Assessment of the Biological Impact of SARS-CoV-2 Genetic Variation Using an Authentic Virus Neutralisation Assay with Convalescent Plasma, Vaccinee Sera, and Standard Reagents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Convalescent Plasma, Vaccinee Sera, and Antibody Standards
2.2. Cell Maintenance and Media
2.3. Virus Isolation from Clinical Material
2.4. Virus Bank Propagation
2.5. Quality Control of Virus Banks
2.6. Focus Reduction Neutralisation Test (FRNT)
2.7. SARS-CoV-2 Pseudotyped Neutralisation Test
2.8. Statistics
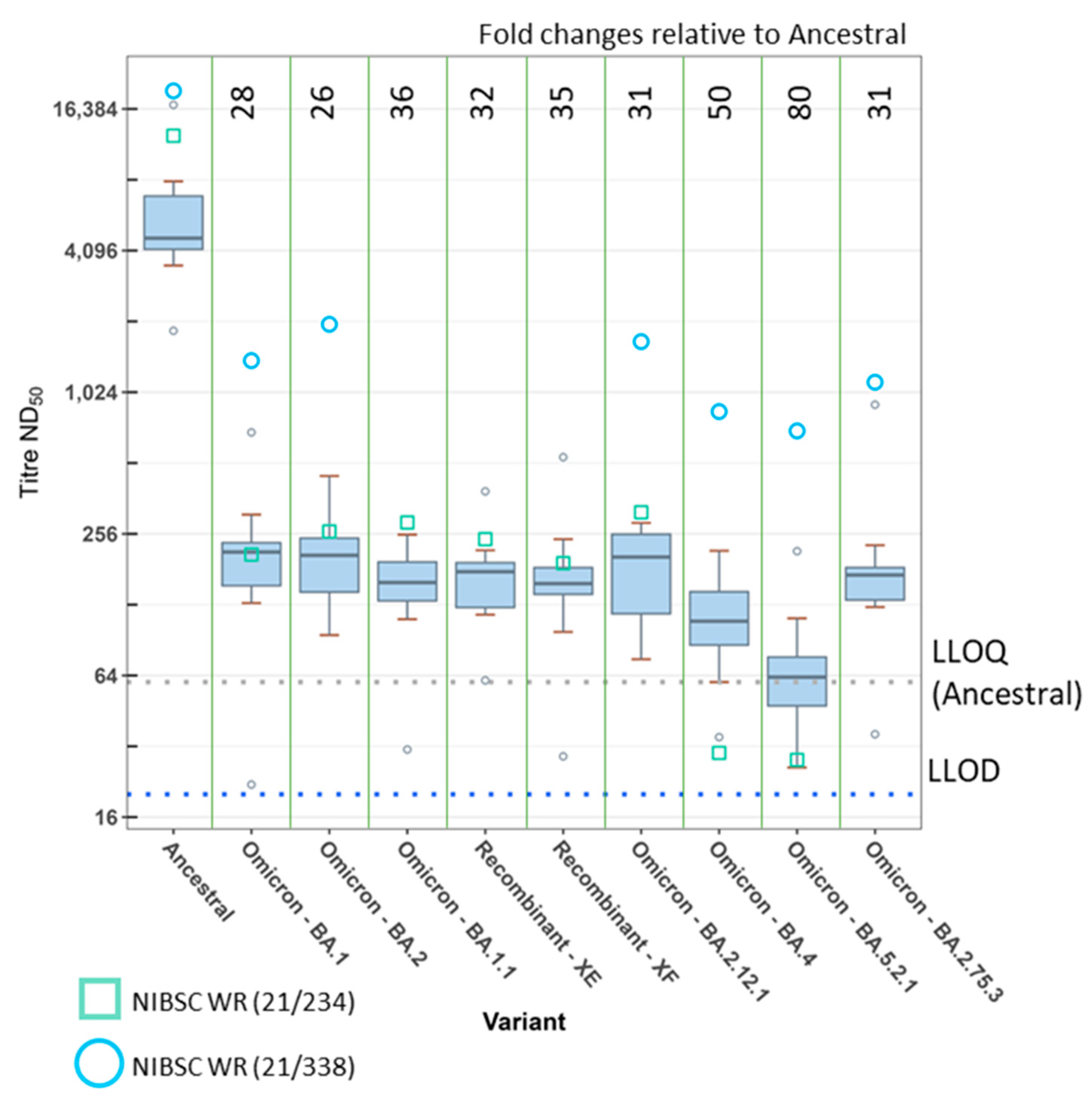
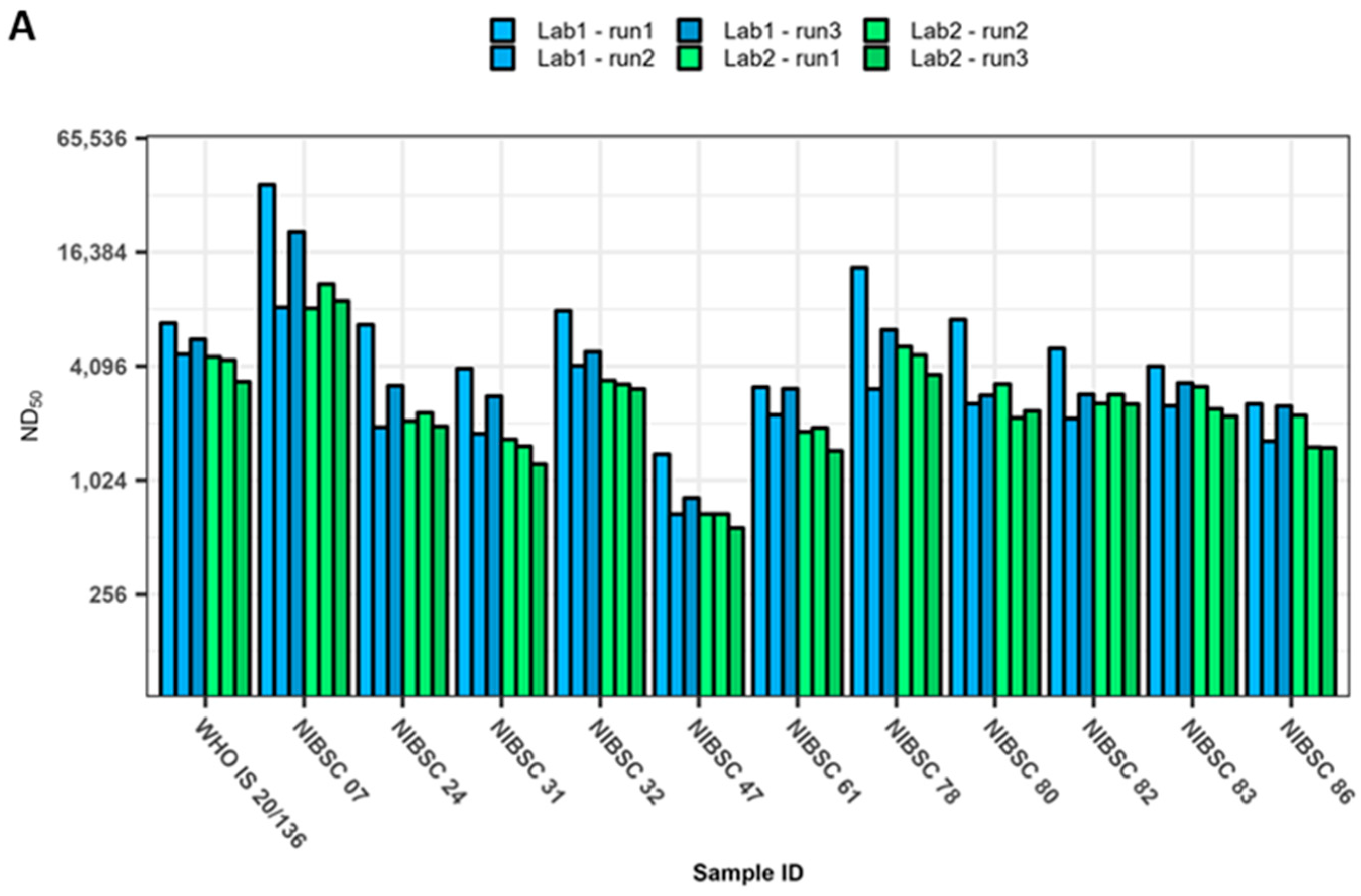
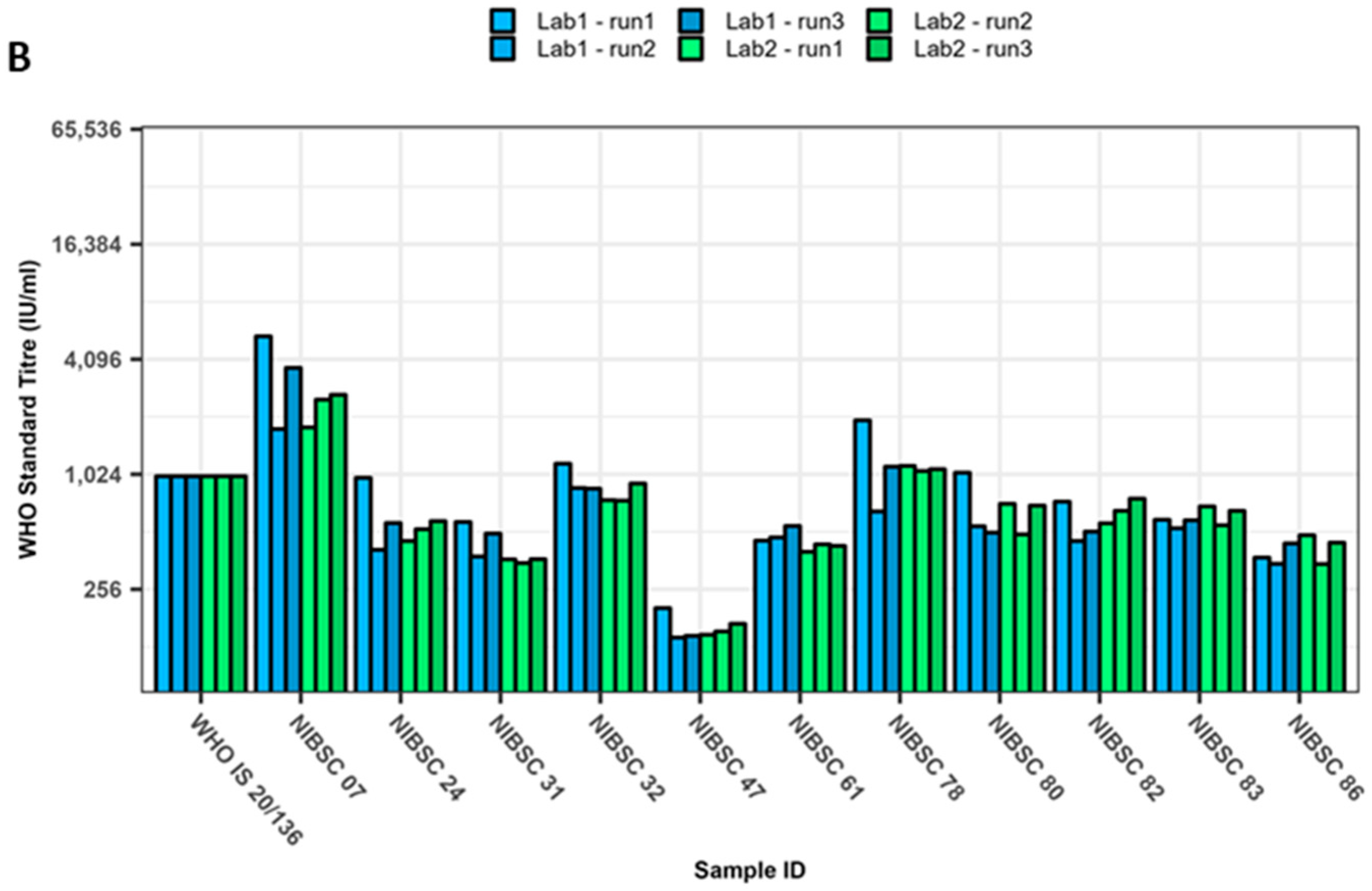
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Peiris, J.; Lai, S.; Poon, L.; Guan, Y.; Yam, L.; Lim, W.; Nicholls, J.; Yee, W.; Yan, W.; Cheung, M.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef] [Green Version]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Del Rio, C.; Malani, P.N. 2019 novel coronavirus—Important information for clinicians. JAMA 2020, 323, 1039. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cucinotta, D.; Vanelli, M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar]
- WHO (World Health Organization). Coronavirus Disease 2019 (COVID-19) Situation Report—121. 2022. Available online: https://apps.who.int/iris/handle/10665/332156 (accessed on 5 December 2022).
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alharbi, N.K.; Padron-Regalado, E.; Thompson, C.P.; Kupke, A.; Wells, D.; Sloan, M.A.; Grehan, K.; Temperton, N.; Lambe, T.; Warimwe, G.; et al. ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine 2017, 35, 3780–3788. [Google Scholar] [CrossRef]
- Pang, N.Y.; Pang, A.S.; Chow, V.T.; Wang, D.Y. Understanding neutralising antibodies against SARS-CoV-2 and their implications in clinical practice. Mil. Med. Res. 2021, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- FDA. Coronavirus (COVID-19) Update: FDA Authorizes Moderna, Pfizer-BioNTech Bivalent COVID-19 Vaccines for Use as a Booster Dose. 2022. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-moderna-pfizer-biontech-bivalent-covid-19-vaccines-use (accessed on 5 December 2022).
- MHRA. First Bivalent COVID-19 Booster Vaccine Approved by UK Medicines Regulator. 2022. Available online: https://www.gov.uk/government/news/first-bivalent-covid-19-booster-vaccine-approved-by-uk-medicines-regulator (accessed on 5 December 2022).
- Rambaut, A.; Loman, N.; Pybus, O.; Barclay, W.; Barrett, J.; Carabelli, A.; Connor, T.; Peacock, T.; Robertston, D.L.; Volz, E. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations. 2020. Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (accessed on 24 February 2023).
- PHE. Public Health England Investigation of Novel SARS-COV-2 Variant 202012/01: Technical Briefing 1. 2020. Available online: https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 (accessed on 24 February 2023).
- WHO. Tracking SARS-CoV-2 Variants. 2022. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 13 June 2022).
- Weisblum, Y.; Schmidt, F.; Zhang, F.; Dasilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021, 433, 166725. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, P.G.; Purkayastha, A.; Quintos, J.; Tam, C.; Lathrop, L.; Tam, K.; Ruiz, M.; Hollis, R.P.; Gomperts, B.N.; Kohn, D.B. Improved SARS-CoV-2 spike glycoproteins for pseudotyping lentiviral vectors. Front. Virol. 2021, 1, 793320. [Google Scholar] [CrossRef]
- Park, W.B.; Kwon, N.-J.; Choi, S.-J.; Kang, C.K.; Choe, P.G.; Kim, J.; Yun, J.; Lee, G.-W.; Seong, M.-W.; Kim, N.J.; et al. Virus isolation from the first patient with SARS-CoV-2 in Korea. J. Korean Med. Sci. 2020, 35, 1142676. [Google Scholar]
- Cureton, D.K.; Massol, R.H.; Whelan, S.P.J.; Kirchhausen, T. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 2010, 6, e1001127. [Google Scholar] [CrossRef] [PubMed]
- Kumru, O.S.; Wang, Y.; Gombotz, C.W.R.; Kelley-Clarke, B.; Cieplak, W.; Kim, T.; Joshi, S.B.; Volkin, D. Physical characterization and stabilization of a lentiviral vector against adsorption and freeze-thaw. J. Pharm. Sci. 2018, 107, 2764–2774. [Google Scholar] [CrossRef]
- Bewley, K.R.; Coombes, N.S.; Gagnon, L.; McInroy, L.; Baker, N.; Shaik, I.; St-Jean, J.R.; St-Amant, N.; Buttigieg, K.R.; Humphries, H.E.; et al. Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat. Protoc. 2021, 16, 3114–3140. [Google Scholar] [CrossRef]
- Ryan, K.A.; Watson, R.; Bewley, K.; Burton, C.; Carnell, O.; Cavell, B.; Challis, A.; Coombes, N.; Emery, K.; Fell, R.; et al. Convalescence from Prototype SARS-CoV-2 Protects Syrian Hamsters from Disease Caused by the Omicron Variant; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2021. [Google Scholar]
- Harris, R.J.; Whitaker, H.; Andrews, N.; Aiano, F.; Amin-Chowdhury, Z.; Flood, J.; Borrow, R.; Linley, E.; Ahmad, S.; Stapley, L.; et al. Serological surveillance of SARS-CoV-2: Six-month trends and antibody response in a cohort of public health workers. J. Infect. 2021, 82, 162–169. [Google Scholar] [CrossRef]
- NIBSC. Working Standard Working Reagent for Anti-SARS-CoV-2 Immunoglobulin NIBSC Code: 21/234. 2022. Available online: https://www.nibsc.org/documents/ifu/21-234.pdf (accessed on 12 December 2022).
- NIBSC. WHO International Standard First WHO International Standard for Anti-SARS-CoV-2 Immunoglobulin (Human) NIBSC Code: 20/136. 2020. Available online: https://www.nibsc.org/documents/ifu/20-136.pdf (accessed on 12 December 2022).
- NIBSC. WHO International Standard 1st International Standard for Antibodies to SARS-CoV-2 Variants of Concern NIBSC Code: 21/338. 2022. Available online: https://www.nibsc.org/documents/ifu/21-338.pdf (accessed on 12 December 2022).
- Rihn, S.J.; Merits, A.; Bakshi, S.; Turnbull, M.L.; Wickenhagen, A.; Alexander, A.J.T.; Baillie, C.; Brennan, B.; Brown, F.; Brunker, K.; et al. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biol. 2021, 19, e3001091. [Google Scholar] [CrossRef]
- Caly, L.; Druce, J.; Roberts, J.; Bond, K.; Tran, T.; Kostecki, R.; Yoga, Y.; Naughton, W.; Taiaroa, G.; Seemann, T.; et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 2020, 212, 459–462. [Google Scholar] [CrossRef] [Green Version]
- Funnell, S.G.P.; Afrough, B.; Baczenas, J.; Berry, N.; Bewley, K.R.; Bradford, R.; Florence, C.; Duff, Y.L.; Lewis, M.; Moriarty, R.V.; et al. A cautionary perspective regarding the isolation and serial propagation of SARS-CoV-2 in Vero cells. NPJ Vaccines 2021, 6, 83. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, K.; Xu, Y.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vaughan, A.; Morgan, M.; Bright, N.; Kavanagh, J.; et al. Metagenomic nanopore sequencing of influenza virus direct from clinical respiratory samples. J. Clin. Microbiol. 2019, 58, e00963-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. Aligning sequence reads, clone sequences and assemblycontigs with BWA-MEM. arXiv 2013, arXiv:1303.3997v2 [q-bio.GN]. [Google Scholar]
- Danecek, P.; Bonfield, J.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.; Whitwham, A.; Keane, T.; McCarthy, S.; Davies, R.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- R. R Core Team: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2014. Available online: http://www.R-project.org/ (accessed on 24 February 2023).
- Canchola, J.A. Correct use of percent coefficient of variation (%CV) formula for log-transformed data. MOJ Proteom. Bioinf. 2017, 6, 316–317. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: Their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev. Med. Virol. 2022, 32, e2270. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Gao, K.; Wei, G.W. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics 2021, 113, 2158–2170. [Google Scholar] [CrossRef] [PubMed]
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020, 10, 14991. [Google Scholar] [CrossRef]
- Brun, J.; Vasiljevic, S.; Gangadharan, B.; Hensen, M.; Chandran, A.V.; Hill, M.L.; Kiappes, J.L.; Dwek, R.A.; Alonzi, D.S.; Struwe, W.B.; et al. Assessing antigen structural integrity through glycosylation analysis of the SARS-CoV-2 viral spike. ACS Cent. Sci. 2021, 7, 586–593. [Google Scholar] [CrossRef]
- Kwilas, S.; Liesman, R.M.; Zhang, L.; Walsh, E.; Pickles, R.J.; Peeples, M.E. Respiratory syncytial virus grown in Vero cells contains a truncated attachment protein that alters its infectivity and dependence on glycosaminoglycans. J. Virol. 2009, 83, 10710–10718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Routhu, N.K.; Lehoux, S.D.; Rouse, E.A.; Bidokhti, M.R.M.; Giron, L.B.; Anzurez, A.; Reid, S.P.; Abdel-Mohsen, M.; Cummings, R.D.; Byrareddy, S.N. Glycosylation of Zika virus is important in host–virus interaction and pathogenic potential. Int. J. Mol. Sci. 2019, 20, 5206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letarov, A.V.; Babenko, V.; Kulikov, E. Free SARS-CoV-2 spike protein S1 particles may play a role in the pathogenesis of COVID-19 infection. Biochemistry 2021, 86, 257–261. [Google Scholar] [CrossRef]
- Girgis, S.; Xu, Z.; Oikonomopoulos, S.; Fedorova, A.D.; Tchesnokov, E.P.; Gordon, C.J.; Schmeing, T.M.; Gotte, M.; Sonenberg, N.; Baranov, P.V.; et al. Evolution of naturally arising SARS-CoV-2 defective interfering particles. Commun. Biol. 2022, 5, 1140. [Google Scholar] [CrossRef]
- Dupont, L.; Snell, L.B.; Graham, C.; Seow, J.; Merrick, B.; Lechmere, T.; Maguire, T.J.A.; Hallett, S.R.; Pickering, S.; Charalampous, T.; et al. Neutralizing antibody activity in convalescent sera from infection in humans with SARS-CoV-2 and variants of concern. Nat. Microbiol. 2021, 6, 1433–1442. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484. [Google Scholar] [CrossRef]
- Wratil, P.R.; Stern, M.; Priller, A.; Willmann, A.; Almanzar, G.; Vogel, E.; Feuerherd, M.; Cheng, C.-C.; Yazici, S.; Christa, C.; et al. Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat. Med. 2022, 28, 496–503. [Google Scholar] [CrossRef]
- Supasa, P.; Zhou, D.; Dejnirattisai, W.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Nutalai, R.; Tuekprakhon, A.; et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 2021, 184, 2201–2211. [Google Scholar] [CrossRef] [PubMed]
- Uriu, K.; Kimura, I.; Shirakawa, K.; Takaori-Kondo, A.; Nakada, T.-A.; Kaneda, A.; Nakagawa, S.; Sato, K. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N. Engl. J. Med. 2021, 385, 2397–2399. [Google Scholar] [CrossRef]
- Bekliz, M.; Adea, K.; Vetter, P.; Eberhardt, C.S.; Hosszu-Fellous, K.; Vu, D.L.; Puhach, O.; Essaidi-Laziosi, M.; Waldvogel-Abramowski, S.; Stephan, C.; et al. Neutralization capacity of antibodies elicited through homologous or heterologous infection or vaccination against SARS-CoV-2 VOCs. Nat. Commun. 2022, 13, 3840. [Google Scholar] [CrossRef]
- Singanallur, N.B.; van Vuren, P.J.; McAuley, A.; Bruce, M.P.; Kuiper, M.J.; Gwini, S.M.; Riddell, S.; Goldie, S.; Drew, T.W.; Blasdell, K.R.; et al. At least three doses of leading vaccines essential for neutralisation of SARS-CoV-2 omicron variant. Front. Immunol. 2022, 13, 883612. [Google Scholar] [CrossRef]
- Tan, C.W.; Lim, B.L.; Young, B.E.; Yeoh, A.Y.; Yung, C.F.; Yap, W.C.; Althaus, T.; Chia, W.N.; Zhu, F.; Lye, D.C.; et al. Comparative neutralisation profile of SARS-CoV-2 omicron subvariants BA.2.75 and BA.5. Lancet Microbe 2022, 3, e898. [Google Scholar] [CrossRef] [PubMed]
- UKHSA. SARS-CoV-2 Variants of Concern and Variants under Investigation in England: Technical Briefing 35. 2022. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1050999/Technical-Briefing-35-28January2022.pdf (accessed on 24 February 2023).
- Althobaity, Y.; Tildesley, M.J. Modelling the impact of non-pharmaceutical interventions on the spread of COVID-19 in Saudi Arabia. Sci. Rep. 2023, 13, 843. [Google Scholar] [CrossRef]
- UKHSA. Press Release: JCVI Advises an Autumn COVID-19 Vaccine Booster. 2023. Available online: https://www.gov.uk/government/news/jcvi-advises-an-autumn-covid-19-vaccine-booster (accessed on 22 February 2023).
- Tada, T.; Zhou, H.; Dcosta, B.; Samanovic, M.; Cornelius, A.; Herati, R.; Mulligan, M.; Landau, N. Neutralization of Mu and C.1.2 SARS-CoV-2 variants by vaccine-elicited antibodies in individuals with and without previous history of infection. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lassauniere, R.; Polacek, C.; Fonager, J.; Bennedbaek, M.; Boding, L.; Rasmussen, M.; Fomsgaard, A. Neutralisation of the SARS-CoV-2 Delta variant sub-lineages AY.4.2 and B.1.617.2 with the mutation E484K by Comirnaty (BNT162b2 mRNA) vaccine-elicited sera, Denmark, 1 to 26 November 2021. Euro Surveill. 2021, 26, 2101059. [Google Scholar] [CrossRef]
- Yadav, P.D.; Sahay, R.; Sapkal, G.; Nyayanit, D.; Shete, A.; Deshpande, G.; Patil, D.; Gupta, N.; Kumar, S.; Abraham, P.; et al. Comparable neutralization of SARS-CoV-2 Delta AY.1 and Delta with individuals sera vaccinated with BBV152. J. Travel Med. 2021, 28, taab154. [Google Scholar] [CrossRef]
- Carreño, J.M.; Alshammary, H.; Singh, G.; Raskin, A.; Amanat, F.; Amoako, A.; Gonzalez-Reiche, A.S.; Van De Guchte, A.; Awawda, M.; Banu, R.; et al. Reduced Neutralizing Activity of Post-SARS-CoV-2 Vaccination Serum against Variants B.1.617.2, B.1.351, B.1.1.7+E484K and a Sub-Variant of C.37; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2021. [Google Scholar]
- Ren, W.; Ju, X.; Gong, M.; Lan, J.; Yu, Y.; Long, Q.; Kenney, D.J.; O’Connell, A.K.; Zhang, Y.; Zhong, J.; et al. Characterization of SARS-CoV-2 variants B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.618 by cell entry and immune evasion. mBio 2022, 13, e0009922. [Google Scholar] [CrossRef] [PubMed]
- Kuzmina, A.; Wattad, S.; Khalaila, Y.; Ottolenghi, A.; Rosental, B.; Engel, S.; Rosenberg, E.; Taube, R. SARS CoV-2 Delta variant exhibits enhanced infectivity and a minor decrease in neutralization sensitivity to convalescent or post-vaccination sera. iScience 2021, 24, 103467. [Google Scholar] [CrossRef]
- Van Gils, M.J.; Lavell, A.; Van Der Straten, K.; Appelman, B.; Bontjer, I.; Poniman, M.; Burger, J.A.; Oomen, M.; Bouhuijs, J.H.; Van Vught, L.A.; et al. Antibody responses against SARS-CoV-2 variants induced by four different SARS-CoV-2 vaccines in health care workers in the Netherlands: A prospective cohort study. PLoS Med. 2022, 19, e1003991. [Google Scholar] [CrossRef]
- Mykytyn, A.Z.; Rissmann, M.; Kok, A.; Rosu, M.E.; Schipper, D.; Breugem, T.I.; Van Den Doel, P.B.; Chandler, F.; Bestebroer, T.; De Wit, M.; et al. Omicron BA.1 and BA.2 are Antigenically Distinct SARS-CoV-2 Variants; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2022. [Google Scholar]
- TGHN. The Global Health Network—Covax Epidemic Preparedness Innovations—Agility. 2023. Available online: https://epi.tghn.org/covax-overview/enabling-sciences/agility_epi/ (accessed on 13 January 2023).


| WHO Designation | Pangolin Lineage | GISAID Clade/Lineage | Nextstrain Clade | Date Assigned | GISAID ID of Isolation Swab (Where Known) and/or Source |
|---|---|---|---|---|---|
| Ancestral virus | B | O | 19A | Jan 2020 | EPI_ISL_406844 [31] |
| Alpha | B.1.1.7 | GRY | 20I (V1) | Dec 2020 | EPI_ISL_683466 |
| Alpha + E484K | B.1.1.7 | GRY | 20I (V1) | Feb 2021 | Gavin Screaton (University of Oxford) |
| Beta | B.1.351 | GH/501Y.V2 | 20H (V2) | Dec 2020 | EPI_ISL_770441 |
| Gamma * | P.1 | GR/5017.V3 | 20J (V3) | Jan 2021 | EPI_ISL_2080492 |
| Gamma–FioCruz * | P.1 | GR/5017.V3 | 20J (V3) | Jan 2021 | FioCruz, Brazil |
| Delta | B.1.617.2 | G/452R.V3 | 21A | May 2021 | EPI_ISL_2742236 |
| Delta | AY.1 | GK | 21A | May 2021 | EPI_ISL_2742878 |
| Delta | AY.4.2 | GK | 21A | May 2021 | EPI_ISL_4306633 |
| Lambda | C.37 | GR/452Q.V1 | 21G | June 2021 | BEI (NR-55654) |
| Kappa | B.1.617.1 | G/452R.V3 | 21B | Apr 2021 | EPI_ISL_2742167 |
| Mu | B.1.621 | GH | 21H | Aug 2021 | Not available |
| Zeta–FioCruz * | P.2 | GR/484K.V2 | 20B/S.484K | Mar 2021 | FioCruz, Brazil |
| Zeta–BEI * | P.2 | GR/484K.V2 | 20B/S.484K | Mar 2021 | BEI (NR-55439) |
| Recombinant–XE | XE | GRA | Recombinant | Mar 2022 | EPI_ISL_11586931 |
| Recombinant–XF | XF | GRA | Recombinant | Mar 2022 | EPI_ISL_10458256 |
| Omicron | BA.1 | GRA | 21K | Dec 2021 | EPI_ISL_7400555 |
| Omicron | BA.1.1 | GRA | 21K | Jan 2022 | EPI_ISL_8165999 |
| Omicron | BA.2 | GRA | 21L | Dec 2021 | Not available |
| Omicron | BA.2.12.1 | GRA | 22C | Apr 2022 | Gavin Screaton (University of Oxford) |
| Omicron | BA.2.75.3 | GRA | 22D | Jun 2022 | EPI_ISL_13882158 |
| Omicron | BA.4 | GRA | 22A | Apr 2022 | EPI_ISL_13157810 |
| Omicron | BA.5.2.1 | GRA | 22B | Apr 2022 | EPI_ISL_12810908 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coombes, N.S.; Bewley, K.R.; Le Duff, Y.; Hurley, M.; Smith, L.J.; Weldon, T.M.; Osman, K.; Pullan, S.; Berry, N.; Hallis, B.; et al. Assessment of the Biological Impact of SARS-CoV-2 Genetic Variation Using an Authentic Virus Neutralisation Assay with Convalescent Plasma, Vaccinee Sera, and Standard Reagents. Viruses 2023, 15, 633. https://doi.org/10.3390/v15030633
Coombes NS, Bewley KR, Le Duff Y, Hurley M, Smith LJ, Weldon TM, Osman K, Pullan S, Berry N, Hallis B, et al. Assessment of the Biological Impact of SARS-CoV-2 Genetic Variation Using an Authentic Virus Neutralisation Assay with Convalescent Plasma, Vaccinee Sera, and Standard Reagents. Viruses. 2023; 15(3):633. https://doi.org/10.3390/v15030633
Chicago/Turabian StyleCoombes, Naomi S., Kevin R. Bewley, Yann Le Duff, Matthew Hurley, Lauren J. Smith, Thomas M. Weldon, Karen Osman, Steven Pullan, Neil Berry, Bassam Hallis, and et al. 2023. "Assessment of the Biological Impact of SARS-CoV-2 Genetic Variation Using an Authentic Virus Neutralisation Assay with Convalescent Plasma, Vaccinee Sera, and Standard Reagents" Viruses 15, no. 3: 633. https://doi.org/10.3390/v15030633
APA StyleCoombes, N. S., Bewley, K. R., Le Duff, Y., Hurley, M., Smith, L. J., Weldon, T. M., Osman, K., Pullan, S., Berry, N., Hallis, B., Charlton, S., Hall, Y., & Funnell, S. G. P. (2023). Assessment of the Biological Impact of SARS-CoV-2 Genetic Variation Using an Authentic Virus Neutralisation Assay with Convalescent Plasma, Vaccinee Sera, and Standard Reagents. Viruses, 15(3), 633. https://doi.org/10.3390/v15030633






