Persistence of a Frameshifting Deletion in SARS-CoV-2 ORF7a for the Duration of a Major Outbreak
Abstract
1. Introduction
2. Materials and Methods
2.1. Re-Analysis of Whole-Genome Sequencing Data
2.2. Analysis of Deletions in Global Delta Genomes
3. Structural Modelling
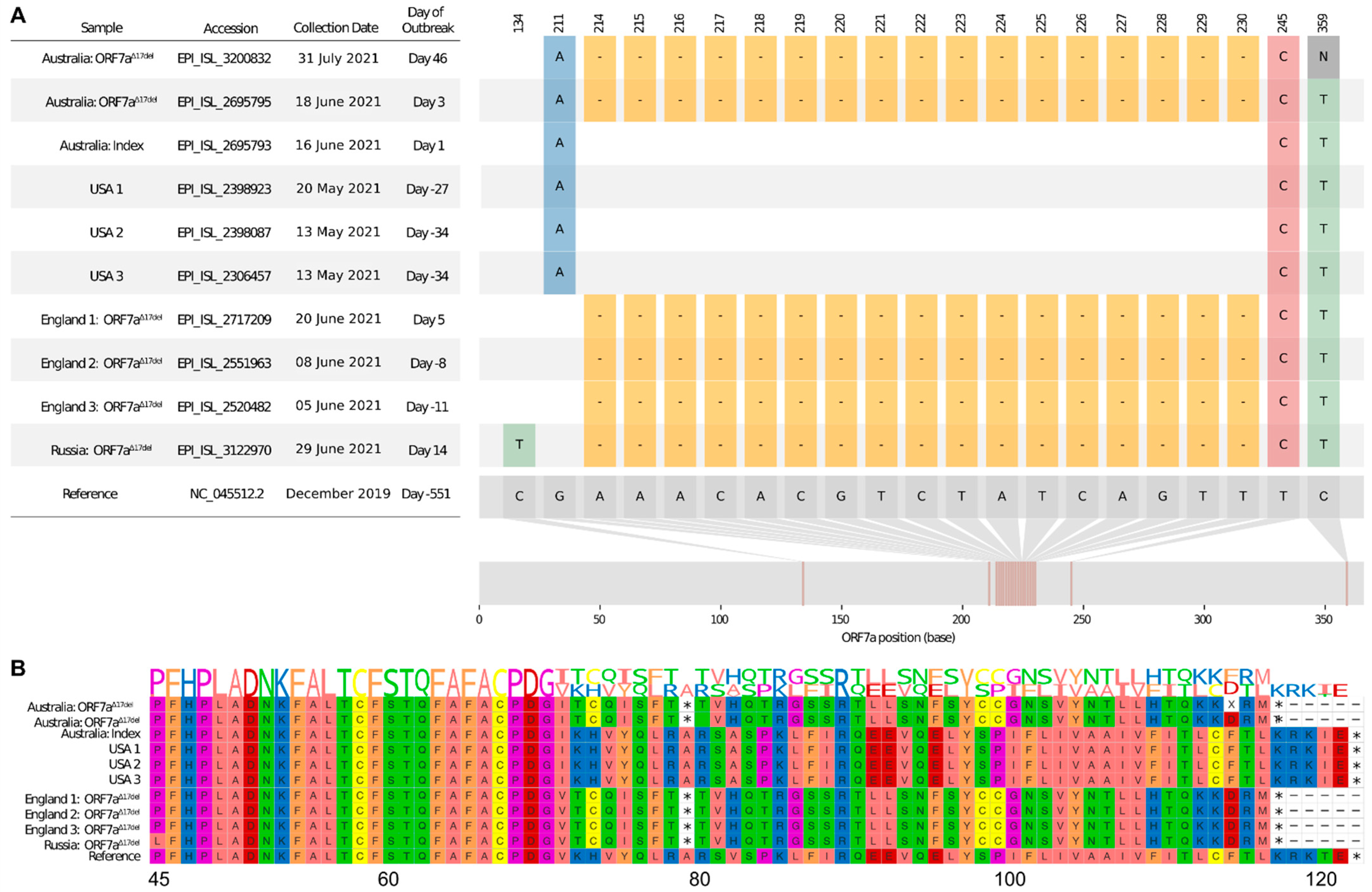
4. Results
4.1. Initial Sub-Consensus Frequency of ORF7aΔ17del
4.2. Frequency of Delta-ORF7aΔ17del
4.3. Structure of ORF7a in Delta-ORF7aΔ17del
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, C.A.; Pekosz, A.; Lee, C.A.; Diamond, M.S.; Fremont, D.H. Structure and Intracellular Targeting of the SARS-Coronavirus Orf7a Accessory Protein. Structure 2005, 13, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Nichols, J.; Snyder, D.T.; Hedges, J.F.; Cicha, C.; Lee, H.; Vanderwood, K.K.; Bimczok, D.; et al. SARS-CoV-2 genomic surveillance identifies naturally occurring truncation of ORF7a that limits immune suppression. Cell Rep. 2021, 35, 109197. [Google Scholar] [CrossRef] [PubMed]
- Holland, L.A.; Kaelin, E.A.; Maqsood, R.; Estifanos, B.; Wu, L.I.; Varsani, A.; Halden, R.U.; Hogue, B.G.; Scotch, M.; Lim, E.S. An 81-Nucleotide Deletion in SARS-CoV-2 ORF7a Identified from Sentinel Surveillance in Arizona (January to March 2020). J. Virol. 2020, 94, e00711-20. [Google Scholar] [CrossRef] [PubMed]
- Martin-Sancho, L.; Lewinski, M.K.; Pache, L.; Stoneham, C.A.; Yin, X.; Becker, M.E.; Pratt, D.; Churas, C.; Rosenthal, S.B.; Liu, S.; et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell 2021, 81, 2656–2668.e8. [Google Scholar] [CrossRef]
- Yi, E.; Oh, J.; Kang, H.-R.; Song, M.J.; Park, S.-H. BST2 inhibits infection of influenza A virus by promoting apoptosis of infected cells. Biochem. Biophys. Res. Commun. 2018, 509, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Huang, C.; Zhou, Z.; Huang, Z.; Su, L.; Kang, S.; Chen, X.; Chen, Q.; He, S.; Rong, X.; et al. Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14+ monocytes. Iscience 2021, 24, 102187. [Google Scholar] [CrossRef] [PubMed]
- Foster, C.S.P.; Rawlinson, W.D. Rapid Spread of a SARS-CoV-2 Delta Variant with a Frameshift Deletion in ORF7a. medRxiv 2021. [Google Scholar] [CrossRef]
- Steinig, E.; Coin, L. Nanoq: Ultra-fast quality control for nanopore reads. J. Open Source Softw. 2022, 7, 2991. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, S.; Su, J.; Leung, A.W.-S.; Lam, T.-W.; Luo, R. Symphonizing pileup and full-alignment for deep learning-based long-read variant calling. Nat. Comput. Sci. 2022, 2, 797–803. [Google Scholar] [CrossRef]
- Foster, C.S.P. deletion_detector: A simple and quick way to detect deletions relative to a reference sequence (v0.2.1). Zenodo 2022. [Google Scholar] [CrossRef]
- Jackson, B. gofasta: Command-line utilities for genomic epidemiology research. Bioinformatics 2022, 38, 4033–4035. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Cao, Z.; Xia, H.; Rajsbaum, R.; Xia, X.; Wang, H.; Shi, P.-Y. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell. Mol. Immunol. 2021, 18, 746–748. [Google Scholar] [CrossRef]
- Addetia, A.; Xie, H.; Roychoudhury, P.; Shrestha, L.; Loprieno, M.; Huang, M.-L.; Jerome, K.R.; Greninger, A.L. Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J. Clin. Virol. 2020, 129, 104523. [Google Scholar] [CrossRef]
- Joonlasak, K.; Batty, E.M.; Kochakarn, T.; Panthan, B.; Kümpornsin, K.; Jiaranai, P.; Wangwiwatsin, A.; Huang, A.; Kotanan, N.; Jaru-Ampornpan, P.; et al. Genomic surveillance of SARS-CoV-2 in Thailand reveals mixed imported populations, a local lineage expansion and a virus with truncated ORF7a. Virus Res. 2020, 292, 198233. [Google Scholar] [CrossRef] [PubMed]
- Panzera, Y.; Ramos, N.; Frabasile, S.; Calleros, L.; Marandino, A.; Tomás, G.; Techera, C.; Grecco, S.; Fuques, E.; Goñi, N.; et al. A deletion in SARS-CoV-2 ORF7 identified in COVID-19 outbreak in Uruguay. Transbound. Emerg. Dis. 2021, 68, 3075–3082. [Google Scholar] [CrossRef] [PubMed]
- Tse, H.; Wong, S.C.-Y.; Ip, K.-F.; Cheng, V.C.-C.; To, K.K.-W.; Lung, D.C.; Choi, G.K.-Y. Genome Sequences of Three SARS-CoV-2 ORF7a Deletion Variants Obtained from Patients in Hong Kong. Genome Announc. 2021, 10, e00251-21. [Google Scholar] [CrossRef]
- Wang, N.; Lysenkov, V.; Orte, K.; Kairisto, V.; Aakko, J.; Khan, S.; Elo, L.L. Variant Calling Tool Evaluation for Variable Size Indel Calling from next Generation Whole Genome and Targeted Sequencing Data. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lobiuc, A.; Șterbuleac, D.; Sturdza, O.; Dimian, M.; Covasa, M. A Conservative Replacement in the Transmembrane Domain of SARS-CoV-2 ORF7a as a Putative Risk Factor in COVID-19. Biology 2021, 10, 1276. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-M.; Huang, K.-J.; Wang, C.-T. BST2/CD317 counteracts human coronavirus 229E productive infection by tethering virions at the cell surface. Virology 2014, 449, 287–296. [Google Scholar] [CrossRef]
- MacArthur, D.G.; Balasubramanian, S.; Frankish, A.; Huang, N.; Morris, J.; Walter, K.; Jostins, L.; Habegger, L.; Pickrell, J.K.; Montgomery, S.B.; et al. A Systematic Survey of Loss-of-Function Variants in Human Protein-Coding Genes. Science 2012, 335, 823–828. [Google Scholar] [CrossRef]
- Ruan, Y.; Luo, Z.; Tang, X.; Li, G.; Wen, H.; He, X.; Lu, X.; Lu, J.; Wu, C.-I. On the founder effect in COVID-19 outbreaks: How many infected travelers may have started them all? Natl. Sci. Rev. 2020, 8, nwaa246. [Google Scholar] [CrossRef] [PubMed]
- Farkas, C.; Fuentes-Villalobos, F.; Garrido, J.L.; Haigh, J.; Barría, M.I. Insights on early mutational events in SARS-CoV-2 virus reveal founder effects across geographical regions. Peerj 2020, 8, e9255. [Google Scholar] [CrossRef]
- Díez-Fuertes, F.; Iglesias-Caballero, M.; García-Pérez, J.; Monzón, S.; Jiménez, P.; Varona, S.; Cuesta, I.; Zaballos, Á.; Jiménez, M.; Checa, L.; et al. A Founder Effect Led Early SARS-CoV-2 Transmission in Spain. J. Virol. 2021, 95, e01583-20. [Google Scholar] [CrossRef]
- Foster, C.S.; Madden, M.; Chan, R.; Agapiou, D.; Bull, R.A.; Rawlinson, W.D.; Van Hal, S.J. SARS-CoV-2 N-gene mutation leading to Xpert Xpress SARS-CoV-2 assay instability. Pathology 2022, 54, 499–501. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Rangarajan, E.S.; Izard, T.; Farzan, M.; Choe, H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Khan, A.; Zia, T.; Suleman, M.; Khan, T.; Ali, S.S.; Abbasi, A.A.; Mohammad, A.; Wei, D. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J. Cell. Physiol. 2021, 236, 7045–7057. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D. Translating virus evolution into epidemiology. Cell Host Microbe 2022, 30, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.H.; Poon, A.F.; Whitlock, M.C. Compensatory mutations are repeatable and clustered within proteins. Proc. R. Soc. B Boil. Sci. 2009, 276, 1823–1827. [Google Scholar] [CrossRef] [PubMed]


| Deletion | First Date Sampled | Last Date Sampled | Date Span (Days) | Genomes | Countries | Frameshift-Inducing |
|---|---|---|---|---|---|---|
| (27607, 27623, 17) | 2 June 2021 | 13 February 2022 | 256 | 28014 | 27 | TRUE |
| (27556, 27619, 64) | 20 March 2021 | 22 February 2022 | 339 | 8945 | 17 | TRUE |
| (27579, 27581, 3) | 22 January 2021 | 4 February 2022 | 378 | 5189 | 68 | FALSE |
| (27551, 27577, 27) | 23 February 2021 | 15 February 2022 | 357 | 3109 | 30 | FALSE |
| (27692, 27697, 6) | 9 March 2021 | 20 February 2022 | 348 | 3108 | 54 | FALSE |
| (27538, 27628, 91) | 4 June 2021 | 10 February 2022 | 251 | 2887 | 44 | TRUE |
| (27588, 27606, 19) | 22 June 2021 | 17 January 2022 | 209 | 2655 | 8 | TRUE |
| (27555, 27578, 24) | 19 May 2021 | 3 February 2022 | 260 | 1916 | 51 | FALSE |
| (27720, 27721, 2) | 12 May 2021 | 22 February 2022 | 286 | 1859 | 46 | TRUE |
| (27553, 27621, 69) | 9 April 2021 | 4 February 2022 | 301 | 1636 | 33 | FALSE |
| (27554, 27621, 68) | 12 April 2021 | 4 January 2022 | 267 | 1585 | 21 | TRUE |
| (27694, 27700, 7) | 9 February 2021 | 12 January 2022 | 337 | 1538 | 38 | TRUE |
| (27548, 27623, 76) | 21 June 2021 | 10 January 2022 | 203 | 1418 | 13 | TRUE |
| (27548, 27554, 7) | 11 January 2021 | 17 January 2022 | 371 | 1290 | 31 | TRUE |
| (27578, 27623, 46) | 4 April 2021 | 31 January 2022 | 302 | 1135 | 34 | TRUE |
| (27555, 27624, 70) | 5 April 2021 | 2 February 2022 | 303 | 1115 | 28 | TRUE |
| (27564, 27586, 23) | 9 June 2021 | 12 January 2022 | 217 | 1044 | 17 | TRUE |
| (27553, 27553, 1) | 22 April 2021 | 18 January 2022 | 271 | 1007 | 35 | TRUE |
| (27555, 27633, 79) | 26 July 2021 | 31 December 2021 | 158 | 1003 | 7 | TRUE |
| (27695, 27700, 6) | 16 April 2021 | 25 January 2022 | 284 | 992 | 35 | FALSE |
| Country | Genomes with ORF7aΔ17del Deletion | Total Genomes in Period | Percentage of Genomes with Deletion | First Date Sampled | Last Date Sampled | Date Span (Days) |
|---|---|---|---|---|---|---|
| Australia | 27912 | 32,048 | 87.09 | 13 June 2021 | 13 February 2022 | 245 |
| USA | 102 | 1,402,035 | 0.01 | 14 July 2021 | 20 December 2021 | 159 |
| England | 76 | 865,173 | 0.01 | 2 June 2021 | 23 December 2021 | 204 |
| Brazil | 24 | 40,686 | 0.06 | 20 September 2021 | 17 January 2022 | 119 |
| France | 17 | 114,469 | 0.01 | 20 October 2021 | 27 December 2021 | 68 |
| India | 7 | 73,032 | 0.01 | 18 September 2021 | 3 January 2022 | 107 |
| Slovenia | 6 | 27,823 | 0.02 | 19 October 2021 | 10 December 2021 | 52 |
| Thailand | 6 | 7775 | 0.08 | 11 November 2021 | 19 December 2021 | 38 |
| Netherlands | 4 | 44,346 | 0.01 | 16 November 2021 | 4 January 2022 | 49 |
| Scotland | 4 | 103,198 | 0.00 | 28 October 2021 | 17 December 2021 | 50 |
| Colombia | 2 | 4525 | 0.04 | 22 November 2021 | 19 December 2021 | 27 |
| Japan | 2 | 95,139 | 0.00 | 30 August 2021 | 28 September 2021 | 29 |
| Mexico | 2 | 23,422 | 0.01 | 12 December 2021 | 12 December 2021 | 0 |
| Romania | 2 | 5884 | 0.03 | 30 September 2021 | 30 September 2021 | 0 |
| Russia | 2 | 7647 | 0.03 | 29 June 2021 | 2 November 2021 | 126 |
| Bahrain | 1 | 1893 | 0.05 | 11 November 2021 | 11 November 2021 | 0 |
| Canada | 1 | 97,883 | 0.00 | 20 October 2021 | 20 October 2021 | 0 |
| Chile | 1 | 8486 | 0.01 | 28 November 2021 | 28 November 2021 | 0 |
| Croatia | 1 | 14,217 | 0.01 | 4 September 2021 | 4 September 2021 | 0 |
| Israel | 1 | 18,019 | 0.01 | 6 November 2021 | 6 November 2021 | 0 |
| Latvia | 1 | 5220 | 0.02 | 13 October 2021 | 13 October 2021 | 0 |
| Lithuania | 1 | 15,449 | 0.01 | 30 October 2021 | 30 October 2021 | 0 |
| Luxembourg | 1 | 9493 | 0.01 | 29 November 2021 | 29 November 2021 | 0 |
| Malta | 1 | 19 | 5.26 | 8 January 2022 | 8 January 2022 | 0 |
| Peru | 1 | 6575 | 0.02 | 28 October 2021 | 28 October 2021 | 0 |
| Vietnam | 1 | 2423 | 0.04 | 16 November 2021 | 16 November 2021 | 0 |
| Wales | 1 | 98,861 | 0.00 | 24 August 2021 | 24 August 2021 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foster, C.S.P.; Bull, R.A.; Tedla, N.; Santiago, F.; Agapiou, D.; Adhikari, A.; Walker, G.J.; Shrestha, L.B.; Van Hal, S.J.; Kim, K.W.; et al. Persistence of a Frameshifting Deletion in SARS-CoV-2 ORF7a for the Duration of a Major Outbreak. Viruses 2023, 15, 522. https://doi.org/10.3390/v15020522
Foster CSP, Bull RA, Tedla N, Santiago F, Agapiou D, Adhikari A, Walker GJ, Shrestha LB, Van Hal SJ, Kim KW, et al. Persistence of a Frameshifting Deletion in SARS-CoV-2 ORF7a for the Duration of a Major Outbreak. Viruses. 2023; 15(2):522. https://doi.org/10.3390/v15020522
Chicago/Turabian StyleFoster, Charles S. P., Rowena A. Bull, Nicodemus Tedla, Fernando Santiago, David Agapiou, Anurag Adhikari, Gregory J. Walker, Lok Bahadur Shrestha, Sebastiaan J. Van Hal, Ki Wook Kim, and et al. 2023. "Persistence of a Frameshifting Deletion in SARS-CoV-2 ORF7a for the Duration of a Major Outbreak" Viruses 15, no. 2: 522. https://doi.org/10.3390/v15020522
APA StyleFoster, C. S. P., Bull, R. A., Tedla, N., Santiago, F., Agapiou, D., Adhikari, A., Walker, G. J., Shrestha, L. B., Van Hal, S. J., Kim, K. W., & Rawlinson, W. D. (2023). Persistence of a Frameshifting Deletion in SARS-CoV-2 ORF7a for the Duration of a Major Outbreak. Viruses, 15(2), 522. https://doi.org/10.3390/v15020522







