana, F.A.; Moulin, C.; Weynand, B.; Fregimilicka, C.; Bouzin, C.; Thurion, P.; et al. Increased IgA Expression in Lung Lymphoid Follicles in Severe Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2019, 199, 592–602. [Google Scholar] [CrossRef]
- Oh, J.E.; Song, E.; Moriyama, M.; Wong, P.; Zhang, S.; Jiang, R.; Strohmeier, S.; Kleinstein, S.H.; Krammer, F.; Iwasaki, A. Intranasal priming induces local lung-resident B cell populations that secrete protective mucosal antiviral IgA. Sci. Immunol. 2021, 6, eabj5129. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Kunitoh-Asari, A.; Hayakawa, K.; Imai, S.; Kasuya, K.; Abe, K.; Adachi, Y.; Fukudome, S.-I.; Takahashi, Y.; Hachimura, S. Oral administration of Lactobacillus plantarum strain AYA enhances IgA secretion and provides survival protection against influenza virus infection in mice. PLoS ONE 2014, 9, e86416. [Google Scholar] [CrossRef] [PubMed]
- Chechushkov, A.; Kozlova, Y.; Baykov, I.; Morozova, V.; Kravchuk, B.; Ushakova, T.; Bardasheva, A.; Zelentsova, E.; Allaf, L.A.; Tikunov, A.; et al. Influence of Caudovirales Phages on Humoral Immunity in Mice. Viruses 2021, 13, 1241. [Google Scholar] [CrossRef]
- Dąbrowska, K.; Miernikiewicz, P.; Piotrowicz, A.; Hodyra, K.; Owczarek, B.; Lecion, D.; Kaźmierczak, Z.; Letarov, A.; Górski, A. Immunogenicity studies of proteins forming the T4 phage head surface. J. Virol. 2014, 88, 12551–12557. [Google Scholar] [CrossRef] [PubMed]
- Hodyra-Stefaniak, K.; Kaźmierczak, Z.; Majewska, J.; Sillankorva, S.; Miernikiewicz, P.; Międzybrodzki, R.; Górski, A.; Azeredo, J.; Lavigne, R.; Lecion, D.; et al. Natural and Induced Antibodies Against Phages in Humans: Induction Kinetics and Immunogenicity for Structural Proteins of PB1-Related Phages. PHAGE 2020, 1, 91–99. [Google Scholar] [CrossRef]
- Nick, J.A.; Dedrick, R.M.; Gray, A.L.; Vladar, E.K.; Smith, B.E.; Freeman, K.G.; Malcolm, K.C.; Epperson, L.E.; Hasan, N.A.; Hendrix, J.; et al. Host and pathogen response to bacteriophage engineered against Mycobacterium abscessus lung infection. Cell 2022, 185, 1860–1874.e12. [Google Scholar] [CrossRef]
- Manrique, P.; Bolduc, B.; Walk, S.T.; van der Oost, J.; de Vos, W.M.; Young, M.J. Healthy human gut phageome. Proc. Natl. Acad. Sci. USA 2016, 113, 10400–10405. [Google Scholar] [CrossRef] [PubMed]
- Viertel, T.M.; Ritter, K.; Horz, H.-P. Viruses versus bacteria-novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J. Antimicrob. Chemother. 2014, 69, 2326–2336. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Tetz, V. Bacteriophages as New Human Viral Pathogens. Microorganisms 2018, 6, 54. [Google Scholar] [CrossRef]
- Duplessis, C.; Warawa, J.M.; Lawrenz, M.B.; Henry, M.; Biswas, B. Successful Intratracheal Treatment of Phage and Antibiotic Combination Therapy of a Multi-Drug Resistant Pseudomonas aeruginosa Murine Model. Antibiotics 2021, 10, 946. [Google Scholar] [CrossRef] [PubMed]
- Prazak, J.; Valente, L.G.; Iten, M.; Federer, L.; Grandgirard, D.; Soto, S.; Resch, G.; Leib, S.L.; Jakob, S.M.; Haenggi, M.; et al. Benefits of Aerosolized Phages for the Treatment of Pneumonia Due to Methicillin-Resistant Staphylococcus aureus: An Experimental Study in Rats. J. Infect. Dis. 2022, 225, 1452–1459. [Google Scholar] [CrossRef] [PubMed]
- Guillon, A.; Pardessus, J.; L’Hostis, G.; Fevre, C.; Barc, C.; Dalloneau, E.; Jouan, Y.; Bodier-Montagutelli, E.; Perez, Y.; Thorey, C.; et al. Inhaled bacteriophage therapy in a porcine model of pneumonia caused by Pseudomonas aeruginosa during mechanical ventilation. Br. J. Pharmacol. 2021, 178, 3829–3842. [Google Scholar] [CrossRef]


| Bacteriophage | Morphology | Phage Titer in Cocktail (PFU/Injection *) | Endotoxin Level (EU/Injection *) | Indicator Strain | Analysis Strain | Reference |
|---|---|---|---|---|---|---|
| DSM 19872 (JG005) | Myovirus | 5 × 107 | 4.30 × 101 | F2230 | DSM 107574 (PA74) | [45] |
| DSM 22045 (JG024) | Myovirus | 5 × 107 | 8.08 × 10−1 | DSM 19882 (PA14) | DSM 107574 (PA74) | [46] |
| 536_P3 | Podovirus | 1 × 108 | 2.93 × 10−2 | 536 | AN33 | - |
| CLB_P2 | Myovirus | 1 × 108 | 9.25 × 10−1 | 55989 | AN33 | [47] |
| LF110_P3 | Myovirus | 1 × 108 | 1.58 × 10−1 | LF110 | AN33 | - |
| LF73_P1 | Myovirus | 1 × 108 | 3.73 × 10−3 | LF73 | AN33 | - |
| DIJ07_P1 | Myovirus | 1 × 108 | 1.49 × 10−3 | DIJ07 | AN33 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weissfuss, C.; Wienhold, S.-M.; Bürkle, M.; Gaborieau, B.; Bushe, J.; Behrendt, U.; Bischoff, R.; Korf, I.H.E.; Wienecke, S.; Dannheim, A.; et al. Repetitive Exposure to Bacteriophage Cocktails against Pseudomonas aeruginosa or Escherichia coli Provokes Marginal Humoral Immunity in Naïve Mice. Viruses 2023, 15, 387. https://doi.org/10.3390/v15020387
Weissfuss C, Wienhold S-M, Bürkle M, Gaborieau B, Bushe J, Behrendt U, Bischoff R, Korf IHE, Wienecke S, Dannheim A, et al. Repetitive Exposure to Bacteriophage Cocktails against Pseudomonas aeruginosa or Escherichia coli Provokes Marginal Humoral Immunity in Naïve Mice. Viruses. 2023; 15(2):387. https://doi.org/10.3390/v15020387
Chicago/Turabian StyleWeissfuss, Chantal, Sandra-Maria Wienhold, Magdalena Bürkle, Baptiste Gaborieau, Judith Bushe, Ulrike Behrendt, Romina Bischoff, Imke H. E. Korf, Sarah Wienecke, Antonia Dannheim, and et al. 2023. "Repetitive Exposure to Bacteriophage Cocktails against Pseudomonas aeruginosa or Escherichia coli Provokes Marginal Humoral Immunity in Naïve Mice" Viruses 15, no. 2: 387. https://doi.org/10.3390/v15020387
APA StyleWeissfuss, C., Wienhold, S.-M., Bürkle, M., Gaborieau, B., Bushe, J., Behrendt, U., Bischoff, R., Korf, I. H. E., Wienecke, S., Dannheim, A., Ziehr, H., Rohde, C., Gruber, A. D., Ricard, J.-D., Debarbieux, L., Witzenrath, M., & Nouailles, G. (2023). Repetitive Exposure to Bacteriophage Cocktails against Pseudomonas aeruginosa or Escherichia coli Provokes Marginal Humoral Immunity in Naïve Mice. Viruses, 15(2), 387. https://doi.org/10.3390/v15020387






