HoBi-like Pestivirus Is Highly Prevalent in Cattle Herds in the Amazon Region (Northern Brazil)
Abstract
:1. Introduction
2. Materials and Methods
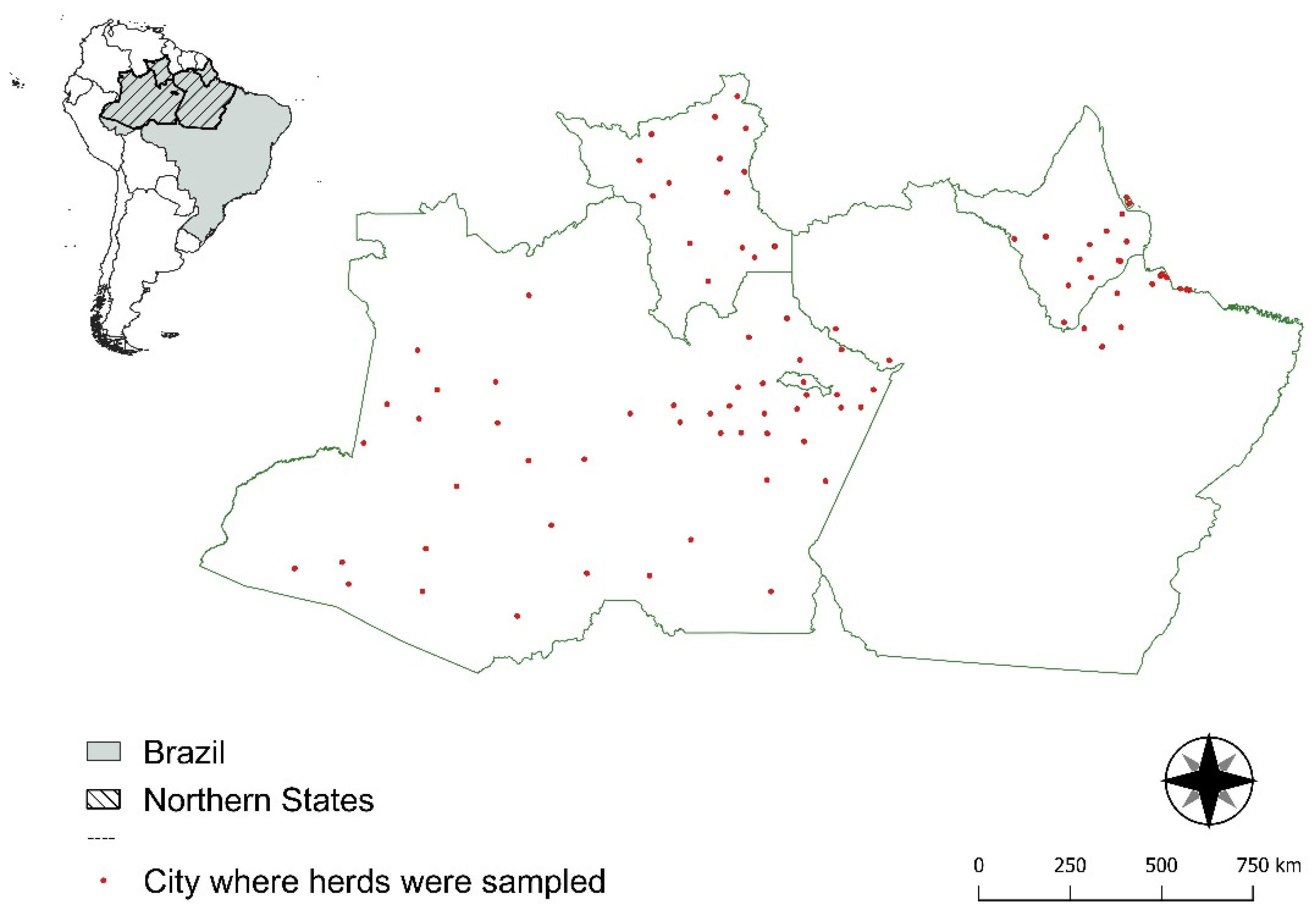
2.1. Target Population and Sample Size
2.2. RNA Isolation and RT–qPCR
2.3. Sequencing and Phylogenetic Analysis
2.4. Virus Neutralization
3. Results
3.1. RT–PCR and Phylogenetic Analyses
3.2. Virus Neutralization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, D.B.; Meyers, G.; Bukh, J.; Gould, E.A.; Monath, T.; Scott Muerhoff, A.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; Simmonds, P.; et al. Proposed Revision to the Taxonomy of the Genus Pestivirus, Family Flaviviridae. J. Gen. Virol. 2017, 98, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef]
- Yeşilbağ, K.; Alpay, G.; Becher, P. Variability and Global Distribution of Subgenotypes of Bovine Viral Diarrhea Virus. Viruses 2017, 9, 128. [Google Scholar] [CrossRef]
- Vilcek, S.; Paton, D.J.; Durkovic, B.; Strojny, L.; Ibata, G.; Moussa, A.; Loitsch, A.; Rossmanith, W.; Vega, S.; Scicluna, M.T.; et al. Bovine Viral Diarrhoea Virus Genotype 1 Can Be Separated into at Least Eleven Genetic Groups. Arch. Virol. 2001, 146, 99–115. [Google Scholar] [CrossRef]
- Deng, M.; Chen, N.; Guidarini, C.; Xu, Z.; Zhang, J.; Cai, L.; Yuan, S.; Sun, Y.; Metcalfe, L. Prevalence and Genetic Diversity of Bovine Viral Diarrhea Virus in Dairy Herds of China. Vet. Microbiol. 2020, 242, 108565. [Google Scholar] [CrossRef]
- Rivas, J.; Hasanaj, A.; Deblon, C.; Gisbert, P.; Garigliany, M.-M. Genetic Diversity of Bovine Viral Diarrhea Virus in Cattle in France between 2018 and 2020. Front. Vet. Sci. 2022, 9, 1028866. [Google Scholar] [CrossRef]
- Ridpath, J.F.; Bolin, S.R.; Dubovi, E.J. Segregation of Bovine Viral Diarrhea Virus into Genotypes. Virology 1994, 205, 66–74. [Google Scholar] [CrossRef]
- Flores, E.F.; Ridpath, J.F.; Weiblen, R.; Vogel, F.S.F.; Gil, L.H.V.G. Phylogenetic Analysis of Brazilian Bovine Viral Diarrhea Virus Type 2 (BVDV-2) Isolates: Evidence for a Subgenotype within BVDV-2. Virus Res. 2002, 87, 51–60. [Google Scholar] [CrossRef]
- de Oliveira, P.S.B.; Silva Júnior, J.V.J.; Weiblen, R.; Flores, E.F. A New (Old) Bovine Viral Diarrhea Virus 2 Subtype: BVDV-2e. Arch. Virol. 2022, 167, 2545–2553. [Google Scholar] [CrossRef]
- Giammarioli, M.; Ridpath, J.F.; Rossi, E.; Bazzucchi, M.; Casciari, C.; De Mia, G.M. Genetic Detection and Characterization of Emerging HoBi-like Viruses in Archival Foetal Bovine Serum Batches. Biologicals 2015, 43, 220–224. [Google Scholar] [CrossRef]
- Kalaiyarasu, S.; Mishra, N.; Jayalakshmi, K.; Selvaraj, P.; Sudhakar, S.B.; Jhade, S.K.; Sood, R.; Premalatha, N.; Singh, V.P. Molecular Characterization of Recent HoBi-like Pestivirus Isolates from Cattle Showing Mucosal Disease-like Signs in India Reveals Emergence of a Novel Genetic Lineage. Transbound Emerg. Dis. 2022, 69, 308–326. [Google Scholar] [CrossRef]
- Baker, J.C. Bovine Viral Diarrhea Virus: A Review. J. Am. Vet. Med. Assoc. 1987, 190, 1449–1458. [Google Scholar] [PubMed]
- Collett, M.S.; Larson, R.; Belzer, S.K.; Retzel, E. Proteins Encoded by Bovine Viral Diarrhea Virus: The Genomic Organization of a Pestivirus. Virology 1988, 165, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Orlich, M.; Thiel, H.-J. Complete Genomic Sequence of Border Disease Virus, a Pestivirus from Sheep. J. Virol. 1998, 72, 5165–5173. [Google Scholar] [CrossRef] [PubMed]
- Tautz, N.; Tews, B.A.; Meyers, G. The Molecular Biology of Pestiviruses. Adv. Virus Res. 2015, 93, 47–160. [Google Scholar] [CrossRef] [PubMed]
- Van Rijn, P.A. Subdivision of the Pestivirus Genus Based on Envelope Glycoprotein E2. Virology 1997, 237, 337–348. [Google Scholar] [CrossRef]
- Vilcek, S.; Durkovic, B.; Kolesarova, M.; Paton, D.J. Genetic Diversity of BVDV: Consequences for Classification and Molecular Epidemiology. Prev. Vet. Med. 2005, 72, 31–35, discussion 215–219. [Google Scholar] [CrossRef]
- Houe, H. Epidemiological Features and Economical Importance of Bovine Virus Diarrhoea Virus (BVDV) Infections. Vet. Microbiol. 1999, 64, 89–107. [Google Scholar] [CrossRef]
- Schweizer, M.; Stalder, H.; Haslebacher, A.; Grisiger, M.; Schwermer, H.; Di Labio, E. Eradication of Bovine Viral Diarrhoea (BVD) in Cattle in Switzerland: Lessons Taught by the Complex Biology of the Virus. Front. Vet. Sci. 2021, 8, 702730. [Google Scholar] [CrossRef] [PubMed]
- Autio, T.; Pohjanvirta, T.; Holopainen, R.; Rikula, U.; Pentikäinen, J.; Huovilainen, A.; Rusanen, H.; Soveri, T.; Sihvonen, L.; Pelkonen, S. Etiology of Respiratory Disease in Non-Vaccinated, Non-Medicated Calves in Rearing Herds. Vet. Microbiol. 2007, 119, 256–265. [Google Scholar] [CrossRef]
- Silveira, S.; Weber, M.N.; Mósena, A.C.S.; da Silva, M.S.; Streck, A.F.; Pescador, C.A.; Flores, E.F.; Weiblen, R.; Driemeier, D.; Ridpath, J.F.; et al. Genetic Diversity of Brazilian Bovine Pestiviruses Detected Between 1995 and 2014. Transbound Emerg. Dis. 2017, 64, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeier, H.; Strebelow, G.; Depner, K.; Hoffmann, B.; Beer, M. Genetic and Antigenic Characterization of an Atypical Pestivirus Isolate, a Putative Member of a Novel Pestivirus Species. J. Gen. Virol. 2004, 85, 3647–3652. [Google Scholar] [CrossRef]
- Stalder, H.P.; Meier, P.; Pfaffen, G.; Wageck-Canal, C.; Rüfenacht, J.; Schaller, P.; Bachofen, C.; Marti, S.; Vogt, H.R.; Peterhans, E. Genetic Heterogeneity of Pestiviruses of Ruminants in Switzerland. Prev. Vet. Med. 2005, 72, 37–41, discussion 215–219. [Google Scholar] [CrossRef] [PubMed]
- Silveira, S.; Baumbach, L.F.; Weber, M.N.; Mósena, A.C.S.; da Silva, M.S.; Cibulski, S.P.; Borba, M.R.; Maia, R.D.; Coimbra, V.C.S.; de Moraes, G.M.; et al. HoBi-like Is the Most Prevalent Ruminant Pestivirus in Northeastern Brazil. Transbound Emerg. Dis. 2018, 65, e113–e120. [Google Scholar] [CrossRef] [PubMed]
- Pecora, A.; Perez Aguirreburualde, M.S.; Ridpath, J.F.; Dus Santos, M.J. Molecular Characterization of Pestiviruses in Fetal Bovine Sera Originating From Argentina: Evidence of Circulation of HoBi-Like Viruses. Front. Vet. Sci. 2019, 6, 359. [Google Scholar] [CrossRef]
- Margineda, C.A.; Ferreyra, F.M.; Masnyj, F.; Audrito, M.; Favaro, P.M.; María José, D.S.; Pecora, A. HoBi-like Pestivirus in 2 Cases of Fatal Respiratory Disease of Feedlot Cattle in Argentina. J. Vet. Diagn. Investig. 2022, 34, 693–698. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, L.; Niu, L.; Shangguan, H.; Huang, C.; Yi, Y.; Zhang, Y.; Gao, M.; Ge, J. Genetic and Evolutionary Analysis of Emerging HoBi-like Pestivirus. Res. Vet. Sci. 2021, 137, 217–225. [Google Scholar] [CrossRef]
- Decaro, N.; Lucente, M.S.; Mari, V.; Cirone, F.; Cordioli, P.; Camero, M.; Sciarretta, R.; Losurdo, M.; Lorusso, E.; Buonavoglia, C. Atypical Pestivirus and Severe Respiratory Disease in Calves, Europe. Emerg. Infect. Dis. 2011, 17, 1549–1552. [Google Scholar] [CrossRef]
- Luzzago, C.; Decaro, N. Epidemiology of Bovine Pestiviruses Circulating in Italy. Front. Vet. Sci. 2021, 8, 669942. [Google Scholar] [CrossRef]
- Dias, R.K.; Cargnelutti, J.F.; Weber, M.N.; Canal, C.W.; Bauermann, F.V.; Ridpath, J.F.; Weiblen, R.; Flores, E.F. Antigenic Diversity of Brazilian Isolates of HoBi-like Pestiviruses. Vet. Microbiol. 2017, 203, 221–228. [Google Scholar] [CrossRef]
- Weber, M.N.; Silveira, S.; Machado, G.; Groff, F.H.S.; Mósena, A.C.S.; Budaszewski, R.F.; Dupont, P.M.; Corbellini, L.G.; Canal, C.W. High Frequency of Bovine Viral Diarrhea Virus Type 2 in Southern Brazil. Virus Res. 2014, 191, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.V.; Konradt, G.; de Souza, S.O.; Bassuino, D.M.; Silveira, S.; Mósena, A.C.S.; Canal, C.W.; Pavarini, S.P.; Driemeier, D. Natural Outbreak of BVDV-1d-Induced Mucosal Disease Lacking Intestinal Lesions. Vet. Pathol. 2017, 54, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Mósena, A.C.S.; Weber, M.N.; Cibulski, S.P.; Silveira, S.; Silva, M.S.; Mayer, F.Q.; Canal, C.W. Genomic Characterization of a Bovine Viral Diarrhea Virus Subtype 1i in Brazil. Arch. Virol. 2017, 162, 1119–1123. [Google Scholar] [CrossRef]
- Ståhl, K.; Alenius, S. BVDV Control and Eradication in Europe—An Update. Jpn. J. Vet. Res. 2012, 60, S31–S39. [Google Scholar] [PubMed]
- Wernike, K.; Schirrmeier, H.; Strebelow, H.-G.; Beer, M. Eradication of Bovine Viral Diarrhea Virus in Germany-Diversity of Subtypes and Detection of Live-Vaccine Viruses. Vet. Microbiol. 2017, 208, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Houe, H.; Lindberg, A.; Moennig, V. Test Strategies in Bovine Viral Diarrhea Virus Control and Eradication Campaigns in Europe. J. Vet. Diagn. Investig. 2006, 18, 427–436. [Google Scholar] [CrossRef]
- IBGE. Rebanho de Bovinos (Bois e Vacas). 2022. Available online: https://www.ibge.gov.br/explica/producao-agropecuaria/bovinos/br (accessed on 17 January 2023).
- USDA-FAS-Brazil: Livestock and Products Annual. 2022. Available online: https://usdabrazil.org.br/wp-content/uploads/2022/10/Livestock-and-Products-Annual_Brasilia_Brazil_BR2022-0054-1.pdf (accessed on 17 January 2023).
- Cameron, A.R.; Baldock, F.C. Two-Stage Sampling in Surveys to Substantiate Freedom from Disease. Prev. Vet. Med. 1998, 34, 19–30. [Google Scholar] [CrossRef]
- Deregt, D.; Gilbert, S.A.; Dudas, S.; Pasick, J.; Baxi, S.; Burton, K.M.; Baxi, M.K. A Multiplex DNA Suspension Microarray for Simultaneous Detection and Differentiation of Classical Swine Fever Virus and Other Pestiviruses. J. Virol. Methods 2006, 136, 17–23. [Google Scholar] [CrossRef]
- Vilcek, S.; Herring, A.J.; Herring, J.A.; Nettleton, P.F.; Lowings, J.P.; Paton, D.J. Pestiviruses Isolated from Pigs, Cattle and Sheep Can Be Allocated into at Least Three Genogroups Using Polymerase Chain Reaction and Restriction Endonuclease Analysis. Arch. Virol. 1994, 136, 309–323. [Google Scholar] [CrossRef]
- Liu, L.; Kampa, J.; Belák, S.; Baule, C. Virus Recovery and Full-Length Sequence Analysis of Atypical Bovine Pestivirus Th/04_KhonKaen. Vet. Microbiol. 2009, 138, 62–68. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Xia, H.; Wahlberg, N.; Belák, S.; Baule, C. Phylogeny, Classification and Evolutionary Insights into Pestiviruses. Virology 2009, 385, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Tajima, M.; Frey, H.R.; Yamato, O.; Maede, Y.; Moennig, V.; Scholz, H.; Greiser-Wilke, I. Prevalence of Genotypes 1 and 2 of Bovine Viral Diarrhea Virus in Lower Saxony, Germany. Virus Res. 2001, 76, 31–42. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- OIE. Bovine Viral Diarrhoea Chapter 3.4.7. Manual of Diagnostic Test and Vaccines for Terrestrial Animals 2019. 2018. Available online: www.oie.int/Standardsetting/Terrestrial-Manual/Access-Online/ (accessed on 15 March 2020).
- Falagan-Lotsch, P.; Lopes, T.S.; Ferreira, N.; Balthazar, N.; Monteiro, A.M.; Borojevic, R.; Granjeiro, J.M. Performance of PCR-Based and Bioluminescent Assays for Mycoplasma Detection. J. Microbiol. Methods 2015, 118, 31–36. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty per Cent Endpointe. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Pinior, B.; Firth, C.L.; Richter, V.; Lebl, K.; Trauffler, M.; Dzieciol, M.; Hutter, S.E.; Burgstaller, J.; Obritzhauser, W.; Winter, P.; et al. A Systematic Review of Financial and Economic Assessments of Bovine Viral Diarrhea Virus (BVDV) Prevention and Mitigation Activities Worldwide. Prev. Vet. Med. 2017, 137, 77–92. [Google Scholar] [CrossRef]
- Moennig, V.; Houe, H.; Lindberg, A. BVD Control in Europe: Current Status and Perspectives. Anim. Health Res. Rev. 2005, 6, 63–74. [Google Scholar] [CrossRef]
- Antos, A. Vaccination Failure in Eradication and Control Programs for Bovine Viral Diarrhea Infection. Front. Vet. Sci. 2021, 8, 688911. [Google Scholar] [CrossRef]
- Mósena, A.C.S.; Cibulski, S.P.; Weber, M.N.; Silveira, S.; Silva, M.S.; Mayer, F.Q.; Roehe, P.M.; Canal, C.W. Genomic and Antigenic Relationships between Two ‘HoBi’-like Strains and Other Members of the Pestivirus Genus. Arch. Virol. 2017, 162, 3025–3034. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.L.; Martins, B.; Cargnelutti, J.F.; Noll, J.G.; Weiblen, R.; Flores, E.F. Genetic Identification of Pestiviruses from Beef Cattle in Southern Brazil. Braz. Microbiol. 2019, 50, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Bauermann, F.V.; Ridpath, J.F. Epidemiology of Pestivirus H in Brazil and Its Control Implications. Front. Vet. Sci. 2021, 8, 693041. [Google Scholar] [CrossRef] [PubMed]
- de Sá, M.E.P.; de Sá, C.V.G.C.; Nicolino, R.R.; Haddad, J.P.A.; McManus, C.; Seixas, L.; de Melo, C.B. Data on Network of Live Cattle Exports from Brazil. Data Brief 2018, 19, 1963–1969. [Google Scholar] [CrossRef]
- Zu Ermgassen, E.K.H.J.; Godar, J.; Lathuillière, M.J.; Löfgren, P.; Gardner, T.; Vasconcelos, A.; Meyfroidt, P. The Origin, Supply Chain, and Deforestation Risk of Brazil’s Beef Exports. Proc. Natl. Acad. Sci. USA 2020, 117, 31770–31779. [Google Scholar] [CrossRef]
- Bauermann, F.V.; Flores, E.F.; Ridpath, J.F. Antigenic Relationships between Bovine Viral Diarrhea Virus 1 and 2 and HoBi Virus: Possible Impacts on Diagnosis and Control. J. Vet. Diagn. Investig. 2012, 24, 253–261. [Google Scholar] [CrossRef]
- Bauermann, F.V.; Flores, E.F.; Falkenberg, S.M.; Weiblen, R.; Ridpath, J.F. Lack of Evidence for the Presence of Emerging HoBi-like Viruses in North American Fetal Bovine Serum Lots. J. Vet. Diagn. Investig. 2014, 26, 10–17. [Google Scholar] [CrossRef]
- Nilnont, T.; Aiumlamai, S.; Kanistanont, K.; Inchaisri, C.; Kampa, J. Bovine Viral Diarrhea Virus (BVDV) Infection in Dairy Cattle Herds in Northeast Thailand. Trop. Anim. Health Prod. 2016, 48, 1201–1208. [Google Scholar] [CrossRef]
- Silveira, S.; Cibulski, S.P.; Junqueira, D.M.; Mósena, A.C.S.; Weber, M.N.; Mayer, F.Q.; Canal, C.W. Phylogenetic and Evolutionary Analysis of HoBi-like Pestivirus: Insights into Origin and Dispersal. Transbound Emerg. Dis. 2020, 67, 1909–1917. [Google Scholar] [CrossRef]
- Bauermann, F.V.; Ridpath, J.F.; Weiblen, R.; Flores, E.F. HoBi-like Viruses: An Emerging Group of Pestiviruses. J. Vet. Diagn. Investig. 2013, 25, 6–15. [Google Scholar] [CrossRef]
- Mosena, A.C.S.; Wolf, J.M.; Paim, W.P.; Baumbach, L.F.; da Silva, M.S.; Silveira, S.; Olegário, J.d.C.; Budaszewski, R.d.F.; Weber, M.N.; Canal, C.W. Temporal Analysis of Bovine Pestivirus Diversity in Brazil. Braz. J. Microbiol. 2022, 53, 1675–1682. [Google Scholar] [CrossRef] [PubMed]
- Spetter, M.J.; Louge Uriarte, E.L.; Verna, A.E.; Odeón, A.C.; González Altamiranda, E.A. Temporal and Geographic Dynamics of Bovine Viral Diarrhea Virus in American Countries. Res. Vet. Sci. 2022, 153, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Tildesley, M.J.; Brand, S.; Brooks Pollock, E.; Bradbury, N.V.; Werkman, M.; Keeling, M.J. The Role of Movement Restrictions in Limiting the Economic Impact of Livestock Infections. Nat. Sustain. 2019, 2, 834–840. [Google Scholar] [CrossRef]
- Weber, M.N.; Mósena, A.C.S.; Simões, S.V.D.; Almeida, L.L.; Pessoa, C.R.M.; Budaszewski, R.F.; Silva, T.R.; Ridpath, J.F.; Riet-Correa, F.; Driemeier, D.; et al. Clinical Presentation Resembling Mucosal Disease Associated with ‘HoBi’-like Pestivirus in a Field Outbreak. Transbound Emerg. Dis. 2016, 63, 92–100. [Google Scholar] [CrossRef]
- Becher, P.; Orlich, M.; Shannon, A.D.; Horner, G.; König, M.; Thiel, H.J. Phylogenetic Analysis of Pestiviruses from Domestic and Wild Ruminants. J. Gen. Virol. 1997, 78, 1357–1366. [Google Scholar] [CrossRef]
- Chernick, A.; Godson, D.L.; van der Meer, F. Metadata beyond the Sequence Enables the Phylodynamic Inference of Bovine Viral Diarrhea Virus Type 1a Isolates from Western Canada. Infect. Genet. Evol. 2014, 28, 367–374. [Google Scholar] [CrossRef]
- Weber, M.N.; Streck, A.F.; Silveira, S.; Mósena, A.C.S.; da Silva, M.S.; Canal, C.W. Homologous Recombination in Pestiviruses: Identification of Three Putative Novel Events between Different Subtypes/Genogroups. Infect. Genet. Evol. 2015, 30, 219–224. [Google Scholar] [CrossRef]
- Scharnböck, B.; Roch, F.-F.; Richter, V.; Funke, C.; Firth, C.L.; Obritzhauser, W.; Baumgartner, W.; Käsbohrer, A.; Pinior, B. A Meta-Analysis of Bovine Viral Diarrhoea Virus (BVDV) Prevalences in the Global Cattle Population. Sci. Rep. 2018, 8, 14420. [Google Scholar] [CrossRef]
- Flores, E.F.; Weiblen, R.; Vogel, F.S.F.; Roehe, P.M.; Alfieri, A.A.; Pituco, E.M. A infecção pelo vírus da diarréia viral bovina (BVDV) no Brasil: Histórico, situação atual e perspectivas.Bovine viral diarrhea virus (BVDV) infection in Brazil: History, current situation and perspectives. Pesq. Vet. Bras. 2005, 25, 125–134. [Google Scholar] [CrossRef]
- de Brito, W.M.E.D.; Alfaia, B.T.; Caixeta, S.P.d.M.B.; Ribeiro, A.C.C.; Miranda, T.d.M.T.; Barbosa, A.C.V.C.; Barthasson, D.L.; Linhares, D.C.; Faria, B.O. Prevalência da infecção pelo Vírus da Diarréia Viral Bovina (BVDV) no Estado de Goiás, Brasil. J. Trop. Pathol. 2010, 39, 07–20. [Google Scholar] [CrossRef]
- Zardo, R. Prevalência e variáveis associadas à infecção por BoHV-1, BVDV, Leptospira spp. e Neospora caninum em bovinos leiteiros no município de Novo Xingu-RS. Prevalência e Variáveis Associadas à Infecção por BoHV-1, BVDV, Leptospira spp. e Neospora Caninum em Bovinos Leiteiros no Município de Novo Xingu-RS. Master’s Thesis, Universidade Federal de Pelotas, Pelotas, Brazil, 2017. [Google Scholar]
- Alves, M.E.M.; Fernandes, F.D.; Monteiro, F.L.; Braunig, P.; Cargnelutti, J.F.; Flores, E.F.; Weiblen, R.; Vogel, F.S.F. Co-Infection by Neopora Caninum and Bovine Viral Diarrhea Virus in Cattle from Rio Grande Do Sul, Brazil, Destined to Exportation. Pesq. Vet. Bras. 2020, 40, 593–597. [Google Scholar] [CrossRef]
- Pecora, A.; Pérez Aguirreburualde, M.S.; Malacari, D.A.; Zabal, O.; Sala, J.M.; Konrad, J.L.; Caspe, S.G.; Bauermann, F.; Ridpath, J.; Dus Santos, M.J. Serologic Evidence of HoBi-like Virus Circulation in Argentinean Water Buffalo. J. Vet. Diagn. Investig. 2017, 29, 926–929. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, S.M.; Bauermann, F.V.; Scoles, G.A.; Bonilla, D.; Dassanayake, R.P. A Serosurvey for Ruminant Pestivirus Exposure Conducted Using Sera From Stray Mexico Origin Cattle Captured Crossing Into Southern Texas. Front. Vet. Sci. 2022, 9, 821247. [Google Scholar] [CrossRef]
- Bauermann, F.V.; Ridpath, J.F.; Dargatz, D.A. A Serosurvey for Ruminant Pestivirus Exposure Conducted Using Cattle Sera Collected for Brucellosis Surveillance in the United States. J. Vet. Diagn. Investig. 2017, 29, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.H.H. Sobre a necessidade de inclusão de pestivírus hobi-like em vacinas respiratórias e reprodutivas para bovinos. About the necessity of including hobi-like pestiviruses in bovine respiratory and reproductive vaccines. Pesq. Vet. Bras. 2021, 41, e06914. [Google Scholar] [CrossRef]
- Flores, E.F.; Cargnelutti, J.F.; Monteiro, F.L.; Bauermann, F.V.; Ridpath, J.F.; Weiblen, R. A Genetic Profile of Bovine Pestiviruses Circulating in Brazil (1998–2018). Anim. Health Res. Rev. 2018, 19, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Bauermann, F.V.; Harmon, A.; Flores, E.F.; Falkenberg, S.M.; Reecy, J.M.; Ridpath, J.F. In Vitro Neutralization of HoBi-like Viruses by Antibodies in Serum of Cattle Immunized with Inactivated or Modified Live Vaccines of Bovine Viral Diarrhea Viruses 1 and 2. Vet. Microbiol. 2013, 166, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Ridpath, J.F. Immunology of BVDV Vaccines. Biologicals 2013, 41, 14–19. [Google Scholar] [CrossRef]
- Moennig, V.; Becher, P. Control of Bovine Viral Diarrhea. Pathogens 2018, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Anziliero, D.; Martins, M.; Weiss, M.; Monteiro, F.L.; Ataide, C.F.; Weiblen, R.; Flores, E.F. Resposta sorológica aos herpesvirus bovino tipos 1 e 5 e vírus da diarreia viral bovina induzida por vacinas comerciais. Serological response to bovine herpesvírus 1 and 5 and bovine viral diarrea virus induced by comercial vaccines. Cienc. Rural. 2015, 45, 58–63. [Google Scholar] [CrossRef] [Green Version]
- Cortese, V.S.; Grooms, D.L.; Ellis, J.; Bolin, S.R.; Ridpath, J.F.; Brock, K.V. Protection of Pregnant Cattle and Their Fetuses against Infection with Bovine Viral Diarrhea Virus Type 1 by Use of a Modified-Live Virus Vaccine. Am. J. Vet. Res. 1998, 59, 1409–1413. [Google Scholar] [PubMed]
- Michel, P.; Thurmond, M.; Hietala, S. Prevaccination Bovine Viral Diarrhea Virus Titers and Subsequent Reproductive Performance in Dairy Heifers. Can. J. Vet. Res. 1993, 57, 236–240. [Google Scholar] [PubMed]
- Brum, M.C.S.; Weiblen, R.; Flores, E.F.; Pituco, E.M.; Tobias, F.L.; Winkelmann, E.R. Proteção fetal frente a desafio com o vírus da Diarréia Viral Bovina (BVDV) em ovelhas imunizadas com duas amostras de vírus modificadas experimentalmente. Fetal protection against challenge with bovine viral diarrhea virus (BVDV) in pregnant ewes immunized with two strains experimentally attenuated. Pesq. Vet. Bras. 2002, 22, 64–72. [Google Scholar] [CrossRef] [Green Version]



| Target | Primer | Pestivirus Species * | Reference |
|---|---|---|---|
| 5′UTR | Pesti F; Pesti R | BVDV-1, BVDV-2 | [40] |
| 324 F; 326 R ** | BVDV-1, BVDV-2, HoBiPeV | [41] | |
| Panpesti F; Panpesti R | BVDV-1, BVDV-2, HoBiPeV | [31] | |
| Npro | TF3; TR3 ** | HoBiPeV | [42] |
| LV Pesti F; LV PestiR | BVDV-2 | [34] | |
| BD1, BD3 | BVDV-1 | [4] | |
| E2 | SF3, SR3 ** | HoBiPeV | [43] |
| E2F, E2R | BVDV-1, BVDV-2 | [44] |
| Federative State | VN Sample | Positive Samples | |||
|---|---|---|---|---|---|
| BVDV-1 | BVDV-2 | HoBiPeV | No Predominant Titer | ||
| AM | 226 | 14 | 2 | 20 | 28 |
| AP | 57 | 5 | 5 | 9 | 13 |
| PA | 29 | 1 | 7 | 4 | 12 |
| RR | 78 | 15 | 11 | 4 | 27 |
| Total | 390 | 35 | 25 | 37 | 80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baumbach, L.F.; Mósena, A.C.S.; Alves, R.S.; Camargo, L.J.; Olegário, J.C.; Lobraico, L.R.; Costa, J.M.N.; Borba, M.R.; Bauermann, F.V.; Weber, M.N.; et al. HoBi-like Pestivirus Is Highly Prevalent in Cattle Herds in the Amazon Region (Northern Brazil). Viruses 2023, 15, 453. https://doi.org/10.3390/v15020453
Baumbach LF, Mósena ACS, Alves RS, Camargo LJ, Olegário JC, Lobraico LR, Costa JMN, Borba MR, Bauermann FV, Weber MN, et al. HoBi-like Pestivirus Is Highly Prevalent in Cattle Herds in the Amazon Region (Northern Brazil). Viruses. 2023; 15(2):453. https://doi.org/10.3390/v15020453
Chicago/Turabian StyleBaumbach, Leticia F., Ana Cristina S. Mósena, Raquel S. Alves, Laura J. Camargo, Juliana C. Olegário, Leonardo R. Lobraico, João Marcos N. Costa, Mauro R. Borba, Fernando V. Bauermann, Matheus N. Weber, and et al. 2023. "HoBi-like Pestivirus Is Highly Prevalent in Cattle Herds in the Amazon Region (Northern Brazil)" Viruses 15, no. 2: 453. https://doi.org/10.3390/v15020453







