Detecting the Neuraminidase R294K Mutation in Avian Influenza A (H7N9) Virus Using Reverse Transcription Droplet Digital PCR Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. R294K Primer Design
2.3. Plasmid Construction and Nucleic Acid Extraction
2.4. Establishing the RT-dd PCR to Detect the R294K Mutation in H7N9 Virus
2.4.1. Droplet Digital PCR Detection of Plasmids and H7N9 Virus RNA
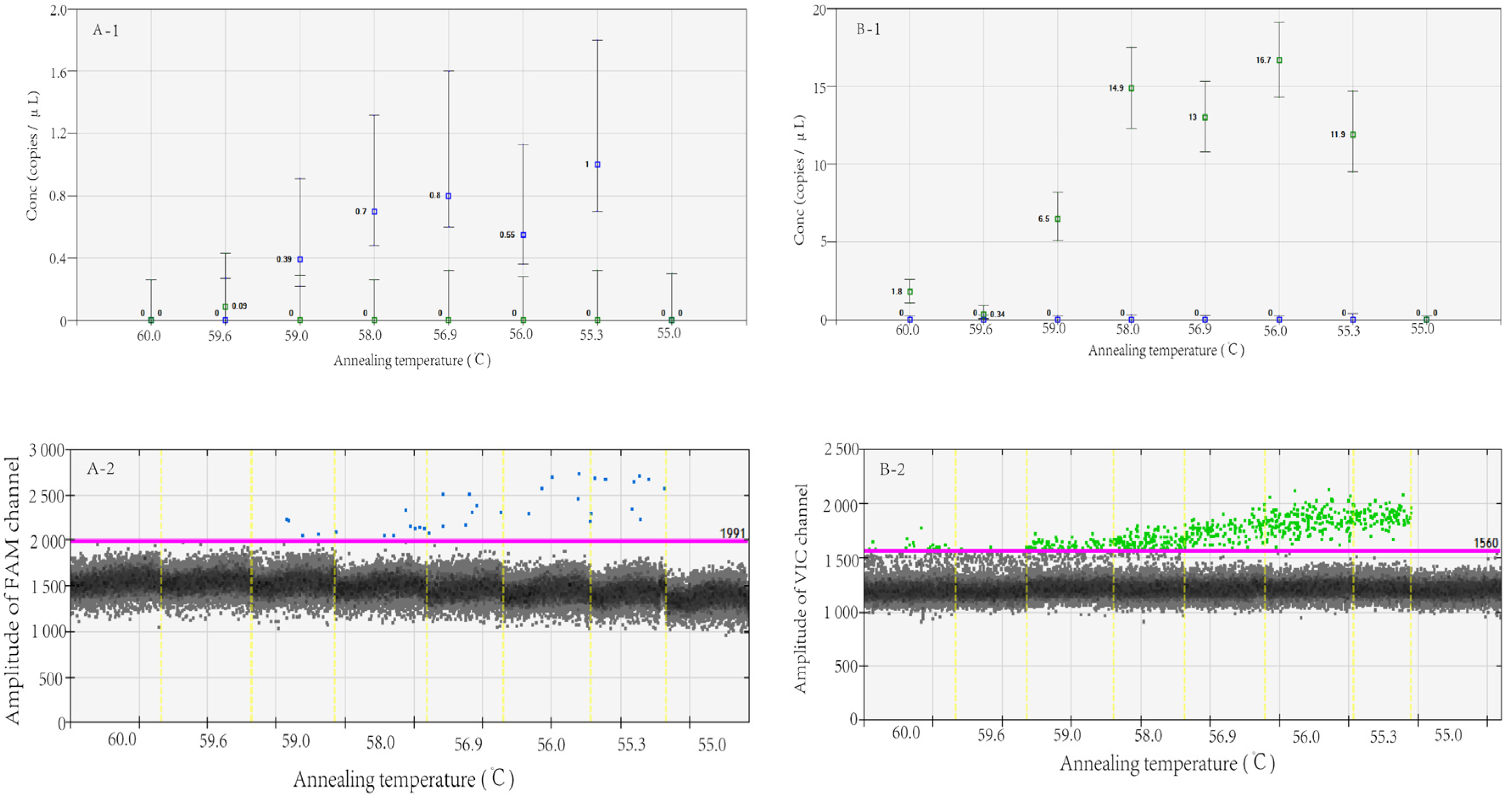
2.4.2. Determining the Optimal Annealing Temperature
2.4.3. Evaluating the Repeatability, Sensitivity, and Specificity
2.4.4. Evaluating the Detection Limit of R294K Mutation
2.5. Application of RT-dd PCR to Detect H7N9 Virus
2.6. R294K Mutation in Genotypic and Phenotypic Analyses
2.7. Statistical and Analysis
3. Results
3.1. Determining the Optimal Annealing Temperature
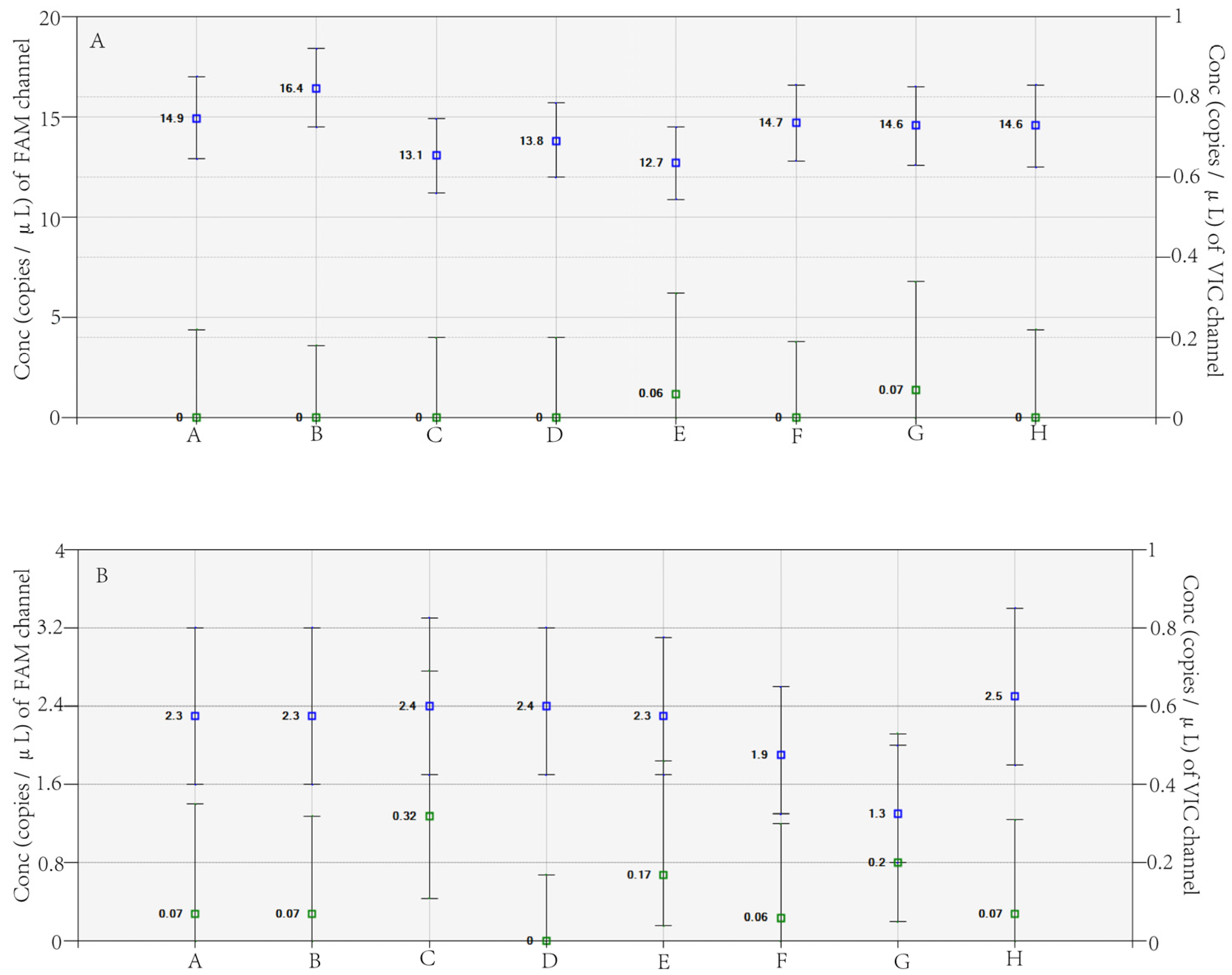
3.2. Evaluating Repeatability, Sensitivity, and Specificity
3.3. Evaluating the Detection Limit of the R294K Mutation
3.4. Using RT-ddPCR to Detect R294K in H7N9 Virus Clinical Samples
3.5. R294K Mutation in the Genotypic and Phenotypic Analyses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, R.; Cao, B.; Hu, Y.; Feng, Z.; Wang, D.; Hu, W.; Chen, J.; Jie, Z.; Qiu, H.; Xu, K.; et al. Human Infection with a Novel Avian-Origin Influenza A (H7N9) Virus. N. Engl. J. Med. 2013, 368, 1888–1897. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, H.; Wu, P.; Uyeki, T.M.; Feng, L.; Lai, S.; Wang, L.; Huo, X.; Xu, K.; Chen, E.; et al. Epidemiology of Avian Influenza A H7N9 Virus in Human Beings across Five Epidemics in Mainland China, 2013–17: An Epidemiological Study of Laboratory-Confirmed Case Series. Lancet Infect. Dis. 2017, 17, 822–832. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, L.; Li, L. Research Progress on Human Infection with Avian Influenza H7N9. Front. Med. 2020, 14, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Bi, Y.; Vavricka, C.J.; Sun, X.; Zhang, Y.; Gao, F.; Zhao, M.; Xiao, H.; Qin, C.; He, J.; et al. Characterization of Two Distinct Neuraminidases from Avian-Origin Human-Infecting H7N9 Influenza Viruses. Cell Res. 2013, 23, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Interim Guidance on the Use of Antiviral Agents for Treatment of Human Infections with Avian Influenza A (H7N9). Available online: https://www.cdc.gov/flu/avianflu/h7n9-antiviral-treatment.htm (accessed on 3 April 2023).
- Hu, Y.; Lu, S.; Song, Z.; Wang, W.; Hao, P.; Li, J.; Zhang, X.; Yen, H.-L.; Shi, B.; Li, T.; et al. Association between Adverse Clinical Outcome in Human Disease Caused by Novel Influenza A H7N9 Virus and Sustained Viral Shedding and Emergence of Antiviral Resistance. Lancet 2013, 381, 2273–2279. [Google Scholar] [CrossRef] [PubMed]
- Marjuki, H.; Mishin, V.P.; Chesnokov, A.P.; De La Cruz, J.A.; Davis, C.T.; Villanueva, J.M.; Fry, A.M.; Gubareva, L.V. Neuraminidase Mutations Conferring Resistance to Oseltamivir in Influenza A(H7N9) Viruses. J. Virol. 2015, 89, 5419–5426. [Google Scholar] [CrossRef]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing–Based Oncology Panels. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Bai, R.; Zhao, Z.; Tao, L.; Ma, M.; Ji, Z.; Jian, M.; Ding, Z.; Dai, X.; Bao, F.; et al. Application of Droplet Digital PCR to Detect the Pathogens of Infectious Diseases. Biosci. Rep. 2018, 38, BSR20181170. [Google Scholar] [CrossRef]
- Mazaika, E.; Homsy, J. Digital Droplet PCR: CNV Analysis and Other Applications. Curr. Protoc. Hum. Genet. 2014, 82, 7–24. [Google Scholar] [CrossRef]
- Huang, W.; Li, X.; Cheng, Y.; Tan, M.; Guo, J.; Wei, H.; Zhao, X.; Lan, Y.; Xiao, N.; Wang, Z.; et al. Characteristics of Oseltamivir-Resistant Influenza A (H1N1) Pdm09 Virus during the 2013–2014 Influenza Season in Mainland China. Virol. J. 2015, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, T.; Chiapponi, C.; Baioni, L.; Cinotti, S.; Ferrari, M. Protein Mutations Following Adaptation of Avian Influenza Viruses in Different Biological Systems. Res. Vet. Sci. 2015, 103, 176–178. [Google Scholar] [CrossRef]
- Pinheiro-de-Oliveira, T.F.; Fonseca, A.A.; Camargos, M.F.; Laguardia-Nascimento, M.; de Oliveira, A.M.; Cottorello, A.C.P.; Goes-Neto, A.; Barbosa-Stancioli, E.F. Development of a Droplet Digital RT-PCR for the Quantification of Foot-and-Mouth Virus RNA. J. Virol. Methods 2018, 259, 129–134. [Google Scholar] [CrossRef]
- Mairiang, D.; Songjaeng, A.; Hansuealueang, P.; Malila, Y.; Lertsethtakarn, P.; Silapong, S.; Poolpanichupatam, Y.; Klungthong, C.; Chin-Inmanu, K.; Thiemmeca, S.; et al. Application of One-Step Reverse Transcription Droplet Digital PCR for Dengue Virus Detection and Quantification in Clinical Specimens. Diagnostics 2021, 11, 639. [Google Scholar] [CrossRef]
- Sun, T.; Jiang, Y.; Gong, P.; Yu, Y. Detection of Microamounts of Novel Coronavirus Residues in Environment by Digital PCR. Chin. Sci. Bull. 2021, 66, 1653–1662. [Google Scholar] [CrossRef]
- Yan, Y.; Jia, X.; Wang, H.; Fu, X.; Ji, J.; He, P.; Chen, L.; Luo, J.; Chen, Z. Dynamic Quantification of Avian Influenza H7N9(A) Virus in a Human Infection during Clinical Treatment Using Droplet Digital PCR. J. Virol. Methods 2016, 234, 22–27. [Google Scholar] [CrossRef]
- Whale, A.S.; Bushell, C.A.; Grant, P.R.; Cowen, S.; Gutierrez-Aguirre, I.; O’Sullivan, D.M.; Žel, J.; Milavec, M.; Foy, C.A.; Nastouli, E.; et al. Detection of Rare Drug Resistance Mutations by Digital PCR in a Human Influenza A Virus Model System and Clinical Samples. J. Clin. Microbiol. 2016, 54, 392–400. [Google Scholar] [CrossRef]
- Rački, N.; Morisset, D.; Gutierrez-Aguirre, I.; Ravnikar, M. One-Step RT-Droplet Digital PCR: A Breakthrough in the Quantification of Waterborne RNA Viruses. Anal. Bioanal. Chem. 2014, 406, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Carbonneau, J.; Shelton, D.N.; Boivin, G. Optimization of Droplet Digital PCR from RNA and DNA Extracts with Direct Comparison to RT-QPCR: Clinical Implications for Quantification of Oseltamivir-Resistant Subpopulations. J. Virol. Methods 2015, 224, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Yen, H.-L.; McKimm-Breschkin, J.L.; Choy, K.-T.; Wong, D.D.Y.; Cheung, P.P.H.; Zhou, J.; Ng, I.H.; Zhu, H.; Webby, R.J.; Guan, Y.; et al. Resistance to Neuraminidase Inhibitors Conferred by an R292K Mutation in a Human Influenza Virus H7N9 Isolate Can Be Masked by a Mixed R/K Viral Population. mBio 2013, 4, e00396-13. [Google Scholar] [CrossRef] [PubMed]
- Pekin, D.; Skhiri, Y.; Baret, J.-C.; Le Corre, D.; Mazutis, L.; Ben Salem, C.; Millot, F.; El Harrak, A.; Hutchison, J.B.; Larson, J.W.; et al. Quantitative and Sensitive Detection of Rare Mutations Using Droplet-Based Microfluidics. Lab. Chip 2011, 11, 2156. [Google Scholar] [CrossRef] [PubMed]
- Longjam, N.; Deb, R.; Sarmah, A.K.; Tayo, T.; Awachat, V.B.; Saxena, V.K. A Brief Review on Diagnosis of Foot-and-Mouth Disease of Livestock: Conventional to Molecular Tools. Vet. Med. Int. 2011, 2011, 905768. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.; Patel, S.; Geyer, J.T.; Racchumi, J.; Chadburn, A.; Simonson, P.; Ouseph, M.M.; Inghirami, G.; Mencia-Trinchant, N.; Guzman, M.L.; et al. Comparison of Multiple Clinical Testing Modalities for Assessment of NPM1-Mutant AML. Front. Oncol. 2021, 11, 701318. [Google Scholar] [CrossRef]
- Kuang, Y.; Xu, P.; Wang, J.; Zheng, Y.; Sun, X.; Li, Z.; Gan, R.; Li, H.; Guo, Y.; Yao, F.; et al. Detecting ALK Rearrangement with RT-PCR: A Reliable Approach Compared with Next-Generation Sequencing in Patients with NSCLC. Mol. Diagn. Ther. 2021, 25, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, S.-M.; Yu, X.-H.; Tang, S.-L.; Tang, C.-K. Coronavirus Disease 2019 (COVID-19): Current Status and Future Perspectives. Int. J. Antimicrob. Agents 2020, 55, 105951. [Google Scholar] [CrossRef] [PubMed]
- Maggi, R.; Breitschwerdt, E.B.; Qurollo, B.; Miller, J.C. Development of a Multiplex Droplet Digital PCR Assay for the Detection of Babesia, Bartonella, and Borrelia Species. Pathogens 2021, 10, 1462. [Google Scholar] [CrossRef]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of Droplet Digital PCR and next Generation Sequencing for Characterizing DNA Reference Material for KRAS Mutation Detection. Sci. Rep. 2018, 8, 9650. [Google Scholar] [CrossRef] [PubMed]

| Primers/Probes | Sequences 5′-3′ | Position |
|---|---|---|
| R294K-F | TGACTGGAACTGCTAAGCAYATTGA | 768–792 |
| R294K-R | TGTCATTGCTACTGGRTCTATCTGA | 874–898 |
| 294K-Pb1 | FAM-CACATGCAAGGACA-MGB | 832–845 |
| R294-Pb2 | VIC-CACATGCAGGGACA-MGB | 832–845 |
| Type | Detection Channel | Number | Results (Copies/μL) | Mean (Copies/μL) | SD | CV (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | VIC | R1 | 8000 | 8500 | 7860 | 7930 | 7980 | 8400 | 8400 | 8100 | 8146.25 | 248.94 | 3.06 |
| R2 | 639 | 660 | 681 | 684 | 636 | 624 | 667 | 680 | 658.88 | 23.20 | 3.52 | ||
| R3 | 47.3 | 47.2 | 47.2 | 46.7 | 48.5 | 48.3 | 47.8 | 47.3 | 47.54 | 0.61 | 1.29 | ||
| R4 | 4.8 | 4.3 | 5.7 | 5.8 | 5.8 | 5.8 | 5.0 | 5.2 | 5.30 | 0.57 | 10.72 | ||
| MT | FAM | K1 | 539 | 520 | 540 | 546 | 510 | 523 | 544 | 532 | 532 | 12.88 | 2.4 |
| K2 | 48.1 | 49.8 | 51.1 | 53.0 | 52 | 49.3 | 42.3 | 46 | 48.95 | 3.48 | 7.1 | ||
| K3 | 10.8 | 10.1 | 11.0 | 10.3 | 11.3 | 11.9 | 10.8 | 10.8 | 10.87 | 0.66 | 6.07 | ||
| K4 | 1.0 | 1.0 | 1.1 | 0.96 | 0.93 | 1.1 | 1.8 | 1.1 | 1.12 | 0.28 | 25.03 | ||
| Number | Detection Channel | Results (Copies/μL) | Mean (Copies/μL) | Mutation Abundance (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | VIC | 8700 | 8160 | 9360 | 8320 | 8860 | 8680 | 9180 | 9200 | 8807.6 | 1.52 |
| FAM | 134 | 144 | 96 | 140 | 140 | 138 | 158 | 134 | 135.6 | ||
| M2 | VIC | 9160 | 8600 | 9060 | 8740 | 8900 | 8840 | 9060 | 9340 | 8962.5 | 0.63 |
| FAM | 44 | 70 | 64 | 52 | 46 | 60 | 62 | 60 | 57.25 | ||
| M3 | VIC | 8980 | 9120 | 8780 | 8700 | 9500 | 9700 | 9560 | 9280 | 9202.5 | 0.29 |
| FAM | 28 | 24.8 | 30.2 | 15.2 | 26.4 | 26.8 | 22.2 | 38 | 26.45 | ||
| M4 | VIC | 10,580 | 10,460 | 11,120 | 10,200 | 13,280 | 10,140 | 10,260 | 10,820 | 10,857.5 | 0.09 |
| FAM | 10 | 8.6 | 9.6 | 10.4 | 6.8 | 12.4 | 8.4 | 12.4 | 9.825 | ||
| M5 | VIC | 10,580 | 10,920 | 11,140 | 11,420 | 11,020 | 11,180 | 10,780 | 11,420 | 11,057.5 | 0.05 |
| FAM | 2.8 | 7.2 | 3.6 | 2.6 | 10.2 | 5.4 | 8.6 | 7.8 | 6.025 | ||
| Strains | NGS | dd PCR | ||||||
|---|---|---|---|---|---|---|---|---|
| R294 | 294K | R294 | 294K | |||||
| A | G | G | A | A | G | |||
| A/Zhejiang/65-1/2017(H7N9 virus) | 0.06% | 99.94% | 0.06% | 99.94% | ||||
| A/Zhejiang/70-2/2017(H7N9 virus) | 8.65% | 91.35% | 4.92% | 95.08% | ||||
| A/Zhejiang/YCHH/2016(H7N9 virus) | 100% | 0 | 100% | 0 | ||||
| Strains | Oseltamivir | Zanamivir | ||
|---|---|---|---|---|
| IC50 (nM) | Inhibition (Multiple) | IC50 (nM) | Inhibition (Multiple) | |
| A/Zhejiang/65-1/2017(H7N9 virus) | 728.30 | >1000.00 | 18.44 | 19.83 |
| A/Zhejiang/70-2/2017(H7N9 virus) | >1000.00 | >1000.00 | 12.05 | 12.96 |
| A/Zhejiang/YCHH/2016(H7N9 virus) | 0.66 | 0.92 | 1.05 | 1.13 |
| GD003 R294 | 0.72 | 1.00 | 0.93 | 1.00 |
| GD003 R294K | >1000.00 | >1000.00 | 10.75 | 11.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, X.; Yan, H.; Su, L.; Sun, Y.; Wang, X.; Gong, L.; Chen, Y.; Li, Z.; Fang, Z.; Mao, H.; et al. Detecting the Neuraminidase R294K Mutation in Avian Influenza A (H7N9) Virus Using Reverse Transcription Droplet Digital PCR Method. Viruses 2023, 15, 983. https://doi.org/10.3390/v15040983
Lou X, Yan H, Su L, Sun Y, Wang X, Gong L, Chen Y, Li Z, Fang Z, Mao H, et al. Detecting the Neuraminidase R294K Mutation in Avian Influenza A (H7N9) Virus Using Reverse Transcription Droplet Digital PCR Method. Viruses. 2023; 15(4):983. https://doi.org/10.3390/v15040983
Chicago/Turabian StyleLou, Xiuyu, Hao Yan, Lingxuan Su, Yi Sun, Xinyin Wang, Liming Gong, Yin Chen, Zhen Li, Zhongbiao Fang, Haiyan Mao, and et al. 2023. "Detecting the Neuraminidase R294K Mutation in Avian Influenza A (H7N9) Virus Using Reverse Transcription Droplet Digital PCR Method" Viruses 15, no. 4: 983. https://doi.org/10.3390/v15040983
APA StyleLou, X., Yan, H., Su, L., Sun, Y., Wang, X., Gong, L., Chen, Y., Li, Z., Fang, Z., Mao, H., Chen, K., & Zhang, Y. (2023). Detecting the Neuraminidase R294K Mutation in Avian Influenza A (H7N9) Virus Using Reverse Transcription Droplet Digital PCR Method. Viruses, 15(4), 983. https://doi.org/10.3390/v15040983





