Immunometabolic Signature during Respiratory Viral Infection: A Potential Target for Host-Directed Therapies
Abstract
1. Introduction
2. Innate Immune Response: The Front Line in the Fight against Infection
2.1. Dendritic Cells
2.2. Macrophages
2.3. Microglia
2.4. Neutrophils
2.5. Natural Killer
2.6. Eosinophils
2.7. Complement System
3. Adaptive Immune Response: The Better Weapon against Infection
3.1. T Lymphocytes
3.2. B Lymphocytes
3.3. Memory Lymphocytes
4. Immunometabolism and Host-Directed Therapies for Respiratory Viruses
| Intervention | Metabolic Target | Proposed Biological Function | Immune Cell | Respiratory Viruses |
|---|---|---|---|---|
| DMF/4-Octyl-Itaconate | GAPDH inhibition | Anti-inflammatory effects [294] | T cells [310] B cells [311] NKs [312] DCs [313] Monocytes/Macrophages [295,296] Microglia [314] Neutrophils [315] | SARS-CoV-2 [295] Influenza [296] |
| Metformin/AICAR | AMPK agonist | Anti-inflammatory effects [316] | T cells [299] B cells [317] Macrophage [318] Monocytes [319] Neutrophils [320] | SARS-CoV-2 [299] |
| Rapamycin | PI3K/mTOR inhibition. | Immunosuppressive effects [321] | T cells [321] Macrophage [321,322] NK [323] DCs [82] | SARS-CoV-2 [324] Rhinovirus [325] Influenza [326] Respiratory syncytial virus [327] |
| LY294002 | PI3K/AKT inhibition | Immunosuppressive effects [321] | T cells [328] Macrophage [329,330] Dendritic cells [331] Neutrophils [332] NKs [333] | Influenza [334] Respiratory syncytial virus [335] |
| HIF prolyl hydroxylase inhibitors | HIF-1α stabilization | Inflammatory response [336] | Macrophage [337] Neutrophils [177] Dendritic cells [78] T cells [338] | SARS-CoV-2 [336] Influenza [339] |
| BAY87-2243/KC7F2 | HIF-1α inhibition | Anti-inflammatory effects [22] | Dendritic cells [340] Macrophage [22] Neutrophils [214] Microglia [341] | SARS-CoV-2 [342] Respiratory syncytial virus [343] |
| 15-Deoxy-Δ12,14-prostaglandin-j2 | PPARy agonist | Induction of resolution profile [308] | Macrophage [307,308,309] Neutrophils [308] Dendritic cells [344] | Influenza [345] Respiratory syncytial virus [346] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Braciale, T.J.; Sun, J.; Kim, T.S. Regulating the adaptive immune response to respiratory virus infection. Nat. Rev. Immunol. 2012, 12, 295–305. [Google Scholar] [CrossRef]
- LeDuc, J.W.; Barry, M.A. SARS, the First Pandemic of the 21st Century1. Emerg. Infect. Dis. 2004, 10, e26. [Google Scholar] [CrossRef]
- Fraser, C.; Donnelly, C.A.; Cauchemez, S.; Hanage, W.P.; Van Kerkhove, M.D.; Hollingsworth, T.D.; Griffin, J.; Baggaley, R.F.; Jenkins, H.E.; Lyons, E.J.; et al. Pandemic Potential of a Strain of Influenza A (H1N1): Early Findings. Science 2009, 324, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Ward, I.L.; Bermingham, C.; Ayoubkhani, D.; Gethings, O.J.; Pouwels, K.B.; Yates, T.; Khunti, K.; Hippisley-Cox, J.; Banerjee, A.; Walker, A.S.; et al. Risk of COVID-19 related deaths for SARS-CoV-2 omicron (B.1.1.529) compared with delta (B.1.617.2): Retrospective cohort study. BMJ 2022, 378, e070695. [Google Scholar] [CrossRef]
- Tazerji, S.S.; Shahabinejad, F.; Tokasi, M.; Rad, M.A.; Khan, M.S.; Safdar, M.; Filipiak, K.J.; Szarpak, L.; Dzieciatkowski, T.; Jurgiel, J.; et al. Global data analysis and risk factors associated with morbidity and mortality of COVID-19. Gene Rep. 2022, 26, 101505. [Google Scholar] [CrossRef] [PubMed]
- Perry, B.L.; Aronson, B.; Pescosolido, B.A. Pandemic precarity: COVID-19 is exposing and exacerbating inequalities in the American heartland. Proc. Natl. Acad. Sci. USA 2021, 118, e2020685118. [Google Scholar] [CrossRef] [PubMed]
- Esseau-Thomas, C.; Galarraga, O.; Khalifa, S. Epidemics, pandemics and income inequality. Health Econ. Rev. 2022, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Troy, N.M.; Bosco, A. Respiratory viral infections and host responses; insights from genomics. Respir. Res. 2016, 17, 156. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Morens, D.M. The Pathology of Influenza Virus Infections. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 499–522. [Google Scholar] [CrossRef]
- Branche, A.R.; Falsey, A.R. Parainfluenza Virus Infection. Semin. Respir. Crit. Care Med. 2016, 37, 538–554. [Google Scholar] [CrossRef]
- Battles, M.B.; McLellan, J.S. Respiratory syncytial virus entry and how to block it. Nat. Rev. Genet. 2019, 17, 233–245. [Google Scholar] [CrossRef]
- Bizot, E.; Bousquet, A.; Charpié, M.; Coquelin, F.; Lefevre, S.; Le Lorier, J.; Patin, M.; Sée, P.; Sarfati, E.; Walle, S.; et al. Rhinovirus: A Narrative Review on Its Genetic Characteristics, Pediatric Clinical Presentations, and Pathogenesis. Front. Pediatr. 2021, 9, 643219. [Google Scholar] [CrossRef] [PubMed]
- House, N.N.C.; Palissery, S.; Sebastian, H. Corona Viruses: A Review on SARS, MERS and COVID-19. Microbiol. Insights 2021, 14, 117863612110024. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.-D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 2004, 101, 4620–4624. [Google Scholar] [CrossRef]
- Okamoto, M.; Tsukamoto, H.; Kouwaki, T.; Seya, T.; Oshiumi, H.; Kato, H.; Oh, S.-W.; Fujita, T.; Takamura, S.; Miyauchi, K.; et al. Recognition of Viral RNA by Pattern Recognition Receptors in the Induction of Innate Immunity and Excessive Inflammation During Respiratory Viral Infections. Viral Immunol. 2017, 30, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Amarante-Mendes, G.P.; Adjemian, S.; Branco, L.M.; Zanetti, L.C.; Weinlich, R.; Bortoluci, K.R. Pattern Recognition Receptors and the Host Cell Death Molecular Machinery. Front. Immunol. 2018, 9, 2379. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Kalia, V.; Sarkar, S.; Gourley, T.S.; Rouse, B.T.; Ahmed, R. Differentiation of memory B and T cells. Curr. Opin. Immunol. 2006, 18, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Stambas, J.; Lu, C.; Tripp, R.A. Innate and adaptive immune responses in respiratory virus infection: Implications for the clinic. Expert Rev. Respir. Med. 2020, 14, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.S.; de Sá, K.S.; Ishimoto, A.Y.; Becerra, A.; Oliveira, S.; Almeida, L.; Gonçalves, A.V.; Perucello, D.B.; Andrade, W.A.; Castro, R.; et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021, 218, e20201707. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; Macary, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Codo, A.C.; Davanzo, G.G.; de Brito Monteiro, L.; de Souza, G.F.; Muraro, S.P.; Virgilio-Da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; de Biagi Junior, C.A.O.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab. 2020, 32, 437–446.e5. [Google Scholar] [CrossRef]
- Gu, Y.; Zuo, X.; Zhang, S.; Ouyang, Z.; Jiang, S.; Wang, F.; Wang, G. The Mechanism behind Influenza Virus Cytokine Storm. Viruses 2021, 13, 1362. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, G.; Paladin, F.; Tonacci, A.; Isola, S.; Allegra, A.; Gangemi, S. The Potential Role of Cytokine Storm Pathway in the Clinical Course of Viral Respiratory Pandemic. Biomedicines 2021, 9, 1688. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Lee, S.-S. A Detailed Overview of Immune Escape, Antibody Escape, Partial Vaccine Escape of SARS-CoV-2 and Their Emerging Variants With Escape Mutations. Front. Immunol. 2022, 13, 801522. [Google Scholar] [CrossRef]
- Zumla, A.; Rao, M.; Wallis, R.S.; Kaufmann, S.H.E.; Rustomjee, R.; Mwaba, P.; Vilaplana, C.; Yeboah-Manu, D.; Chakaya, J.; Ippolito, G.; et al. Host-directed therapies for infectious diseases: Current status, recent progress, and future prospects. Lancet Infect. Dis. 2016, 16, e47–e63. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.E.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 2018, 17, 35–56. [Google Scholar] [CrossRef]
- Palmer, C.S. Innate metabolic responses against viral infections. Nat. Metab. 2022, 4, 1245–1259. [Google Scholar] [CrossRef]
- Liu, Z.; Le, Y.; Chen, H.; Zhu, J.; Lu, D. Role of PKM2-Mediated Immunometabolic Reprogramming on Development of Cytokine Storm. Front. Immunol. 2021, 12, 748573. [Google Scholar] [CrossRef]
- Batabyal, R.; Freishtat, N.; Hill, E.; Rehman, M.; Freishtat, R.; Koutroulis, I. Metabolic dysfunction and immunometabolism in COVID-19 pathophysiology and therapeutics. Int. J. Obes. 2021, 45, 1163–1169. [Google Scholar] [CrossRef]
- Xiao, N.; Nie, M.; Pang, H.; Wang, B.; Hu, J.; Meng, X.; Li, K.; Ran, X.; Long, Q.; Deng, H.; et al. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021, 12, 1618. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, F.J.; Divangahi, M. Targeting immunometabolism in host defence against Mycobacterium tuberculosis. Immunology 2021, 162, 145–159. [Google Scholar] [CrossRef]
- Kleinehr, J.; Wilden, J.J.; Boergeling, Y.; Ludwig, S.; Hrincius, E.R. Metabolic Modifications by Common Respiratory Viruses and Their Potential as New Antiviral Targets. Viruses 2021, 13, 2068. [Google Scholar] [CrossRef]
- Gonçalves, S.M.; Ferreira, A.V.; Cunha, C.; Carvalho, A. Targeting immunometabolism in host-directed therapies to fungal disease. Clin. Exp. Immunol. 2022, 208, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Blank, F.; Wehrli, M.; Lehmann, A.; Baum, O.; Gehr, P.; von Garnier, C.; Rothen-Rutishauser, B.M. Macrophages and dendritic cells express tight junction proteins and exchange particles in an in vitro model of the human airway wall. Immunobiology 2011, 216, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Jahnsen, F.L.; Strickland, D.H.; Thomas, J.A.; Tobagus, I.T.; Napoli, S.; Zosky, G.R.; Turner, D.J.; Sly, P.D.; Stumbles, P.A.; Holt, P.G. Accelerated Antigen Sampling and Transport by Airway Mucosal Dendritic Cells following Inhalation of a Bacterial Stimulus. J. Immunol. 2006, 177, 5861–5867. [Google Scholar] [CrossRef]
- Vermaelen, K.; Pauwels, R. Accurate and simple discrimination of mouse pulmonary dendritic cell and macrophage populations by flow cytometry: Methodology and new insights. Cytometry 2004, 61, 170–177. [Google Scholar] [CrossRef]
- Jakubzick, C.; Tacke, F.; Llodra, J.; van Rooijen, N.; Randolph, G.J. Modulation of Dendritic Cell Trafficking to and from the Airways. J. Immunol. 2006, 176, 3578–3584. [Google Scholar] [CrossRef] [PubMed]
- Jakubzick, C.; Tacke, F.; Ginhoux, F.; Wagers, A.J.; van Rooijen, N.; Mack, M.; Merad, M.; Randolph, G.J. Blood Monocyte Subsets Differentially Give Rise to CD103+ and CD103− Pulmonary Dendritic Cell Populations. J. Immunol. 2008, 180, 3019–3027. [Google Scholar] [CrossRef]
- Worbs, T.; Hammerschmidt, S.I.; Förster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2017, 17, 30–48. [Google Scholar] [CrossRef]
- Liu, K.; Victora, G.D.; Schwickert, T.A.; Guermonprez, P.; Meredith, M.M.; Yao, K.; Chu, F.-F.; Randolph, G.J.; Rudensky, A.Y.; Nussenzweig, M. In Vivo Analysis of Dendritic Cell Development and Homeostasis. Science 2009, 324, 392–397. [Google Scholar] [CrossRef]
- Cabeza-Cabrerizo, M.; van Blijswijk, J.; Wienert, S.; Heim, D.; Jenkins, R.P.; Chakravarty, P.; Rogers, N.; Frederico, B.; Acton, S.; Beerling, E.; et al. Tissue clonality of dendritic cell subsets and emergency DCpoiesis revealed by multicolor fate mapping of DC progenitors. Sci. Immunol. 2019, 4, eaaw1941. [Google Scholar] [CrossRef]
- Nakano, H.; Lyons-Cohen, M.R.; Whitehead, G.S.; Nakano, K.; Cook, D.N. Distinct functions of CXCR4, CCR2, and CX3CR1 direct dendritic cell precursors from the bone marrow to the lung. J. Leukoc. Biol. 2017, 101, 1143–1153. [Google Scholar] [CrossRef]
- Saeki, H.; Moore, A.M.; Brown, M.J.; Hwang, S.T. Cutting edge: Secondary lymphoid-tissue chemokine (SLC) and CC chemokine receptor 7 (CCR7) participate in the emigration pathway of mature dendritic cells from the skin to regional lymph nodes. J. Immunol. 1999, 162, 2472–2475. [Google Scholar] [CrossRef] [PubMed]
- De Jong, E.C.; Smits, H.H.; Kapsenberg, M.L. Dendritic cell-mediated T cell polarization. Springer Semin. Immunopathol. 2004, 26, 289–307. [Google Scholar] [CrossRef] [PubMed]
- Sawai, C.; Sisirak, V.; Ghosh, H.S.; Hou, E.Z.; Ceribelli, M.; Staudt, L.M.; Reizis, B. Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J. Exp. Med. 2013, 210, 2151–2159. [Google Scholar] [CrossRef] [PubMed]
- Swiecki, M.; Miller, H.; Sesti-Costa, R.; Cella, M.; Gilfillan, S.; Colonna, M. Microbiota induces tonic CCL2 systemic levels that control pDC trafficking in steady state. Mucosal Immunol. 2017, 10, 936–945. [Google Scholar] [CrossRef]
- Reizis, B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 2019, 50, 37–50. [Google Scholar] [CrossRef]
- Toor, S.M.; Saleh, R.; Nair, V.S.; Taha, R.Z.; Elkord, E. T-cell responses and therapies against SARS-CoV-2 infection. Immunology 2020, 162, 30–43. [Google Scholar] [CrossRef]
- Ballesteros-Tato, A.; León, B.; Lund, F.E.; Randall, T.D. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8+ T cell responses to influenza. Nat. Immunol. 2010, 11, 216–224. [Google Scholar] [CrossRef]
- Channappanavar, R.; Zhao, J.; Perlman, S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014, 59, 118–128. [Google Scholar] [CrossRef]
- Geurtsvan Kessel, C.H.; Willart, M.; Van Rijt, L.; Muskens, F.; Kool, M.; Baas, C.; Thielemans, K.; Bennett, C.; Clausen, B.; Hoogsteden, H.; et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b− but not plasmacytoid dendritic cells. J. Exp. Med. 2008, 205, 1621–1634. [Google Scholar] [CrossRef]
- Del Rio, M.-L.; Rodriguez-Barbosa, J.-I.; Kremmer, E.; Förster, R. CD103− and CD103+ Bronchial Lymph Node Dendritic Cells Are Specialized in Presenting and Cross-Presenting Innocuous Antigen to CD4+ and CD8+ T Cells. J. Immunol. 2007, 178, 6861–6866. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.W.S.; Prabhu, N.; Betts, R.J.; Ge, M.Q.; Dai, X.; Hutchinson, P.E.; Lew, F.C.; Wong, K.L.; Hanson, B.J.; Macary, P.A.; et al. Lung CD103+ Dendritic Cells Efficiently Transport Influenza Virus to the Lymph Node and Load Viral Antigen onto MHC Class I for Presentation to CD8 T Cells. J. Immunol. 2011, 187, 6011–6021. [Google Scholar] [CrossRef]
- Lukens, M.V.; Kruijsen, D.; Coenjaerts, F.E.J.; Kimpen, J.L.L.; van Bleek, G.M. Respiratory Syncytial Virus-Induced Activation and Migration of Respiratory Dendritic Cells and Subsequent Antigen Presentation in the Lung-Draining Lymph Node. J. Virol. 2009, 83, 7235–7243. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; To, K.K.-W.; Wong, Y.-C.; Liu, L.; Zhou, B.; Li, X.; Huang, H.; Mo, Y.; Luk, T.-Y.; Lau, T.T.-K.; et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity 2020, 53, 864–877.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, J.; Van Rooijen, N.; Perlman, S. Evasion by Stealth: Inefficient Immune Activation Underlies Poor T Cell Response and Severe Disease in SARS-CoV-Infected Mice. PLoS Pathog. 2009, 5, e1000636. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O’Driscoll, L. The relevance of using 3D cell cultures, in addition to 2D monolayer cultures, when evaluating breast cancer drug sensitivity and resistance. Oncotarget 2016, 7, 45745–45756. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, J.; Legge, K.; Perlman, S. Age-related increases in PGD2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J. Clin. Investig. 2011, 121, 4921–4930. [Google Scholar] [CrossRef]
- Wong, L.-Y.R.; Zheng, J.; Wilhelmsen, K.; Li, K.; Ortiz, M.E.; Schnicker, N.J.; Thurman, A.; Pezzulo, A.A.; Szachowicz, P.J.; Li, P.; et al. Eicosanoid signalling blockade protects middle-aged mice from severe COVID-19. Nature 2022, 605, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peters, N.; Schwarze, J. Plasmacytoid Dendritic Cells Limit Viral Replication, Pulmonary Inflammation, and Airway Hyperresponsiveness in Respiratory Syncytial Virus Infection. J. Immunol. 2006, 177, 6263–6270. [Google Scholar] [CrossRef]
- Smit, J.J.; Rudd, B.D.; Lukacs, N.W. Plasmacytoid dendritic cells inhibit pulmonary immunopathology and promote clearance of respiratory syncytial virus. J. Exp. Med. 2006, 203, 1153–1159. [Google Scholar] [CrossRef]
- Langlois, R.A.; Legge, K.L. Plasmacytoid Dendritic Cells Enhance Mortality during Lethal Influenza Infections by Eliminating Virus-Specific CD8 T Cells. J. Immunol. 2010, 184, 4440–4446. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.; Crotta, S.; McCabe, T.M.; Wack, A. Pathogenic potential of interferon αβ in acute influenza infection. Nat. Commun. 2014, 5, 3864. [Google Scholar] [CrossRef] [PubMed]
- Severa, M.; Diotti, R.A.; Etna, M.P.; Rizzo, F.; Fiore, S.; Ricci, D.; Iannetta, M.; Sinigaglia, A.; Lodi, A.; Mancini, N.; et al. Differential plasmacytoid dendritic cell phenotype and type I Interferon response in asymptomatic and severe COVID-19 infection. PLoS Pathog. 2021, 17, e1009878. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Lokugamage, K.G.; Hage, A.; de Vries, M.; Valero-Jimenez, A.M.; Schindewolf, C.; Dittmann, M.; Rajsbaum, R.; Menachery, V.D. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020, 94, e01410-20. [Google Scholar] [CrossRef]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Swiecki, M.; Colonna, M. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol. Rev. 2010, 234, 142–162. [Google Scholar] [CrossRef]
- Cervantes-Barragan, L.; Züst, R.; Weber, F.; Spiegel, M.; Lang, K.S.; Akira, S.; Thiel, V.; Ludewig, B. Control of coronavirus infection through plasmacytoid dendritic-cell–derived type I interferon. Blood 2006, 109, 1131–1137. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef]
- Everts, B.; Amiel, E.; Huang, S.C.-C.; Smith, A.M.; Chang, C.-H.; Lam, W.Y.; Redmann, V.; Freitas, T.C.; Blagih, J.; van der Windt, G.J.W.; et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKε supports the anabolic demands of dendritic cell activation. Nat. Immunol. 2014, 15, 323–332. [Google Scholar] [CrossRef]
- Krawczyk, C.M.; Holowka, T.; Sun, J.; Blagih, J.; Amiel, E.; De Berardinis, R.J.; Cross, J.R.; Jung, E.; Thompson, C.B.; Jones, R.G.; et al. Toll-like receptor–induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 2010, 115, 4742–4749. [Google Scholar] [CrossRef]
- Thwe, P.M.; Fritz, D.I.; Snyder, J.P.; Smith, P.R.; Curtis, K.D.; O’Donnell, A.; Galasso, N.A.; Sepaniac, L.A.; Adamik, B.J.; Hoyt, L.R.; et al. Syk-dependent glycolytic reprogramming in dendritic cells regulates IL-1β production to β-glucan ligands in a TLR-independent manner. J. Leukoc. Biol. 2019, 106, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Guak, H.; Al Habyan, S.; Ma, E.H.; Aldossary, H.; Al-Masri, M.; Won, S.Y.; Ying, T.; Fixman, E.D.; Jones, R.G.; McCaffrey, L.M.; et al. Glycolytic metabolism is essential for CCR7 oligomerization and dendritic cell migration. Nat. Commun. 2018, 9, 2463. [Google Scholar] [CrossRef]
- Jantsch, J.; Chakravortty, D.; Turza, N.; Prechtel, A.T.; Buchholz, B.; Gerlach, R.G.; Volke, M.; Gläsner, J.; Warnecke, C.; Wiesener, M.S.; et al. Hypoxia and Hypoxia-Inducible Factor-1α Modulate Lipopolysaccharide-Induced Dendritic Cell Activation and Function. J. Immunol. 2008, 180, 4697–4705. [Google Scholar] [CrossRef] [PubMed]
- Everts, B.; Amiel, E.; Van Der Windt, G.J.W.; Freitas, T.C.; Chott, R.; Yarasheski, K.E.; Pearce, E.L.; Pearce, E.J. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood 2012, 120, 1422–1431. [Google Scholar] [CrossRef]
- Fekete, T.; Sütö, M.I.; Bencze, D.; Mázló, A.; Szabo, A.; Biro, T.; Bacsi, A.; Pazmandi, K. Human Plasmacytoid and Monocyte-Derived Dendritic Cells Display Distinct Metabolic Profile Upon RIG-I Activation. Front. Immunol. 2018, 9, 3070. [Google Scholar] [CrossRef]
- Amiel, E.; Everts, B.; Freitas, T.C.; King, I.L.; Curtis, J.D.; Pearce, E.L.; Pearce, E.J. Inhibition of Mechanistic Target of Rapamycin Promotes Dendritic Cell Activation and Enhances Therapeutic Autologous Vaccination in Mice. J. Immunol. 2012, 189, 2151–2158. [Google Scholar] [CrossRef] [PubMed]
- Boor, P.P.C.; Metselaar, H.J.; Mancham, S.; van der Laan, L.J.W.; Kwekkeboom, J. Rapamycin has suppressive and stimulatory effects on human plasmacytoid dendritic cell functions. Clin. Exp. Immunol. 2013, 174, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Hussaarts, L.; Smits, H.H.; Schramm, G.; Ham, A.J.; Zon, G.C.; Haas, H.; Guigas, B.; Yazdanbakhsh, M. Rapamycin and omega-1: mTOR-dependent and -independent Th2 skewing by human dendritic cells. Immunol. Cell Biol. 2013, 91, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Bajwa, G.; DeBerardinis, R.J.; Shao, B.; Hall, B.; Farrar, J.D.; Gill, M.A. Cutting Edge: Critical Role of Glycolysis in Human Plasmacytoid Dendritic Cell Antiviral Responses. J. Immunol. 2016, 196, 2004–2009. [Google Scholar] [CrossRef]
- Wu, D.; Sanin, D.E.; Everts, B.; Chen, Q.; Qiu, J.; Buck, M.D.; Patterson, A.; Smith, A.M.; Chang, C.H.; Liu, Z.; et al. Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function. Immunity 2016, 44, 1325–1336. [Google Scholar] [CrossRef]
- Haidinger, M.; Poglitsch, M.; Geyeregger, R.; Kasturi, S.; Zeyda, M.; Zlabinger, G.J.; Pulendran, B.; Hörl, W.H.; Säemann, M.D.; Weichhart, T. A Versatile Role of Mammalian Target of Rapamycin in Human Dendritic Cell Function and Differentiation. J. Immunol. 2010, 185, 3919–3931. [Google Scholar] [CrossRef]
- Cao, W.; Manicassamy, S.; Tang, H.; Kasturi, S.P.; Pirani, A.; Murthy, N.; Pulendran, B. Toll-like receptor–mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 2008, 9, 1157–1164. [Google Scholar] [CrossRef]
- Wolf, Y.; Boura-Halfon, S.; Cortese, N.; Haimon, Z.; Shalom, H.S.; Kuperman, Y.; Kalchenko, V.; Brandis, A.; David, E.; Segal-Hayoun, Y.; et al. Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat. Immunol. 2017, 18, 665–674. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.-A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Ginhoux, F.; Jung, S. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014, 14, 392–404. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Marchesi, F.; Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 2022, 21, 799–820. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef] [PubMed]
- Olefsky, J.M.; Glass, C.K. Macrophages, Inflammation, and Insulin Resistance. Annu. Rev. Physiol. 2009, 72, 219–246. [Google Scholar] [CrossRef]
- Wang, Y.; Stancliffe, E.; Fowle-Grider, R.; Wang, R.; Wang, C.; Schwaiger-Haber, M.; Shriver, L.P.; Patti, G.J. Saturation of the mitochondrial NADH shuttles drives aerobic glycolysis in proliferating cells. Mol. Cell 2022, 82, 3270–3283.e9. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Däbritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470.e13. [Google Scholar] [CrossRef] [PubMed]
- Vats, D.; Mukundan, L.; Odegaard, J.I.; Zhang, L.; Smith, K.L.; Morel, C.R.; Wagner, R.A.; Greaves, D.R.; Murray, P.J.; Chawla, A. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 2006, 4, 13–24. [Google Scholar] [CrossRef]
- Kopf, M.; Schneider, C.; Nobs, S.P. The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 2015, 16, 36–44. [Google Scholar] [CrossRef]
- Westphalen, K.; Gusarova, G.A.; Islam, M.N.; Subramanian, M.; Cohen, T.S.; Prince, A.S.; Bhattacharya, J. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature 2014, 506, 503–506. [Google Scholar] [CrossRef]
- Hussell, T.; Bell, T.J. Alveolar macrophages: Plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014, 14, 81–93. [Google Scholar] [CrossRef]
- Shaykhiev, R.; Krause, A.; Salit, J.; Strulovici-Barel, Y.; Harvey, B.-G.; O’Connor, T.P.; Crystal, R.G. Smoking-Dependent Reprogramming of Alveolar Macrophage Polarization: Implication for Pathogenesis of Chronic Obstructive Pulmonary Disease. J. Immunol. 2009, 183, 2867–2883. [Google Scholar] [CrossRef]
- Soroosh, P.; Doherty, T.; Duan, W.; Mehta, A.K.; Choi, H.; Adams, Y.F.; Mikulski, Z.; Khorram, N.; Rosenthal, P.; Broide, D.H.; et al. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med. 2013, 210, 775–788. [Google Scholar] [CrossRef]
- Fernandez, S.; Jose, P.; Avdiushko, M.G.; Kaplan, A.M.; Cohen, D.A. Inhibition of IL-10 Receptor Function in Alveolar Macrophages by Toll-Like Receptor Agonists. J. Immunol. 2004, 172, 2613–2620. [Google Scholar] [CrossRef]
- Gao, X.; Dong, Y.; Liu, Z.; Niu, B. Silencing of triggering receptor expressed on myeloid cells-2 enhances the inflammatory responses of alveolar macrophages to lipopolysaccharide. Mol. Med. Rep. 2013, 7, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M. TREMs in the immune system and beyond. Nat. Rev. Immunol. 2003, 3, 445–453. [Google Scholar] [CrossRef]
- Steele, C.; Marrero, L.; Swain, S.D.; Harmsen, A.G.; Zheng, M.; Brown, G.; Gordon, S.; Shellito, J.E.; Kolls, J.K. Alveolar Macrophage–mediated Killing of Pneumocystis carinii f. sp. muris Involves Molecular Recognition by the Dectin-1 β-Glucan Receptor. J. Exp. Med. 2003, 198, 1677–1688. [Google Scholar] [CrossRef]
- Snelgrove, R.J.; Goulding, J.; Didierlaurent, A.M.; Lyonga, D.; Vekaria, S.; Edwards, L.; Gwyer, E.; Sedgwick, J.D.; Barclay, A.N.; Hussell, T. A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat. Immunol. 2008, 9, 1074–1083. [Google Scholar] [CrossRef]
- Pechkovsky, D.V.; Prasse, A.; Kollert, F.; Engel, K.M.; Dentler, J.; Luttmann, W.; Friedrich, K.; Müller-Quernheim, J.; Zissel, G. Alternatively activated alveolar macrophages in pulmonary fibrosis—Mediator production and intracellular signal transduction. Clin. Immunol. 2010, 137, 89–101. [Google Scholar] [CrossRef]
- Draijer, C.; Robbe, P.; Boorsma, C.E.; Hylkema, M.N.; Melgert, B.N. Characterization of Macrophage Phenotypes in Three Murine Models of House-Dust-Mite-Induced Asthma. Mediat. Inflamm. 2013, 2013, 632049. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.; Martinez-Pomares, L.; Stacey, M.; Lin, H.-H.; Brown, G.; Gordon, S. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 2005, 23, 901–944. [Google Scholar] [CrossRef] [PubMed]
- Gardai, S.; Xiao, Y.-Q.; Dickinson, M.; Nick, J.; Voelker, D.; Greene, K.; Henson, P. By Binding SIRP or Calreticulin/CD91, Lung Collectins Act as Dual Function Surveillance Molecules to Suppress or Enhance Inflammation. Cell 2003, 115, 13–23. [Google Scholar] [CrossRef]
- Janssen, W.J.; McPhillips, K.A.; Dickinson, M.G.; Linderman, D.J.; Morimoto, K.; Xiao, Y.Q.; Oldham, K.M.; Vandivier, R.W.; Henson, P.M.; Gardai, S.J. Surfactant Proteins A and D Suppress Alveolar Macrophage Phagocytosis via Interaction with SIRPα. Am. J. Respir. Crit. Care Med. 2008, 178, 158–167. [Google Scholar] [CrossRef]
- Schyns, J.; Bureau, F.; Marichal, T. Lung Interstitial Macrophages: Past, Present, and Future. J. Immunol. Res. 2018, 2018, 5160794. [Google Scholar] [CrossRef]
- Sabatel, C.; Radermecker, C.; Fievez, L.; Paulissen, G.; Chakarov, S.; Fernandes, C.; Olivier, S.; Toussaint, M.; Pirottin, D.; Xiao, X.; et al. Exposure to Bacterial CpG DNA Protects from Airway Allergic Inflammation by Expanding Regulatory Lung Interstitial Macrophages. Immunity 2017, 46, 457–473. [Google Scholar] [CrossRef]
- Gibbings, S.L.; Thomas, S.M.; Atif, S.M.; McCubbrey, A.L.; Desch, A.N.; Danhorn, T.; Leach, S.M.; Bratton, D.L.; Henson, P.M.; Janssen, W.J.; et al. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. Am. J. Respir. Cell Mol. Biol. 2017, 57, 66–76. [Google Scholar] [CrossRef]
- Maus, U.A.; Janzen, S.; Wall, G.; Srivastava, M.; Blackwell, T.S.; Christman, J.W.; Seeger, W.; Welte, T.; Lohmeyer, J. Resident Alveolar Macrophages Are Replaced by Recruited Monocytes in Response to Endotoxin-Induced Lung Inflammation. Am. J. Respir. Cell Mol. Biol. 2006, 35, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.S.; Krasnow, M.A. Developmental origin of lung macrophage diversity. Development 2016, 143, 1318–1327. [Google Scholar] [CrossRef]
- Crowell, R.E.; Heaphy, E.; Valdez, Y.E.; Mold, C.; Lehnert, B.E. Alveolar nd Interstitial Macrophage Populations in the Murine Lung. Exp. Lung Res. 1992, 18, 435–446. [Google Scholar] [CrossRef]
- Hou, F.; Xiao, K.; Tang, L.; Xie, L. Diversity of Macrophages in Lung Homeostasis and Diseases. Front. Immunol. 2021, 12, 753940. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, B.; Mims, C.A. Interaction of influenza virus with mouse macrophages. Infect. Immun. 1981, 31, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Pribul, P.K.; Harker, J.; Wang, B.; Wang, H.; Tregoning, J.S.; Schwarze, J.; Openshaw, P.J.M. Alveolar Macrophages Are a Major Determinant of Early Responses to Viral Lung Infection but Do Not Influence Subsequent Disease Development. J. Virol. 2008, 82, 4441–4448. [Google Scholar] [CrossRef] [PubMed]
- Miller, L. Macrophage Polarization in Virus-Host Interactions. J. Clin. Cell. Immunol. 2015, 6, 311. [Google Scholar] [CrossRef]
- Yip, M.S.; Leung, N.H.L.; Cheung, C.Y.; Li, P.H.; Lee, H.H.Y.; Daëron, M.; Peiris, J.S.M.; Bruzzone, R.; Jaume, M. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014, 11, 82. [Google Scholar] [CrossRef] [PubMed]
- Tate, M.D.; Pickett, D.L.; van Rooijen, N.; Brooks, A.; Reading, P.C. Critical Role of Airway Macrophages in Modulating Disease Severity during Influenza Virus Infection of Mice. J. Virol. 2010, 84, 7569–7580. [Google Scholar] [CrossRef]
- Costers, S.; Lefebvre, D.J.; Delputte, P.L.; Nauwynck, H.J. Porcine reproductive and respiratory syndrome virus modulates apoptosis during replication in alveolar macrophages. Arch. Virol. 2008, 153, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Kreijtz, J.; Fouchier, R.; Rimmelzwaan, G. Immune responses to influenza virus infection. Virus Res. 2011, 162, 19–30. [Google Scholar] [CrossRef] [PubMed]
- McGill, J.; Van Rooijen, N.; Legge, K.L. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J. Exp. Med. 2008, 205, 1635–1646. [Google Scholar] [CrossRef]
- Wijburg, O.L.; DiNatale, S.; Vadolas, J.; van Rooijen, N.; Strugnell, R.A. Alveolar macrophages regulate the induction of primary cytotoxic T-lymphocyte responses during influenza virus infection. J. Virol. 1997, 71, 9450–9457. [Google Scholar] [CrossRef]
- Huang, F.-F.; Barnes, P.F.; Feng, Y.; Donis, R.; Chroneos, Z.C.; Idell, S.; Allen, T.; Perez, D.R.; Whitsett, J.A.; Dunussi-Joannopoulos, K.; et al. GM-CSF in the Lung Protects against Lethal Influenza Infection. Am. J. Respir. Crit. Care Med. 2011, 184, 259–268. [Google Scholar] [CrossRef]
- Purnama, C.; Ng, S.L.; Tetlak, P.; Setiagani, Y.A.; Kandasamy, M.; Baalasubramanian, S.; Karjalainen, K.; Ruedl, C. Transient ablation of alveolar macrophages leads to massive pathology of influenza infection without affecting cellular adaptive immunity. Eur. J. Immunol. 2014, 44, 2003–2012. [Google Scholar] [CrossRef]
- Knoll, R.; Schultze, J.L.; Schulte-Schrepping, J. Monocytes and Macrophages in COVID-19. Front. Immunol. 2021, 12, 720109. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Common Terminology Criteria for Adverse Events (CTCAE) Common Terminology Criteria for Adverse Events (CTCAE) v5.0. 2017. Available online: https://www.meddra.org/ (accessed on 5 November 2022).
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Convertini, P.; Cucci, L.; Panaro, M.A.; Di Noia, M.A.; Calvello, R.; Palmieri, F.; Iacobazzi, V. The mitochondrial citrate carrier: A new player in inflammation. Biochem. J. 2011, 438, 433–436. [Google Scholar] [CrossRef]
- Lampropoulou, V.; Sergushichev, A.; Bambouskova, M.; Nair, S.; Vincent, E.E.; Loginicheva, E.; Cervantes-Barragan, L.; Ma, X.; Huang, S.C.-C.; Griss, T.; et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016, 24, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Pathmaperuma, A.; Maña, P.; Cheung, S.; Kugathas, K.; Josiah, A.; Koina, M.; Broomfield, A.; Delghingaro-Augusto, V.; Ellwood, D.; Dahlstrom, J. Fatty acids alter glycerolipid metabolism and induce lipid droplet formation, syncytialisation and cytokine production in human trophoblasts with minimal glucose effect or interaction. Placenta 2010, 31, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Galván-Peña, S.; Carroll, R.G.; Newman, C.; Hinchy, E.C.; Palsson-McDermott, E.; Robinson, E.K.; Covarrubias, S.; Nadin, A.; James, A.M.; Haneklaus, M.; et al. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat. Commun. 2019, 10, 338. [Google Scholar] [CrossRef]
- Chan, D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. 2019, 15, 235–259. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, X.; Nie, X.; Sun, L.; Tang, T.-S.; Chen, D.; Sun, Q. COX5B Regulates MAVS-mediated Antiviral Signaling through Interaction with ATG5 and Repressing ROS Production. PLoS Pathog. 2012, 8, e1003086. [Google Scholar] [CrossRef]
- Wu, B.; Hur, S. How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 2015, 12, 91–98. [Google Scholar] [CrossRef]
- Yoshizumi, T.; Ichinohe, T.; Sasaki, O.; Otera, H.; Kawabata, S.-I.; Mihara, K.; Koshiba, T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat. Commun. 2014, 5, 4713. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, K.; Oshiumi, H.; Takeda, M.; Ishihara, N.; Yanagi, Y.; Seya, T.; Kawabata, S.-I.; Koshiba, T. Immunology Mitofusin 2 Inhibits Mitochondrial Antiviral Signaling. Sci. Signal. 2009, 2, ra47. [Google Scholar] [CrossRef]
- Castanier, C.; Garcin, D.; Vazquez, A.; Arnoult, D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2009, 11, 133–138. [Google Scholar] [CrossRef]
- Prinz, M.; Jung, S.; Priller, J. Microglia Biology: One Century of Evolving Concepts. Cell 2019, 179, 292–311. [Google Scholar] [CrossRef]
- Li, Q.; Barres, B.A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef]
- Paolicelli, R.C.; Bolasco, G.; Pagani, F.; Maggi, L.; Scianni, M.; Panzanelli, P.; Giustetto, M.; Ferreira, T.A.; Guiducci, E.; Dumas, L.; et al. Synaptic Pruning by Microglia Is Necessary for Normal Brain Development. Science 2011, 333, 1456–1458. [Google Scholar] [CrossRef]
- Borst, K.; Schwabenland, M.; Prinz, M. Microglia metabolism in health and disease. Neurochem. Int. 2019, 130, 104331. [Google Scholar] [CrossRef] [PubMed]
- Barbosa-Silva, M.C.; Santos, L.E.; Rangel, B. The Impact of Non-Neurotropic Influenza Strains on the Brain: A Role for Microglial Priming? J. Neurosci. 2018, 38, 7758–7760. [Google Scholar] [CrossRef] [PubMed]
- Polonio, C.M.; da Silva, P.; Russo, F.B.; Hyppolito, B.R.N.; Zanluqui, N.G.; Benazzato, C.; Beltrão-Braga, P.C.B.; Muxel, S.M.; Peron, J.P.S. microRNAs Control Antiviral Immune Response, Cell Death and Chemotaxis Pathways in Human Neuronal Precursor Cells (NPCs) during Zika Virus Infection. Int. J. Mol. Sci. 2022, 23, 10282. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, R.; Thapa, S. (Eds.) Infectious Diseases and Nanomedicine I; Springer: New Delhi, India, 2014; Volume 807. [Google Scholar] [CrossRef]
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Silva, L.S.; Nogueira, M.H.; Antunes, A.S.L.M.; Vendramini, P.H.; Valença, A.G.F.; Brandão-Teles, C.; Zuccoli, G.D.S.; et al. Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc. Natl. Acad. Sci. USA 2022, 119, e2200960119. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, L.G.; Angelo, Y.D.S.; Yamamoto, P.; Carregari, V.C.; Crunfli, F.; Reis-De-Oliveira, G.; Costa, L.; Vendramini, P.H.; Duque, A.; dos Santos, N.B.; et al. SARS-CoV -2 infection impacts carbon metabolism and depends on glutamine for replication in Syrian hamster astrocytes. J. Neurochem. 2022, 163, 113–132. [Google Scholar] [CrossRef]
- Arbour, N.; Day, R.; Newcombe, J.; Talbot, P.J. Neuroinvasion by Human Respiratory Coronaviruses. J. Virol. 2000, 74, 8913–8921. [Google Scholar] [CrossRef]
- Mascia, L. Acute Lung Injury in Patients with Severe Brain Injury: A Double Hit Model. Neurocrit. Care 2009, 11, 417–426. [Google Scholar] [CrossRef]
- McGavern, D.B.; Kang, S.S. Illuminating viral infections in the nervous system. Nat. Rev. Immunol. 2011, 11, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Swanson, P.; McGavern, D. Portals of Viral Entry into the Central Nervous System. In The Blood-Brain Barrier in Health and Disease; CRC Press: Boca Raton, FL, USA, 2015; Volume 2, pp. 23–47. [Google Scholar] [CrossRef]
- Furr, S.R.; Marriott, I. Viral CNS infections: Role of glial pattern recognition receptors in neuroinflammation. Front. Microbiol. 2012, 3, 201. [Google Scholar] [CrossRef]
- Ferraro, E.; Germanò, M.; Mollace, R.; Mollace, V.; Malara, N. HIF-1, the Warburg Effect, and Macrophage/Microglia Polarization Potential Role in COVID-19 Pathogenesis. Oxidative Med. Cell. Longev. 2021, 2021, 8841911. [Google Scholar] [CrossRef]
- Düsedau, H.P.; Steffen, J.; Figueiredo, C.A.; Boehme, J.D.; Schultz, K.; Erck, C.; Korte, M.; Faber-Zuschratter, H.; Smalla, K.-H.; Dieterich, D.; et al. Influenza A Virus (H1N1) Infection Induces Microglial Activation and Temporal Dysbalance in Glutamatergic Synaptic Transmission. mBio 2021, 12, e0177621. [Google Scholar] [CrossRef] [PubMed]
- Jurgens, H.A.; Amancherla, K.; Johnson, R.W. Influenza Infection Induces Neuroinflammation, Alters Hippocampal Neuron Morphology, and Impairs Cognition in Adult Mice. J. Neurosci. 2012, 32, 3958–3968. [Google Scholar] [CrossRef]
- Melo, H.M.; Silva, G.D.S.S.D.; Sant’Ana, M.R.; Teixeira, C.V.L.; Clarke, J.R.; Coreixas, V.S.M.; de Melo, B.C.; Fortuna, J.T.; Forny-Germano, L.; Ledo, J.H.; et al. Palmitate Is Increased in the Cerebrospinal Fluid of Humans with Obesity and Induces Memory Impairment in Mice via Pro-inflammatory TNF-α. Cell Rep. 2020, 30, 2180–2194.e8. [Google Scholar] [CrossRef]
- Lahon, A.; Arya, R.P.; Banerjea, A.C. Dengue Virus Dysregulates Master Transcription Factors and PI3K/AKT/mTOR Signaling Pathway in Megakaryocytes. Front. Cell. Infect. Microbiol. 2021, 11, 715208. [Google Scholar] [CrossRef]
- Devanney, N.A.; Stewart, A.N.; Gensel, J.C. Microglia and macrophage metabolism in CNS injury and disease: The role of immunometabolism in neurodegeneration and neurotrauma. Exp. Neurol. 2020, 329, 113310. [Google Scholar] [CrossRef] [PubMed]
- Ley, K.; Hoffman, H.M.; Kubes, P.; Cassatella, M.A.; Zychlinsky, A.; Hedrick, C.C.; Catz, S.D. Neutrophils: New insights and open questions. Sci. Immunol. 2018, 3, eaat4579. [Google Scholar] [CrossRef] [PubMed]
- Cossío, I.; Lucas, D.; Hidalgo, A. Neutrophils as regulators of the hematopoietic niche. Blood 2019, 133, 2140–2148. [Google Scholar] [CrossRef]
- Mócsai, A.; Walzog, B.; Lowell, C.A. Intracellular signalling during neutrophil recruitment. Cardiovasc. Res. 2015, 107, 373–385. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, K.; Rankin, S.M. CXCR4, the master regulator of neutrophil trafficking in homeostasis and disease. Eur. J. Clin. Investig. 2018, 48, e12949. [Google Scholar] [CrossRef] [PubMed]
- Eash, K.J.; Greenbaum, A.; Gopalan, P.K.; Link, D.C. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Investig. 2010, 120, 2423–2431. [Google Scholar] [CrossRef] [PubMed]
- Semerad, C.L.; Liu, F.; Gregory, A.D.; Stumpf, K.; Link, D.C. G-CSF Is an Essential Regulator of Neutrophil Trafficking from the Bone Marrow to the Blood. Immunity 2002, 17, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Burdon, P.C.; Bridger, G.; Gutierrez-Ramos, J.-C.; Williams, T.J.; Rankin, S.M. Chemokines Acting via CXCR2 and CXCR4 Control the Release of Neutrophils from the Bone Marrow and Their Return following Senescence. Immunity 2003, 19, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Dikshit, M. Metabolic Insight of Neutrophils in Health and Disease. Front. Immunol. 2019, 10, 2099. [Google Scholar] [CrossRef] [PubMed]
- Injarabian, L.; Devin, A.; Ransac, S.; Marteyn, B.S. Neutrophil Metabolic Shift during Their Lifecycle: Impact on Their Survival and Activation. Int. J. Mol. Sci. 2019, 21, 287. [Google Scholar] [CrossRef] [PubMed]
- Maianski, N.A.; Geissler, J.; Srinivasula, S.M.; Alnemri, E.S.; Roos, D.; Kuijpers, T.W. Functional characterization of mitochondria in neutrophils: A role restricted to apoptosis. Cell Death Differ. 2003, 11, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Simsek, T.; Kocabas, F.; Zheng, J.; DeBerardinis, R.J.; Mahmoud, A.I.; Olson, E.N.; Schneider, J.W.; Zhang, C.C.; Sadek, H.A. The Distinct Metabolic Profile of Hematopoietic Stem Cells Reflects Their Location in a Hypoxic Niche. Cell Stem Cell 2010, 7, 380–390. [Google Scholar] [CrossRef]
- Bainton, D.F.; Ullyot, J.L.; Farquhar, M.G. The development of neutrophilic polymorphonuclear leukocytes in human bone marrow. J. Exp. Med. 1971, 134, 907–934. [Google Scholar] [CrossRef] [PubMed]
- Riffelmacher, T.; Clarke, A.; Richter, F.C.; Stranks, A.; Pandey, S.; Danielli, S.; Hublitz, P.; Yu, Z.; Johnson, E.; Schwerd, T.; et al. Autophagy-Dependent Generation of Free Fatty Acids Is Critical for Normal Neutrophil Differentiation. Immunity 2017, 47, 466–480.e5. [Google Scholar] [CrossRef]
- Walmsley, S.R.; Print, C.; Farahi, N.; Peyssonnaux, C.; Johnson, R.S.; Cramer, T.; Sobolewski, A.; Condliffe, A.M.; Cowburn, A.S.; Johnson, N.; et al. Hypoxia-induced neutrophil survival is mediated by HIF-1α–dependent NF-κB activity. J. Exp. Med. 2005, 201, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Ledderose, C.; Seier, T.; Graf, A.F.; Brix, B.; Chong, E.; Junger, W.G. Mitochondria Regulate Neutrophil Activation by Generating ATP for Autocrine Purinergic Signaling. J. Biol. Chem. 2014, 289, 26794–26803. [Google Scholar] [CrossRef]
- Chen, Y.; Yao, Y.; Sumi, Y.; Li, A.; To, U.K.; Elkhal, A.; Inoue, Y.; Woehrle, T.; Zhang, Q.; Hauser, C.; et al. Purinergic Signaling: A Fundamental Mechanism in Neutrophil Activation. Sci. Signal. 2010, 3, ra45. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Ledderose, C.; Graf, A.F.; Brix, B.; Birsak, T.; Lee, A.; Zhang, J.; Junger, W.G. mTOR and differential activation of mitochondria orchestrate neutrophil chemotaxis. J. Cell Biol. 2015, 210, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.C.A.; Hebeba, C.B.; Hastreiter, A.A.; de Oliveira, D.C.; Makiyama, E.N.; Farsky, S.H.P.; Borelli, P.; Fock, R.A. Exogenous glutamine impairs neutrophils migration into infections sites elicited by lipopolysaccharide by a multistep mechanism. Amino Acids 2019, 51, 451–462. [Google Scholar] [CrossRef]
- Weisdorf, D.J.; Craddock, P.R.; Jacob, H.S. Granulocytes utilize different energy sources for movement and phagocytosis. Inflammation 1982, 6, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.M.; Karnovsky, M.L. Glycogen accumulation in polymorphonuclear leukocytes, and other intracellular alterations that occur during inflammation. J. Cell Biol. 1982, 95, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Sadiku, P.; Willson, J.A.; Ryan, E.M.; Sammut, D.; Coelho, P.; Watts, E.R.; Grecian, R.; Young, J.M.; Bewley, M.; Arienti, S.; et al. Neutrophils Fuel Effective Immune Responses through Gluconeogenesis and Glycogenesis. Cell Metab. 2021, 33, 411–423.e4. [Google Scholar] [CrossRef]
- Sadiku, P.; Willson, J.A.; Dickinson, R.S.; Murphy, F.; Harris, A.J.; Lewis, A.; Sammut, D.; Mirchandani, A.S.; Ryan, E.; Watts, E.R.; et al. Prolyl hydroxylase 2 inactivation enhances glycogen storage and promotes excessive neutrophilic responses. J. Clin. Investig. 2017, 127, 3407–3420. [Google Scholar] [CrossRef]
- Sbarra, A.J.; Karnovsky, M.L. The biochemical basis of phagocytosis. I. Metabolic changes during the ingestion of particles by polymorphonuclear leukocytes. J. Biol. Chem. 1959, 234, 1355–1362. [Google Scholar] [CrossRef]
- Borregaard, N.; Herlin, T. Energy Metabolism of Human Neutrophils during Phagocytosis. J. Clin. Investig. 1982, 70, 550–557. [Google Scholar] [CrossRef]
- Fossati, G.; Moulding, D.A.; Spiller, D.G.; Moots, R.J.; White, M.R.H.; Edwards, S.W. The Mitochondrial Network of Human Neutrophils: Role in Chemotaxis, Phagocytosis, Respiratory Burst Activation, and Commitment to Apoptosis. J. Immunol. 2003, 170, 1964–1972. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Espinosa, O.; Rojas-Espinosa, O.; Moreno-Altamirano, M.M.B.; López-Villegas, E.O.; Sánchez-García, F.J. Metabolic requirements for neutrophil extracellular traps formation. Immunology 2015, 145, 213–224. [Google Scholar] [CrossRef]
- Azevedo, E.P.; Rochael, N.C.; Guimarães-Costa, A.B.; de Souza-Vieira, T.S.; Ganilho, J.; Saraiva, E.M.; Palhano, F.L.; Foguel, D. A Metabolic Shift toward Pentose Phosphate Pathway Is Necessary for Amyloid Fibril- and Phorbol 12-Myristate 13-Acetate-induced Neutrophil Extracellular Trap (NET) Formation. J. Biol. Chem. 2015, 290, 22174–22183. [Google Scholar] [CrossRef] [PubMed]
- Britt, E.C.; Lika, J.; Giese, M.A.; Schoen, T.J.; Seim, G.L.; Huang, Z.; Lee, P.Y.; Huttenlocher, A.; Fan, J. Switching to the cyclic pentose phosphate pathway powers the oxidative burst in activated neutrophils. Nat. Metab. 2022, 4, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Pithon-Curi, T.C.; Levada, A.C.; Lopes, L.; Doi, S.Q.; Curi, R. Glutamine plays a role in superoxide production and the expression of p47phox, p22phox and gp91phox in rat neutrophils. Clin. Sci. 2002, 103, 403–408. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Zhu, L. Role of neutrophils in acute viral infection. Immun. Inflamm. Dis. 2021, 9, 1186–1196. [Google Scholar] [CrossRef] [PubMed]
- Nicolás-Ávila, J.Á.; Adrover, J.M.; Hidalgo, A. Neutrophils in Homeostasis, Immunity, and Cancer. Immunity 2017, 46, 15–28. [Google Scholar] [CrossRef]
- Giacalone, V.D.; Margaroli, C.; Mall, M.A.; Tirouvanziam, R. Neutrophil Adaptations upon Recruitment to the Lung: New Concepts and Implications for Homeostasis and Disease. Int. J. Mol. Sci. 2020, 21, 851. [Google Scholar] [CrossRef] [PubMed]
- Johansson, C.; Kirsebom, F.C.M. Neutrophils in respiratory viral infections. Mucosal Immunol. 2021, 14, 815–827. [Google Scholar] [CrossRef]
- Camp, J.V.; Jonsson, C.B. A Role for Neutrophils in Viral Respiratory Disease. Front. Immunol. 2017, 8, 550. [Google Scholar] [CrossRef]
- Soto, J.A.; Gálvez, N.M.S.; Benavente, F.M.; Pizarro-Ortega, M.S.; Lay, M.K.; Riedel, C.; Bueno, S.M.; Gonzalez, P.A.; Kalergis, A.M. Human Metapneumovirus: Mechanisms and Molecular Targets Used by the Virus to Avoid the Immune System. Front. Immunol. 2018, 9, 2466. [Google Scholar] [CrossRef]
- Kirsebom, F.C.M.; Kausar, F.; Nuriev, R.; Makris, S.; Johansson, C. Neutrophil recruitment and activation are differentially dependent on MyD88/TRIF and MAVS signaling during RSV infection. Mucosal Immunol. 2019, 12, 1244–1255. [Google Scholar] [CrossRef]
- Tang, B.M.; Shojaei, M.; Teoh, S.; Meyers, A.; Ho, J.; Ball, T.B.; Keynan, Y.; Pisipati, A.; Kumar, A.; Eisen, D.P.; et al. Neutrophils-related host factors associated with severe disease and fatality in patients with influenza infection. Nat. Commun. 2019, 10, 3422. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E.A.; He, X.-Y.; Denorme, F.; Campbell, R.A.; Ng, D.; Salvatore, S.P.; Mostyka, M.; Baxter-Stoltzfus, A.; Borczuk, A.C.; Loda, M.; et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood 2020, 136, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.K.; Zhu, L.; Procario, M.C.; Weinberg, J.B. IL-17 contributes to neutrophil recruitment but not to control of viral replication during acute mouse adenovirus type 1 respiratory infection. Virology 2014, 456–457, 259–267. [Google Scholar] [CrossRef]
- Habibi, M.S.; Thwaites, R.S.; Chang, M.; Jozwik, A.; Paras, A.; Kirsebom, F.; Varese, A.; Owen, A.; Cuthbertson, L.; James, P.; et al. Neutrophilic inflammation in the respiratory mucosa predisposes to RSV infection. Science 2020, 370, 6513. [Google Scholar] [CrossRef]
- Santos, A.F.; Póvoa, P.; Paixão, P.; Mendonça, A.; Taborda-Barata, L. Changes in Glycolytic Pathway in SARS-COV 2 Infection and Their Importance in Understanding the Severity of COVID-19. Front. Chem. 2021, 9, 685196. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hook, J.S.; Ding, Q.; Chung, S.; Moreland, J.; Agathocleous, M. Neutrophil Metabolic Rewiring in Severe and Mild COVID-19. Blood 2021, 138 (Suppl. S1), 3129. [Google Scholar] [CrossRef]
- Li, S.; Zhao, F.; Ye, J.; Li, K.; Wang, Q.; Du, Z.; Yue, Q.; Wang, S.; Wu, Q.; Chen, H. Cellular metabolic basis of altered immunity in the lungs of patients with COVID-19. Med. Microbiol. Immunol. 2022, 211, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Giam, Y.H.; Shoemark, A.; Chalmers, J.D. Neutrophil dysfunction in bronchiectasis: An emerging role for immunometabolism. Eur. Respir. J. 2021, 58, 2003157. [Google Scholar] [CrossRef] [PubMed]
- Mirchandani, A.S.; Jenkins, S.J.; Bain, C.C.; Sanchez-Garcia, M.A.; Lawson, H.; Coelho, P.; Murphy, F.; Griffith, D.M.; Zhang, A.; Morrison, T.; et al. Hypoxia shapes the immune landscape in lung injury and promotes the persistence of inflammation. Nat. Immunol. 2022, 23, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Faden, H.; Kaul, T.N.; Ogra, P.L. Activation of Oxidative and Arachidonic Acid Metabolism in Neutrophils by Respiratory Syncytial Virus Antibody Complexes: Possible Role in Disease. J. Infect. Dis. 1983, 148, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Kaul, T.N.; Faden, H.; Ogra, P.L. Effect of respiratory syncytial virus and virus-antibody complexes on the oxidative metabolism of human neutrophils. Infect. Immun. 1981, 32, 649–654. [Google Scholar] [CrossRef]
- Watts, E.R.; Howden, A.J.; Morrison, T.; Sadiku, P.; Hukelmann, J.L.; von Kriegsheim, A.; Ghesquière, B.; Murphy, F.; Mirchandani, A.S.; Humphries, D.C.; et al. Hypoxia drives murine neutrophil protein scavenging to maintain central carbon metabolism. J. Clin. Investig. 2021, 131, e134073. [Google Scholar] [CrossRef] [PubMed]
- Schimke, L.F.; Marques, A.H.C.; Baiocchi, G.C.; Prado, C.A.d.S.; Fonseca, D.L.M.; Freire, P.P.; Plaça, D.R.; Filgueiras, I.S.; Salgado, R.C.; Jansen-Marques, G.; et al. Severe COVID-19 Shares a Common Neutrophil Activation Signature with Other Acute Inflammatory States. Cells 2022, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Veras, F.P.; Pontelli, M.C.; Silva, C.M.; Toller-Kawahisa, J.E.; de Lima, M.; Nascimento, D.C.; Schneider, A.H.; Caetité, D.; Tavares, L.A.; Paiva, I.M.; et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020, 217, e20201129. [Google Scholar] [CrossRef] [PubMed]
- Borella, R.; De Biasi, S.; Paolini, A.; Boraldi, F.; Tartaro, D.L.; Mattioli, M.; Fidanza, L.; Neroni, A.; Caro-Maldonado, A.; Meschiari, M.; et al. Metabolic reprograming shapes neutrophil functions in severe COVID-19. Eur. J. Immunol. 2022, 52, 484–502. [Google Scholar] [CrossRef] [PubMed]
- Neufeldt, C.J.; Cerikan, B.; Cortese, M.; Frankish, J.; Lee, J.-Y.; Plociennikowska, A.; Heigwer, F.; Prasad, V.; Joecks, S.; Burkart, S.S.; et al. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. Commun. Biol. 2022, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Baillet, A.; Hograindleur, M.; El Benna, J.; Grichine, A.; Berthier, S.; Morel, F.; Paclet, M. Unexpected function of the phagocyte NADPH oxidase in supporting hyperglycolysis in stimulated neutrophils: Key role of 6-phosphofructo-2-kinase. FASEB J. 2017, 31, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Theresine, M.; Patil, N.D.; Zimmer, J. Airway Natural Killer Cells and Bacteria in Health and Disease. Front. Immunol. 2020, 11, 585048. [Google Scholar] [CrossRef]
- Carrega, P.; Ferlazzo, G. Natural killer cell distribution and trafficking in human tissues. Front. Immunol. 2012, 3, 347. [Google Scholar] [CrossRef]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef]
- Nguyen, K.B.; Salazar-Mather, T.P.; Dalod, M.Y.; Van Deusen, J.B.; Wei, X.-Q.; Liew, F.Y.; Caligiuri, M.A.; Durbin, J.E.; Biron, C.A. Coordinated and Distinct Roles for IFN-αβ, IL-12, and IL-15 Regulation of NK Cell Responses to Viral Infection. J. Immunol. 2002, 169, 4279–4287. [Google Scholar] [CrossRef]
- Biron, C.A.; Nguyen, K.B.; Pien, G.C.; Cousens, L.P.; Salazar-Mather, T.P. Natural killer cells in antiviral defense: Function and Regulation by Innate Cytokines. Annu. Rev. Immunol. 1999, 17, 189–220. [Google Scholar] [CrossRef] [PubMed]
- Littwitz-Salomon, E.; Moreira, D.; Frost, J.N.; Choi, C.; Liou, K.T.; Ahern, D.K.; O’Shaughnessy, S.; Wagner, B.; Biron, C.A.; Drakesmith, H.; et al. Metabolic requirements of NK cells during the acute response against retroviral infection. Nat. Commun. 2021, 12, 5376. [Google Scholar] [CrossRef]
- Assmann, N.; O’Brien, K.L.; Donnelly, R.P.; Dyck, L.; Zaiatz-Bittencourt, V.; Loftus, R.M.; Heinrich, P.; Oefner, P.J.; Lynch, L.; Gardiner, C.M.; et al. Srebp-controlled glucose metabolism is essential for NK cell functional responses. Nat. Immunol. 2017, 18, 1197–1206. [Google Scholar] [CrossRef]
- Cong, J. Metabolism of Natural Killer Cells and Other Innate Lymphoid Cells. Front. Immunol. 2020, 11, 1989. [Google Scholar] [CrossRef] [PubMed]
- Loftus, R.M.; Assmann, N.; Kedia-Mehta, N.; O’Brien, K.L.; Garcia, A.; Gillespie, C.; Hukelmann, J.L.; Oefner, P.J.; Lamond, A.I.; Gardiner, C.M.; et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat. Commun. 2018, 9, 2341. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, R.P.; Loftus, R.M.; Keating, S.E.; Liou, K.T.; Biron, C.A.; Gardiner, C.M.; Finlay, D.K. mTORC1-Dependent Metabolic Reprogramming Is a Prerequisite for NK Cell Effector Function. J. Immunol. 2014, 193, 4477–4484. [Google Scholar] [CrossRef]
- Victorino, F.; Bigley, T.M.; Park, E.; Yao, C.-H.; Benoit, J.; Yang, L.-P.; Piersma, S.J.; Lauron, E.J.; Davidson, R.M.; Patti, G.J.; et al. HIF1α is required for NK cell metabolic adaptation during virus infection. eLife 2021, 10, e68484. [Google Scholar] [CrossRef]
- Abdala-Valencia, H.; Coden, M.E.; Chiarella, S.E.; Jacobsen, E.A.; Bochner, B.S.; Lee, J.J.; Berdnikovs, S. Shaping eosinophil identity in the tissue contexts of development, homeostasis, and disease. J. Leukoc. Biol. 2018, 104, 95–108. [Google Scholar] [CrossRef]
- Klion, A.D.; Ackerman, S.J.; Bochner, B.S. Contributions of Eosinophils to Human Health and Disease. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 179–209. [Google Scholar] [CrossRef]
- Acharya, K.R.; Ackerman, S.J. Eosinophil Granule Proteins: Form and Function. J. Biol. Chem. 2014, 289, 17406–17415. [Google Scholar] [CrossRef]
- Harrison, A.M.; Bonville, C.A.; Rosenberg, H.F.; Domachowske, J.B. Respiratory Syncytical Virus–induced Chemokine Expression in the Lower Airways. Am. J. Respir. Crit. Care Med. 1999, 159, 1918–1924. [Google Scholar] [CrossRef] [PubMed]
- Weller, P.F.; Spencer, L.A. Functions of tissue-resident eosinophils. Nat. Rev. Immunol. 2017, 17, 746–760. [Google Scholar] [CrossRef] [PubMed]
- Ravin, K.A.; Loy, M. The Eosinophil in Infection. Clin. Rev. Allergy Immunol. 2016, 50, 214–227. [Google Scholar] [CrossRef]
- Michaeloudes, C.; Bhavsar, P.K.; Mumby, S.; Xu, B.; Hui, C.K.M.; Chung, K.F.; Adcock, I.M. Role of Metabolic Reprogramming in Pulmonary Innate Immunity and Its Impact on Lung Diseases. J. Innate Immun. 2020, 12, 31–46. [Google Scholar] [CrossRef]
- Jones, N.; Vincent, E.E.; Felix, L.C.; Cronin, J.G.; Scott, L.M.; Hole, P.S.; Lacy, P.; Thornton, C.A. Interleukin-5 drives glycolysis and reactive oxygen species-dependent citric acid cycling by eosinophils. Allergy 2020, 75, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Phipps, S.; Lam, C.E.; Mahalingam, S.; Newhouse, M.; Ramirez, R.; Rosenberg, H.F.; Foster, P.S.; Matthaei, K.I. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 2007, 110, 1578–1586. [Google Scholar] [CrossRef]
- Rodrigo-Muñoz, J.; Sastre, B.; Cañas, J.; Gil-Martínez, M.; Redondo, N.; del Pozo, V. Eosinophil Response Against Classical and Emerging Respiratory Viruses: COVID-19. J. Investig. Allergol. Clin. Immunol. 2021, 31, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Ricklin, D.; Hajishengallis, G.; Yang, K.; Lambris, J.D. Complement: A key system for immune surveillance and homeostasis. Nat. Immunol. 2010, 11, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Dunkelberger, J.R.; Song, W.-C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef]
- Killick, J.; Morisse, G.; Sieger, D.; Astier, A.L. Complement as a regulator of adaptive immunity. Semin. Immunopathol. 2018, 40, 37–48. [Google Scholar] [CrossRef]
- Csomor, E.; Bajtay, Z.; Sándor, N.; Kristóf, K.; Arlaud, G.J.; Thiel, S.; Erdei, A. Complement protein C1q induces maturation of human dendritic cells. Mol. Immunol. 2007, 44, 3389–3397. [Google Scholar] [CrossRef]
- Agrawal, P.; Nawadkar, R.; Ojha, H.; Kumar, J.; Sahu, A. Complement Evasion Strategies of Viruses: An Overview. Front. Microbiol. 2017, 8, 1117. [Google Scholar] [CrossRef]
- Mellors, J.; Tipton, T.; Longet, S.; Carroll, M. Viral Evasion of the Complement System and Its Importance for Vaccines and Therapeutics. Front. Immunol. 2020, 11, 1450. [Google Scholar] [CrossRef]
- Ghannam, A.; Fauquert, J.-L.; Thomas, C.; Kemper, C.; Drouet, C. Human complement C3 deficiency: Th1 induction requires T cell-derived complement C3a and CD46 activation. Mol. Immunol. 2014, 58, 98–107. [Google Scholar] [CrossRef]
- Carroll, M.C. The complement system in B cell regulation. Mol. Immunol. 2004, 41, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Weaver, D.J., Jr.; Reis, E.S.; Pandey, M.K.; Köhl, G.; Harris, N.; Gerard, C.; Köhl, J. C5a receptor-deficient dendritic cells promote induction of Treg and Th17 cells. Eur. J. Immunol. 2010, 40, 710–721. [Google Scholar] [CrossRef]
- Ma, L.; Sahu, S.K.; Cano, M.; Kuppuswamy, V.; Bajwa, J.; McPhatter, J.; Pine, A.; Meizlish, M.L.; Goshua, G.; Chang, C.H.; et al. Increased complement activation is a distinctive feature of severe SARS-CoV-2 infection. Sci. Immunol. 2021, 6, eabh2259. [Google Scholar] [CrossRef] [PubMed]
- Afzali, B.; Noris, M.; Lambrecht, B.N.; Kemper, C. The state of complement in COVID-19. Nat. Rev. Immunol. 2022, 22, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Santiesteban-Lores, L.E.; Amamura, T.A.; da Silva, T.F.; Midon, L.M.; Carneiro, M.C.; Isaac, L.; Bavia, L. A double edged-sword—The Complement System during SARS-CoV-2 infection. Life Sci. 2021, 272, 119245. [Google Scholar] [CrossRef] [PubMed]
- Holter, J.C.; Pischke, S.E.; de Boer, E.; Lind, A.; Jenum, S.; Holten, A.R.; Tonby, K.; Barratt-Due, A.; Sokolova, M.; Schjalm, C.; et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc. Natl. Acad. Sci. USA 2020, 117, 25018–25025. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.B.; da Silva, Z.E.V.; Gomes, C.; Reis, C.A.; Amorim, M.J. Complement Decay-Accelerating Factor is a modulator of influenza A virus lung immunopathology. PLoS Pathog. 2021, 17, e1009381. [Google Scholar] [CrossRef]
- Wu, X.; Bao, L.; Hu, Z.; Yao, D.; Li, F.; Li, H.; Xu, X.; An, Y.; Wang, X.; Bin Cao, B.; et al. Ficolin A exacerbates severe H1N1 influenza virus infection-induced acute lung immunopathological injury via excessive complement activation. Cell. Mol. Immunol. 2021, 18, 2278–2280. [Google Scholar] [CrossRef]
- Kolev, M.; Kemper, C. Keeping It All Going—Complement Meets Metabolism. Front. Immunol. 2017, 8, 1. [Google Scholar] [CrossRef]
- Liszewski, M.K.; Kolev, M.; Le Friec, G.; Leung, M.; Bertram, P.G.; Fara, A.F.; Subias, M.; Pickering, M.C.; Drouet, C.; Meri, S.; et al. Intracellular Complement Activation Sustains T Cell Homeostasis and Mediates Effector Differentiation. Immunity 2013, 39, 1143–1157. [Google Scholar] [CrossRef] [PubMed]
- Arbore, G.; West, E.E.; Spolski, R.; Robertson, A.A.B.; Klos, A.; Rheinheimer, C.; Dutow, P.; Woodruff, T.M.; Yu, Z.X.; O’Neill, L.A.; et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4+ T cells. Science 2016, 352, aad1210. [Google Scholar] [CrossRef]
- Mancini, N.; Solforosi, L.; Clementi, N.; De Marco, D.; Clementi, M.; Burioni, R. A potential role for monoclonal antibodies in prophylactic and therapeutic treatment of influenza. Antivir. Res. 2011, 92, 15–26. [Google Scholar] [CrossRef]
- Ayres, J.S. Immunometabolism of infections. Nat. Rev. Immunol. 2020, 20, 79–80. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.; Openshaw, P.J. Antiviral B cell and T cell immunity in the lungs. Nat. Immunol. 2015, 16, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Frauwirth, K.A.; Riley, J.L.; Harris, M.H.; Parry, R.V.; Rathmell, J.C.; Plas, D.R.; Elstrom, R.L.; June, C.H.; Thompson, C.B. The CD28 signaling pathway regulates glucose metabolism. Immunity 2002, 16, 769–777. [Google Scholar] [CrossRef]
- Makowski, L.; Chaib, M.; Rathmell, J.C. Immunometabolism: From basic mechanisms to translation. Immunol. Rev. 2020, 295, 5–14. [Google Scholar] [CrossRef]
- Bantug, G.R.; Fischer, M.; Grählert, J.; Balmer, M.L.; Unterstab, G.; Develioglu, L.; Steiner, R.; Zhang, L.; Costa, A.S.H.; Gubser, P.M.; et al. Mitochondria-Endoplasmic Reticulum Contact Sites Function as Immunometabolic Hubs that Orchestrate the Rapid Recall Response of Memory CD8+ T Cells. Immunity 2018, 48, 542–555.e6. [Google Scholar] [CrossRef]
- Pearce, E.L.; Walsh, M.C.; Cejas, P.J.; Harms, G.M.; Shen, H.; Wang, L.-S.; Jones, R.G.; Choi, Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 2009, 460, 103–107. [Google Scholar] [CrossRef]
- Nakaya, M.; Xiao, Y.; Zhou, X.; Chang, J.-H.; Chang, M.; Cheng, X.; Blonska, M.; Lin, X.; Sun, S.-C. Inflammatory T Cell Responses Rely on Amino Acid Transporter ASCT2 Facilitation of Glutamine Uptake and mTORC1 Kinase Activation. Immunity 2014, 40, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Angelin, A.; Gil-De-Gómez, L.; Dahiya, S.; Jiao, J.; Guo, L.; Levine, M.H.; Wang, Z.; Quinn, W.J., III; Kopinski, P.K.; Wang, L.; et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 2017, 25, 1282–1293.e7. [Google Scholar] [CrossRef]
- Gerriets, V.A.; Kishton, R.J.; Nichols, A.G.; Macintyre, A.; Inoue, M.; Ilkayeva, O.; Winter, P.S.; Liu, X.; Priyadharshini, B.; Slawinska, M.E.; et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Investig. 2015, 125, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Macintyre, A.N.; Gerriets, V.A.; Nichols, A.G.; Michalek, R.D.; Rudolph, M.C.; DeOliveira, D.; Anderson, S.M.; Abel, E.D.; Chen, B.J.; Hale, L.P.; et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014, 20, 61–72. [Google Scholar] [CrossRef]
- Gerriets, V.A.; Kishton, R.J.; Johnson, M.O.; Cohen, S.; Siska, P.J.; Nichols, A.G.; Warmoes, M.O.; De Cubas, A.A.; Maciver, N.J.; Locasale, J.W.; et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 2016, 17, 1459–1466. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, G.; Xu, Z.-G.; Tu, H.; Hu, F.; Dai, J.; Chang, Y.; Chen, Y.; Lu, Y.; Zeng, H.; et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell 2019, 178, 176–189.e15. [Google Scholar] [CrossRef]
- Baazim, H.; Schweiger, M.; Moschinger, M.; Xu, H.; Scherer, T.; Popa, A.; Gallage, S.; Ali, A.; Khamina, K.; Kosack, L.; et al. CD8+ T cells induce cachexia during chronic viral infection. Nat. Immunol. 2019, 20, 701–710. [Google Scholar] [CrossRef]
- Hewitt, E.W. The MHC class I antigen presentation pathway: Strategies for viral immune evasion. Immunology 2003, 110, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, X.-P.; Zhu, X.-L.; Li, T.; Liu, Y. IRG1 increases MHC class I level in macrophages through STAT-TAP1 axis depending on NADPH oxidase mediated reactive oxygen species. Int. Immunopharmacol. 2017, 48, 76–83. [Google Scholar] [CrossRef]
- Alés-Martínez, J.; Cuende, E.; Martínez-A, C.; Parkhouse, R.; Pezzi, L.; Scott, D.W. Signalling in B cells. Immunol. Today 1991, 12, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Parker, D.C. T Cell-Dependent B Cell Activation. Annu. Rev. Immunol. 1993, 11, 331–360. [Google Scholar] [CrossRef]
- Luo, W.; Yin, Q. B Cell Response to Vaccination. Immunol. Investig. 2021, 50, 780–801. [Google Scholar] [CrossRef]
- Guthmiller, J.; Utset, H.; Wilson, P. B Cell Responses against Influenza Viruses: Short-Lived Humoral Immunity against a Life-Long Threat. Viruses 2021, 13, 965. [Google Scholar] [CrossRef] [PubMed]
- Laubreton, D.; Drajac, C.; Eléouët, J.-F.; Rameix-Welti, M.-A.; Lo-Man, R.; Riffault, S.; Descamps, D. Regulatory B Lymphocytes Colonize the Respiratory Tract of Neonatal Mice and Modulate Immune Responses of Alveolar Macrophages to RSV Infection in IL-10-Dependant Manner. Viruses 2020, 12, 822. [Google Scholar] [CrossRef] [PubMed]
- Scheid, J.F.; Barnes, C.O.; Eraslan, B.; Hudak, A.; Keeffe, J.R.; Cosimi, L.A.; Brown, E.M.; Muecksch, F.; Weisblum, Y.; Zhang, S.; et al. B cell genomics behind cross-neutralization of SARS-CoV-2 variants and SARS-CoV. Cell 2021, 184, 3205–3221.e24. [Google Scholar] [CrossRef]
- Woodruff, M.C.; Ramonell, R.P.; Haddad, N.S.; Anam, F.A.; Rudolph, M.E.; Walker, T.A.; Truong, A.D.; Dixit, A.N.; Han, J.E.; Cabrera-Mora, M.; et al. Dysregulated naive B cells and de novo autoreactivity in severe COVID-19. Nature 2022, 611, 139–147. [Google Scholar] [CrossRef]
- Desai, P.; Stanfield, J.; Tahiliani, V.; Abboud, G.; Salek-Ardakani, S. Lack of B Lymphocytes Enhances CD8 T Cell-Mediated Resistance against Respiratory Viral Infection but Compromises Memory Cell Formation. J. Virol. 2020, 94, e01877-19. [Google Scholar] [CrossRef]
- Çölkesen, F.; Kurt, E.K.; Vatansev, H.; Korkmaz, C.; Çölkesen, F.; Yücel, F.; Yıldız, E.; Evcen, R.; Aykan, F.S.; Kılınç, M.; et al. Memory B cells and serum immunoglobulins are associated with disease severity and mortality in patients with COVID-19. Postgrad. Med. J. 2022, 98, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Taliani, G.; Follini, E.; Guglielmetti, L.; Bernuzzi, P.; Faggi, A.; Ferrante, P.; Fronti, E.; Gerna, L.; Leoni, M.C.; Paolillo, F.; et al. Case Report: B Lymphocyte Disorders Under COVID-19 Inflammatory Pressure. Front. Oncol. 2021, 10, 582901. [Google Scholar] [CrossRef] [PubMed]
- Caro-Maldonado, A.; Wang, R.; Nichols, A.G.; Kuraoka, M.; Milasta, S.; Sun, L.D.; Gavin, A.L.; Abel, E.D.; Kelsoe, G.; Green, D.R.; et al. Metabolic Reprogramming Is Required for Antibody Production That Is Suppressed in Anergic but Exaggerated in Chronically BAFF-Exposed B Cells. J. Immunol. 2014, 192, 3626–3636. [Google Scholar] [CrossRef]
- Dufort, F.J.; Bleiman, B.F.; Gumina, M.R.; Blair, D.; Wagner, D.J.; Roberts, M.F.; Abu-Amer, Y.; Chiles, T.C. Cutting Edge: IL-4-Mediated Protection of Primary B Lymphocytes from Apoptosis via Stat6-Dependent Regulation of Glycolytic Metabolism. J. Immunol. 2007, 179, 4953–4957. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Chan, L.N.; Klemm, L.; Braas, D.; Chen, Z.; Geng, H.; Zhang, Q.C.; Aghajanirefah, A.; Cosgun, K.; Sadras, T.; et al. B-Cell-Specific Diversion of Glucose Carbon Utilization Reveals a Unique Vulnerability in B Cell Malignancies. Cell 2018, 173, 470–484.e18. [Google Scholar] [CrossRef] [PubMed]
- Akkaya, M.; Traba, J.; Roesler, A.S.; Miozzo, P.; Akkaya, B.; Theall, B.P.; Sohn, H.; Pena, M.; Smelkinson, M.; Kabat, J.; et al. Second signals rescue B cells from activation-induced mitochondrial dysfunction and death. Nat. Immunol. 2018, 19, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Burrows, N.; Bashford-Rogers, R.J.M.; Bhute, V.J.; Peñalver, A.; Ferdinand, J.R.; Stewart, B.J.; Smith, J.E.G.; Deobagkar-Lele, M.; Giudice, G.; Connor, T.M.; et al. Dynamic regulation of hypoxia-inducible factor-1α activity is essential for normal B cell development. Nat. Immunol. 2020, 21, 1408–1420. [Google Scholar] [CrossRef] [PubMed]
- Kotagiri, P.; Mescia, F.; Hanson, A.L.; Turner, L.; Bergamaschi, L.; Peñalver, A.; Richoz, N.; Moore, S.D.; Ortmann, B.M.; Dunmore, B.J.; et al. The impact of hypoxia on B cells in COVID-19. Ebiomedicine 2022, 77, 103878. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, D.; van der Windt, G.J.; Huang, S.C.-C.; Curtis, J.D.; Chang, C.-H.; Buck, M.D.; Qiu, J.; Smith, A.M.; Lam, W.Y.; DiPlato, L.M.; et al. Memory CD8+ T Cells Use Cell-Intrinsic Lipolysis to Support the Metabolic Programming Necessary for Development. Immunity 2014, 41, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Tian, T.; Park, C.O.; Lofftus, S.Y.; Mei, S.; Liu, X.; Luo, C.; O’Malley, J.T.; Gehad, A.; Teague, J.E.; et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 2017, 543, 252–256. [Google Scholar] [CrossRef]
- Wallis, R.S.; O’Garra, A.; Sher, A.; Wack, A. Host-directed immunotherapy of viral and bacterial infections: Past, present and future. Nat. Rev. Immunol. 2022, 23, 121–133. [Google Scholar] [CrossRef]
- Kim, J.-S.; Kim, Y.-R.; Yang, C.-S. Host-Directed Therapy in Tuberculosis: Targeting Host Metabolism. Front. Immunol. 2020, 11, 1790. [Google Scholar] [CrossRef]
- Rao, M.; Dodoo, E.; Zumla, A.; Maeurer, M. Immunometabolism and Pulmonary Infections: Implications for Protective Immune Responses and Host-Directed Therapies. Front. Microbiol. 2019, 10, 962. [Google Scholar] [CrossRef]
- Hoyle, C.; Green, J.P.; Allan, S.M.; Brough, D.; Lemarchand, E. Itaconate and fumarate derivatives inhibit priming and activation of the canonical NLRP3 inflammasome in macrophages. Immunology 2022, 165, 460–480. [Google Scholar] [CrossRef]
- Liao, S.-T.; Han, C.; Xu, D.-Q.; Fu, X.-W.; Wang, J.-S.; Kong, L.-Y. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nat. Commun. 2019, 10, 5091. [Google Scholar] [CrossRef]
- Olagnier, D.; Farahani, E.; Thyrsted, J.; Blay-Cadanet, J.; Herengt, A.; Idorn, M.; Hait, A.; Hernaez, B.; Knudsen, A.; Iversen, M.B.; et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020, 11, 4938. [Google Scholar] [CrossRef] [PubMed]
- Sohail, A.; Iqbal, A.A.; Sahini, N.; Chen, F.; Tantawy, M.; Waqas, S.F.; Winterhoff, M.; Ebensen, T.; Schultz, K.; Geffers, R.; et al. Itaconate and derivatives reduce interferon responses and inflammation in influenza A virus infection. PLoS Pathog. 2022, 18, e1010219. [Google Scholar] [CrossRef]
- Kheirollahi, V.; Wasnick, R.M.; Biasin, V.; Vazquez-Armendariz, A.I.; Chu, X.; Moiseenko, A.; Weiss, A.; Wilhelm, J.; Zhang, J.-S.; Kwapiszewska, G.; et al. Metformin induces lipogenic differentiation in myofibroblasts to reverse lung fibrosis. Nat. Commun. 2019, 10, 2987. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.E.; Hossain, A.T.; Nair, H.; Chisti, M.J.; Dockrell, D.; El Arifeen, S.; Campbell, H. Prevalence of hypoxaemia in children with pneumonia in low-income and middle-income countries: A systematic review and meta-analysis. Lancet Glob. Health 2022, 10, e348–e359. [Google Scholar] [CrossRef]
- Kamyshnyi, O.; Matskevych, V.; Lenchuk, T.; Strilbytska, O.; Storey, K.; Lushchak, O. Metformin to decrease COVID-19 severity and mortality: Molecular mechanisms and therapeutic potential. Biomed. Pharmacother. 2021, 144, 112230. [Google Scholar] [CrossRef]
- Weichhart, T.; Saemann, M.D. The PI3K/Akt/mTOR pathway in innate immune cells: Emerging therapeutic applications. Ann. Rheum. Dis. 2008, 67 (Suppl. S3), iii70–iii74. [Google Scholar] [CrossRef]
- Smallwood, H.S.; Duan, S.; Morfouace, M.; Rezinciuc, S.; Shulkin, B.L.; Shelat, A.; Zink, E.E.; Milasta, S.; Bajracharya, R.; Oluwaseum, A.J.; et al. Targeting Metabolic Reprogramming by Influenza Infection for Therapeutic Intervention. Cell Rep. 2017, 19, 1640–1653. [Google Scholar] [CrossRef]
- Matthay, M.A.; Zemans, R.L.; Zimmerman, G.A.; Arabi, Y.M.; Beitler, J.R.; Mercat, A.; Herridge, M.; Randolph, A.G.; Calfee, C.S. Acute respiratory distress syndrome. Nat. Rev. Dis. Prim. 2019, 5, 18. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Agarwal, S.; Shrimali, N.M.; Guchhait, P. Interplay between hypoxia and inflammation contributes to the progression and severity of respiratory viral diseases. Mol. Asp. Med. 2021, 81, 101000. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.T.; Scholz, C.C. The effect of HIF on metabolism and immunity. Nat. Rev. Nephrol. 2022, 18, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.B.; Pugin, J.; Lee, J.S.; Matthay, M.A. Cytokine-mediated inflammation in acute lung injury. Cytokine Growth Factor Rev. 2003, 14, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Straus, D.S.; Glass, C.K. Anti-inflammatory actions of PPAR ligands: New insights on cellular and molecular mechanisms. Trends Immunol. 2007, 28, 551–558. [Google Scholar] [CrossRef]
- Kim, W.; Lee, H.-N.; Jang, J.-H.; Kim, S.H.; Lee, Y.-H.; Hahn, Y.-I.; Ngo, H.-K.; Choi, Y.; Joe, Y.; Chung, H.T.; et al. 15-Deoxy-Δ12,14-Prostaglandin J2Exerts Proresolving Effects Through Nuclear Factor E2-Related Factor 2-Induced Expression of CD36 and Heme Oxygenase-1. Antioxid. Redox Signal. 2017, 27, 1412–1431. [Google Scholar] [CrossRef]
- Kim, W.; Jang, J.-H.; Zhong, X.; Seo, H.; Surh, Y.-J. 15-Deoxy-△12,14-Prostaglandin J2 Promotes Resolution of Experimentally Induced Colitis. Front. Immunol. 2021, 12, 615803. [Google Scholar] [CrossRef]
- Han, Z.; Zhu, T.; Liu, X.; Li, C.; Yue, S.; Liu, X.; Yang, L.; Yang, L.; Li, L. 15-deoxy-Δ12,14-prostaglandin J2reduces recruitment of bone marrow-derived monocyte/macrophages in chronic liver injury in mice. Hepatology 2012, 56, 350–360. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, Q.; Mao, G.; Dowling, C.A.; Lundy, S.K.; Mao-Draayer, Y. Dimethyl Fumarate Selectively Reduces Memory T Cells and Shifts the Balance between Th1/Th17 and Th2 in Multiple Sclerosis Patients. J. Immunol. 2017, 198, 3069–3080. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Rezk, A.; Ghadiri, M.; Luessi, F.; Zipp, F.; Li, H.; Giacomini, P.S.; Antel, J.; Bar-Or, A. Dimethyl Fumarate Treatment Mediates an Anti-Inflammatory Shift in B Cell Subsets of Patients with Multiple Sclerosis. J. Immunol. 2017, 198, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Calabresi, P.; Bhargava, P. Dimethyl fumarate treatment alters NK cell function in multiple sclerosis. Eur. J. Immunol. 2018, 48, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.K.; Sandey, M.; Suryawanshi, A.; Cattley, R.C.; Mishra, A. Dimethyl fumarate abrogates dust mite-induced allergic asthma by altering dendritic cell function. Immun. Inflamm. Dis. 2019, 7, 201–213. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Gupta, D.P.; Park, S.H.; Yang, H.-J.; Song, G.J. Anti-Inflammatory Effects of Dimethyl Fumarate in Microglia via an Autophagy Dependent Pathway. Front. Pharmacol. 2021, 12, 612981. [Google Scholar] [CrossRef]
- Müller, S.; Behnen, M.; Bieber, K.; Möller, S.; Hellberg, L.; Witte, M.; Hänsel, M.; Zillikens, D.; Solbach, W.; Laskay, T.; et al. Dimethylfumarate Impairs Neutrophil Functions. J. Investig. Dermatol. 2016, 136, 117–126. [Google Scholar] [CrossRef]
- Salminen, A.; Hyttinen, J.M.T.; Kaarniranta, K. AMP-activated protein kinase inhibits NF-κB signaling and inflammation: Impact on healthspan and lifespan. J. Mol. Med. 2011, 89, 667–676. [Google Scholar] [CrossRef]
- Xiao, N.; Wang, J.; Wang, T.; Xiong, X.; Zhou, J.; Su, X.; Peng, J.; Yang, C.; Li, X.; Lin, G.; et al. Metformin abrogates pathological TNF-α-producing B cells through mTOR-dependent metabolic reprogramming in polycystic ovary syndrome. eLife 2022, 11, e74713. [Google Scholar] [CrossRef] [PubMed]
- Xian, H.; Liu, Y.; Nilsson, A.R.; Gatchalian, R.; Crother, T.R.; Tourtellotte, W.G.; Zhang, Y.; Aleman-Muench, G.R.; Lewis, G.; Chen, W.; et al. Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammation. Immunity 2021, 54, 1463–1477.e11. [Google Scholar] [CrossRef] [PubMed]
- Cory, T.J.; Emmons, R.S.; Yarbro, J.R.; Davis, K.L.; Pence, B.D. Metformin Suppresses Monocyte Immunometabolic Activation by SARS-CoV-2 Spike Protein Subunit 1. Front. Immunol. 2021, 12, 733921. [Google Scholar] [CrossRef] [PubMed]
- Menegazzo, L.; Scattolini, V.; Cappellari, R.; Bonora, B.M.; Albiero, M.; Bortolozzi, M.; Romanato, F.; Ceolotto, G.; de Kreutzeberg, S.V.; Avogaro, A.; et al. The antidiabetic drug metformin blunts NETosis in vitro and reduces circulating NETosis biomarkers in vivo. Acta Diabetol. 2018, 55, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Janes, M.R.; Fruman, D.A. Immune Regulation by Rapamycin: Moving Beyond T Cells. Sci. Signal. 2009, 2, pe25. [Google Scholar] [CrossRef]
- Floto, R.A.; Sarkar, S.; Perlstein, E.O.; Kampmann, B.; Schreiber, S.L.; Rubinsztein, D.C. Small Molecule Enhancers of Rapamycin-Induced TOR Inhibition Promote Autophagy, Reduce Toxicity in Huntington’s Disease Models and Enhance Killing of Mycobacteria by Macrophages. Autophagy 2007, 3, 620–622. [Google Scholar] [CrossRef] [PubMed]
- Frank, C.; Zhang, B.; Felice, M.; Blazar, B.R.; Miller, J.S. mTORC1 Signaling and Metabolism Are Dynamically Regulated during Human NK Cell Maturation and in Adaptive NK Cell Responses to Viral Infection. Blood 2015, 126, 209. [Google Scholar] [CrossRef]
- Mullen, P.J.; Garcia, G.; Purkayastha, A.; Matulionis, N.; Schmid, E.W.; Momcilovic, M.; Sen, C.; Langerman, J.; Ramaiah, A.; Shackelford, D.B.; et al. SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition. Nat. Commun. 2021, 12, 1876. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.; Stokes, C.A.; Prestwich, E.C.; Roberts, R.L.; Juss, J.K.; Sabroe, I.; Parker, L.C. Phosphoinositide-3 Kinase Inhibition Modulates Responses to Rhinovirus by Mechanisms that Are Predominantly Independent of Autophagy. PLoS ONE 2014, 9, e116055. [Google Scholar] [CrossRef]
- Alsuwaidi, A.R.; George, J.A.; Almarzooqi, S.; Hartwig, S.M.; Varga, S.M.; Souid, A.-K. Sirolimus alters lung pathology and viral load following influenza A virus infection. Respir. Res. 2017, 18, 136. [Google Scholar] [CrossRef] [PubMed]
- De Souza, A.P.D.; de Freitas, D.N.; Fernandes, K.E.A.; da Cunha, M.D.; Fernandes, J.L.A.; Gassen, R.B.; Fazolo, T.; Pinto, L.A.; Scotta, M.; Mattiello, R.; et al. Respiratory syncytial virus induces phosphorylation of mTOR at ser2448 in CD8 T cells from nasal washes of infected infants. Clin. Exp. Immunol. 2016, 183, 248–257. [Google Scholar] [CrossRef]
- Breslin, E.M.; White, P.C.; Shore, A.M.; Clement, M.; Brennan, P. LY294002 and rapamycin co-operate to inhibit T-cell proliferation. Br. J. Pharmacol. 2005, 144, 791–800. [Google Scholar] [CrossRef]
- García-Fojeda, B.; Minutti, C.M.; Montero-Fernández, C.; Stamme, C.; Casals, C. Signaling Pathways That Mediate Alveolar Macrophage Activation by Surfactant Protein A and IL-4. Front. Immunol. 2022, 13, 860262. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.-J.; Liu, T.; Yang, H.-H.; Duan, J.-X.; Yang, J.-T.; Guan, X.-X.; Xiong, J.-B.; Zhang, Y.-F.; Zhang, C.-Y.; Zhou, Y.; et al. TREM-1 governs NLRP3 inflammasome activation of macrophages by firing up glycolysis in acute lung injury. Int. J. Biol. Sci. 2023, 19, 242–257. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Sen, P.; Wallet, M.; Long, B.; Baldwin, A.S.; Tisch, R. Immunoregulation of dendritic cells by IL-10 is mediated through suppression of the PI3K/Akt pathway and of IκB kinase activity. Blood 2004, 104, 1100–1109. [Google Scholar] [CrossRef]
- Kinkead, L.C.; Krysa, S.J.; Allen, L.-A.H. Neutrophil Survival Signaling During Francisella tularensis Infection. Front. Cell. Infect. Microbiol. 2022, 12, 889290. [Google Scholar] [CrossRef]
- Cluff, E.; Magdaleno, C.C.; Fernandez, E.; House, T.; Swaminathan, S.; Varadaraj, A.; Rajasekaran, N. Hypoxia-inducible factor-1 alpha expression is induced by IL-2 via the PI3K/mTOR pathway in hypoxic NK cells and supports effector functions in NKL cells and ex vivo expanded NK cells. Cancer Immunol. Immunother. 2022, 71, 1989–2005. [Google Scholar] [CrossRef]
- Shin, Y.-K.; Liu, Q.; Tikoo, S.K.; Babiuk, L.A.; Zhou, Y. Effect of the phosphatidylinositol 3-kinase/Akt pathway on influenza A virus propagation. J. Gen. Virol. 2007, 88, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.W.; Monick, M.M.; Staber, J.; Yarovinsky, T.; Carter, A.B.; Hunninghake, G.W. Respiratory Syncytial Virus Inhibits Apoptosis and Induces NF-κB Activity through a Phosphatidylinositol 3-Kinase-dependent Pathway. J. Biol. Chem. 2002, 277, 492–501. [Google Scholar] [CrossRef]
- Wing, P.A.; Keeley, T.P.; Zhuang, X.; Lee, J.Y.; Prange-Barczynska, M.; Tsukuda, S.; Morgan, S.B.; Harding, A.C.; Argles, I.L.; Kurlekar, S.; et al. Hypoxic and pharmacological activation of HIF inhibits SARS-CoV-2 infection of lung epithelial cells. Cell Rep. 2021, 35, 109020. [Google Scholar] [CrossRef] [PubMed]
- Strowitzki, M.J.; Kimmer, G.; Wehrmann, J.; Ritter, A.S.; Radhakrishnan, P.; Opitz, V.M.; Tuffs, C.; Biller, M.; Kugler, J.; Keppler, U.; et al. Inhibition of HIF-prolyl hydroxylases improves healing of intestinal anastomoses. J. Clin. Investig. 2021, 6, e139191. [Google Scholar] [CrossRef] [PubMed]
- McGettrick, A.F.; O’Neill, L.A. The Role of HIF in Immunity and Inflammation. Cell Metab. 2020, 32, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chen, J.; Cheng, L.; Xu, K.; Yang, Y.; Su, X. Deficiency of HIF-1α enhances influenza A virus replication by promoting autophagy in alveolar type II epithelial cells. Emerg. Microbes Infect. 2020, 9, 691–706. [Google Scholar] [CrossRef]
- Tran, C.W.; Gold, M.J.; Garcia-Batres, C.; Tai, K.; Elford, A.R.; Himmel, M.E.; Elia, A.J.; Ohashi, P.S. Hypoxia-inducible factor 1 alpha limits dendritic cell stimulation of CD8 T cell immunity. PLoS ONE 2020, 15, e0244366. [Google Scholar] [CrossRef]
- Rathnasamy, G.; Ling, E.-A.; Kaur, C. Hypoxia inducible factor-1α mediates iron uptake which induces inflammatory response in amoeboid microglial cells in developing periventricular white matter through MAP kinase pathway. Neuropharmacology 2014, 77, 428–440. [Google Scholar] [CrossRef]
- Tian, M.; Liu, W.; Li, X.; Zhao, P.; Shereen, M.A.; Zhu, C.; Huang, S.; Liu, S.; Yu, X.; Yue, M.; et al. HIF-1α promotes SARS-CoV-2 infection and aggravates inflammatory responses to COVID-19. Signal Transduct. Target. Ther. 2021, 6, 308. [Google Scholar] [CrossRef]
- Morris, D.R.; Qu, Y.; Agrawal, A.; Garofalo, R.P.; Casola, A. HIF-1α Modulates Core Metabolism and Virus Replication in Primary Airway Epithelial Cells Infected with Respiratory Syncytial Virus. Viruses 2020, 12, 1088. [Google Scholar] [CrossRef]
- Appel, S.; Mirakaj, V.; Bringmann, A.; Weck, M.M.; Grünebach, F.; Brossart, P. PPAR-γ agonists inhibit toll-like receptor-mediated activation of dendritic cells via the MAP kinase and NF-κB pathways. Blood 2005, 106, 3888–3894. [Google Scholar] [CrossRef]
- Cloutier, A.; Marois, I.; Cloutier, D.; Verreault, C.; Cantin, A.M.; Richter, M.V. The Prostanoid 15-Deoxy-Δ12,14-Prostaglandin-J2 Reduces Lung Inflammation and Protects Mice Against Lethal Influenza Infection. J. Infect. Dis. 2012, 205, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Arnold, R.; König, W. Peroxisome proliferator-activated receptor-γ agonists inhibit the replication of respiratory syncytial virus (RSV) in human lung epithelial cells. Virology 2006, 350, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.H.; Cardani, A.; Braciale, T.J. The host immune response in respiratory virus infection: Balancing virus clearance and immunopathology. Semin. Immunopathol. 2016, 38, 471–482. [Google Scholar] [CrossRef] [PubMed]
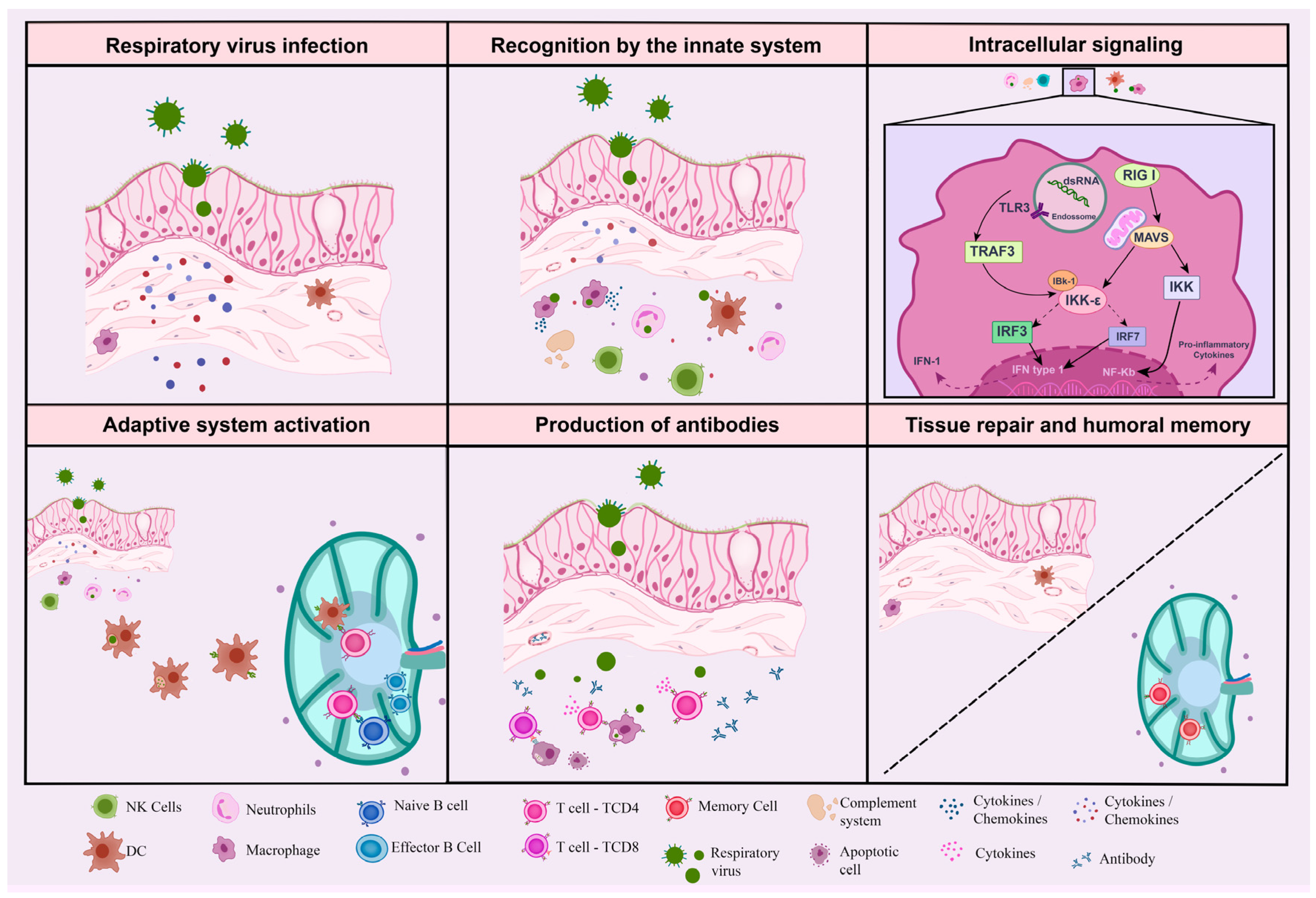
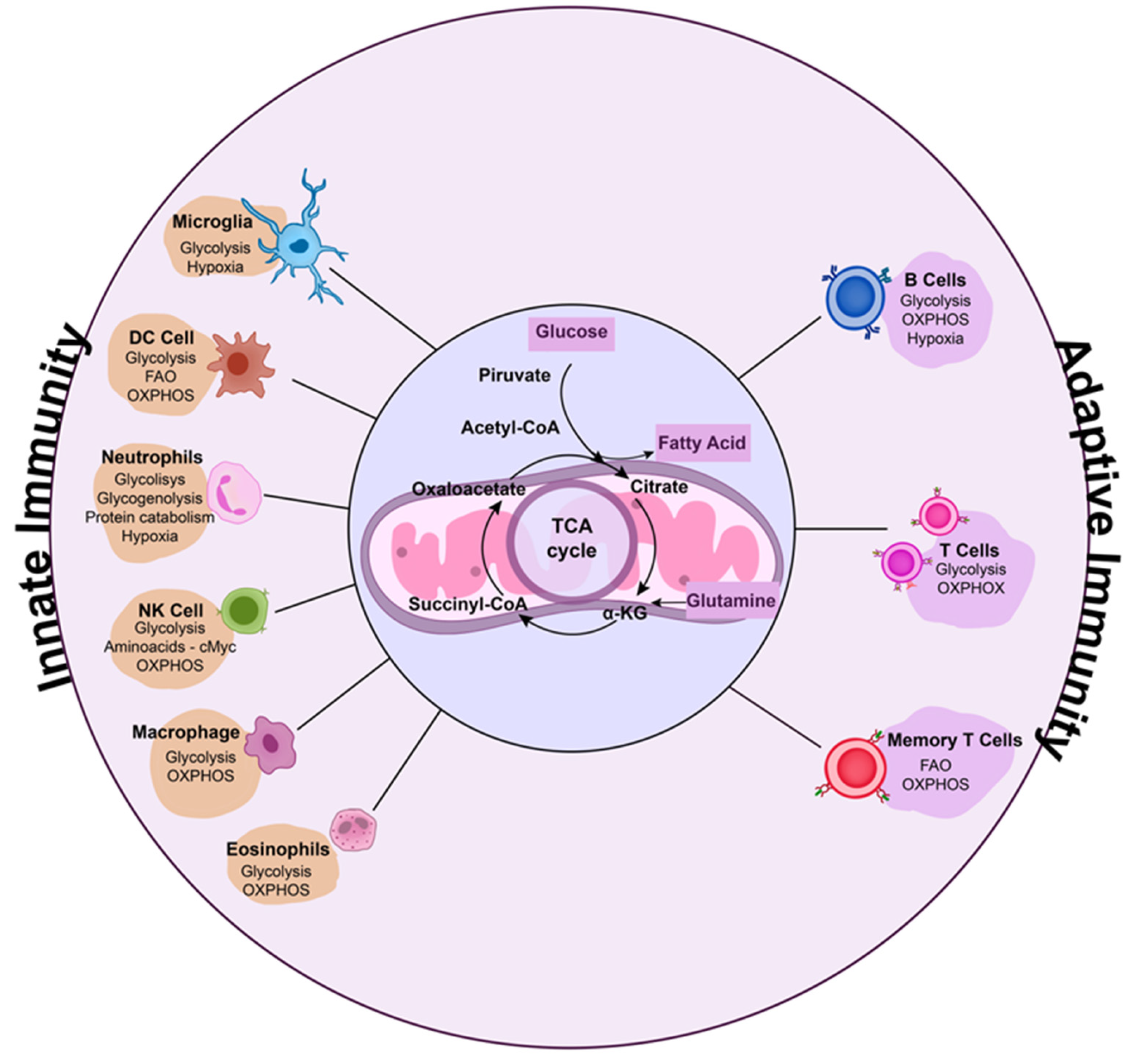
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menezes dos Reis, L.; Berçot, M.R.; Castelucci, B.G.; Martins, A.J.E.; Castro, G.; Moraes-Vieira, P.M. Immunometabolic Signature during Respiratory Viral Infection: A Potential Target for Host-Directed Therapies. Viruses 2023, 15, 525. https://doi.org/10.3390/v15020525
Menezes dos Reis L, Berçot MR, Castelucci BG, Martins AJE, Castro G, Moraes-Vieira PM. Immunometabolic Signature during Respiratory Viral Infection: A Potential Target for Host-Directed Therapies. Viruses. 2023; 15(2):525. https://doi.org/10.3390/v15020525
Chicago/Turabian StyleMenezes dos Reis, Larissa, Marcelo Rodrigues Berçot, Bianca Gazieri Castelucci, Ana Julia Estumano Martins, Gisele Castro, and Pedro M. Moraes-Vieira. 2023. "Immunometabolic Signature during Respiratory Viral Infection: A Potential Target for Host-Directed Therapies" Viruses 15, no. 2: 525. https://doi.org/10.3390/v15020525
APA StyleMenezes dos Reis, L., Berçot, M. R., Castelucci, B. G., Martins, A. J. E., Castro, G., & Moraes-Vieira, P. M. (2023). Immunometabolic Signature during Respiratory Viral Infection: A Potential Target for Host-Directed Therapies. Viruses, 15(2), 525. https://doi.org/10.3390/v15020525








