Molecular Characterization of Avian Rotaviruses F and G Detected in Brazilian Poultry Flocks
Abstract
1. Introduction
2. Materials and Methods
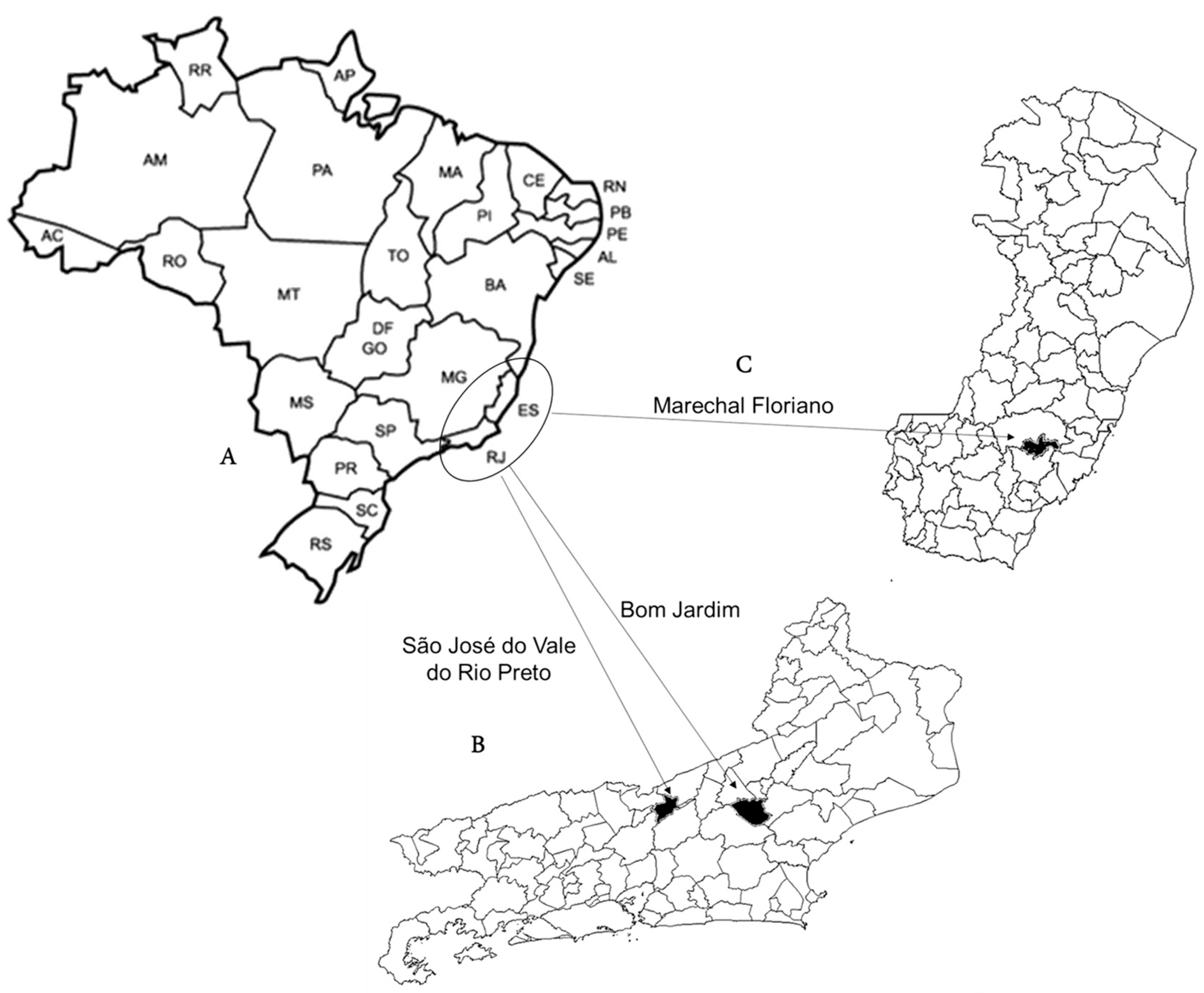
2.1. Samples
2.2. RNA Extraction, Amplification, and Sequencing
3. Results
3.1. Analysis of RVF Strains
3.1.1. Genomic Segment Encoding VP1
3.1.2. Genomic Segment Encoding VP2
3.1.3. Genomic Segment Encoding VP4
3.1.4. Genomic Segment Encoding VP6
3.1.5. Genomic Segment Encoding VP7
3.1.6. Genomic Segment Encoding NSP1
3.1.7. Genomic Segment Encoding NSP2
3.1.8. Genomic Segment Encoding NSP4
3.1.9. Genomic Segment Encoding NSP5
4. Analysis of RVG Strains
4.1. Genomic Segment Encoding VP6
4.2. Genomic Segment Encoding NSP2
4.3. Genomic Segment Encoding NSP5
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Primer | Primer Sequence 5′ → 3′ | Gene | Position † |
|---|---|---|---|
| RFVP1.F1 | ATTAATTGCGTACGATGGGGA | VP1 | 5–25 |
| RFVP1.F2 | GGATCCTGCAATATCTACATC | 429–449 | |
| RFVP1.F3 | GCAAATGTWTATTCATGGTC | 991–1010 | |
| RFVP1.F4 | TGCTTAAAGATGAYGTTATTA | 1145–1165 | |
| RFVP1.F5 | GGTAGAAGRGATGTACCAGGT | 1372–1392 | |
| RFVP1.R1 | TTAATYATTTTTGATGAATA | 835–854 | |
| RFVP1.R2 | ACTTCCAAATTTTATTTCC | 1242–1260 | |
| RFVP1.R3 | GCTAAATTTGCAATAGAATTA | 1807–1827 | |
| RFVP1.R4 | TAACRTTCATAAGTGCATAAG | 1985–2005 | |
| RFVP1.R5 | CCTGATATYTTAACTGATGTC | 2229–2249 | |
| RFVP1.R6 | GATARTAAATCATAAACATATG | 2984–3005 | |
| RFVP1.R7 | ATTATTGCAGCTTACTCTTGG | 3270–3290 | |
| RFVP2.F1 | GGCTATTATTGGCAGGATGT | VP2 | 1–20 |
| RFVP2.F2 | GCAGARATGAGRCATAGAGT | 764–783 | |
| RFVP2.F3 | GGGAAACATTAACGTTAACT | 1041–1060 | |
| RFVP2.F4 | GTAATGAATCAGCAATATGC | 1772–1791 | |
| RFVP2.R1 | CAGATTATAAGGAACATATC | 978–997 | |
| RFVP2.R2 | CTAACTATATTAGCTTCAGC | 788–807 | |
| RFVP2.R3 | AGAGCTAATACAAACATTCC | 1307–1326 | |
| RFVP2.R4 | GATGCATCAGGATATACAGC | 1862–1881 | |
| RFVP2.R5 | GGAGATAATACTCCTCCAGC | 2741–2760 | |
| RFVP4.F1 | GATGGCTTCTCGCTTTTGGG | VP4 | 11–30 |
| RFVP4.F2 | AAATGCGGTAGTTTTGGAATAG | 352–372 | |
| RFVP4.F3 | GGTTATCAATTTTCAACGTCT | 787–809 | |
| RFVP4.F4 | GTAGGAAGTATAACTCCATA | 1104–1123 | |
| RFVP4.F5 | ATCAAGRTGGCAGAAAARTT | 1500–1519 | |
| RFVP4.R1 | CATATACCATGTTTCTGAAT | 1010–1029 | |
| RFVP4.R2 | AATCCAGCTTGTGACACGAC | 690–709 | |
| RFVP4.R3 | AGGATAATATGGAGTTATACT | 1110–1130 | |
| RFVP4.R4 | TTGAAT TCCTGTCGTAATTGG | 1480–1500 | |
| RFVP4.R5 | TATGCTATATTAGCATCCTTA | 1980–2000 | |
| RFVP4.R6 | ATACTGTTTCTCGCTCAAAGT | 2250–2270 | |
| RFVP6.F1 | GGCTTATAAAAGTCAATCAG | VP6 | 1–20 |
| RFVP6.F2 | AAGTCAATCAGTCGCAATGG | 10–29 | |
| RFVP6.F3 | CGTACAGAACCAGGTCAAATGTG | 620–642 | |
| RFVP6.F4 | GCCAGTGCCAATATCTGATGC | 667–687 | |
| F5.RVF | ATGTGAAACTGAAATGTGTCTTGAATC | 310–336 | |
| F6.RVF | TACAAAAAGTTAGAGTTAGAACAGCTTA | 447–474 | |
| RFVP6.F7 | GGTCTACTTAATGATCAARTAC | 730–737 | |
| RFVP6.F8 | CAATACAAGTTGATACTGATGC | 690–710 | |
| RFVP6.R1 | CCTGCYACATCATCCATAGC | 539–558 | |
| R2.RVF | TTGATCATTAAGTAGACCAGAYGCATC | 707–733 | |
| RFVP6.R3 | AGCTGTTCTAACTCTAACTT | 453–472 | |
| RFVP6.R4 | CAGTGATACTATCGGAATAAACCA | 1241–1264 | |
| RFVP6.R5 | CACAGTTGCCCGGCCAAACG | 1268–1287 | |
| RFVP6.R6 | GCATATTATCTTGTCTAGAT | 1150–1169 | |
| RFVP7.F1 | CGAACAGCCTCCATCAGCTCGTGT | VP7 | 17–40 |
| RFVP7.F2 | CCTGTAATTCAGGATGTTTGCTG | 57–79 | |
| RFVP7.F2 | GTARCCTGTAATTCAGGATGTT | 53–74 | |
| RFVP7.F3 | GAAGTAAATGAGTTATTTCAAT | 355–376 | |
| RFVP7.F4 | TCGACTGATGTGAAAACATATG | 601–622 | |
| RFVP7.F4 | TCAACTGATGCTACTACATATG | 601–622 | |
| RFVP7.F5 | TATAGAATGAATATTACTGG | 673–692 | |
| RFVP7.R1 | GGTCATAATGTTGTTCGCAAC | 970–990 | |
| RFVP7.R2 | CAACGTTAATGATTATTTATC | 953–973 | |
| RFVP7.R3 | GAGGAATAATCTGGTCCAAC | 733–752 | |
| RFVP7.R4 | GATAAGTCATCTGGTCCATG | 412–431 | |
| RFVP7.R4 | GATAAATCACTCGGCCCAT | 413–431 | |
| RFNSP1.F1 | GTG TGC CGA TTC AGA GAT GG | NSP1 | 23–42 |
| RFNSP1.F2 | GGT ATG AGT GTA GTW CCA GC | 711–730 | |
| RFNSP1.F3 | CAA TTY CGT GAT TGG AA | 1176–1192 | |
| RFNSP1.R1 | CCG TGT GCG ATG CTG AAT CGG | 1745–1765 | |
| RFNSP1.R2 | GTA TAC TCA CAA TCT TAT CAT | 792–812 | |
| RFNSP1.R3 | ACT AAT CAR TCA CCT CTT AT | 1401–1420 | |
| RFNSP2.F1 | TTRTTTTTGATATAGAGCAGT | NSP2 | 10–30 |
| RFNSP2.F3 | GGTCCGGCAAAATCCTGCCTGCCG | 28–51 | |
| RFNSP2.F4 | GAATGATGCAGAAGATAGAC | 412–431 | |
| RFNSP2.F5 | GAGTATAAAATTACATTCAA | 608–627 | |
| RFNSP2.R1 | GGTCGTAGTATGATATAGAT | 1049–1068 | |
| RFNSP2.R2 | TTGCTTTAATAGCTTTRAGAG | 786–806 | |
| RFNSP2.R3 | GTACTTATTCATATATTCAATA | 529–550 | |
| RFNSP2.R4 | GTTTCAGGATTAAGTTATTGGG | 1019–1040 | |
| RFNSP4.F1 | CCTCATCTTAGTTATACGTAC | NSP4 | 11–31 |
| RFNSP4.F2 | CCCTCAGTGGTTTTGACAAG | 31–50 | |
| RFNSP4.R1 | GGTCATAACTCATCCGTTAG | 659–678 | |
| RFNSP4.R2 | GCTGATCGCTGCACTCTGG | 641–659 | |
| RFNSP4.R3 | GGTACAGTATGTTATACGC | 607–621 | |
| RFNSP4.R4 | ACCACTAGCATCTTCAGTTC | 406–425 | |
| RFNSP5.F1 | GAGCATGGATCTTGATATAGAC | NSP5 | 20–42 |
| RFNSP5.F2 | ATGGAATCTGTAAATAAT | 303–320 | |
| RFNSP5.F3 | TCAGTTAAGTCATCAAATTC | 357–376 | |
| RFNSP5.R1 | CATGATTATAGATCGGATATAAGC | 656–679 | |
| RFNSP5.R2 | GCTGAGTAATGCTTTGCACTGC | 411–432 | |
| RGVP6.F1 | GGAAAGAAATCTCCAACCTAG | VP6 | 1–21 |
| RGVP6.F2 | GCTAAGCTCGAACCTCAAATT | 459–479 | |
| 772F-RVG | CAGATATGGCGAGRGGTGAT | 772–791 | |
| RGVP6.R1 | AATTCTATTACTATATCACC | 786–805 | |
| RGVP6.R2 | AGCTTAGCTGTATCATATGC | 447–466 | |
| 1229R-RVG | AAACTCTCCTCCACAGCCGA | 1229–1248 | |
| RGNSP2.F1 | GAGTGCGTCGTGAGAAGGGAG | NSP2 | 20–40 |
| RGNSP2.F3 | GTTTTTGARGATGTATTTGAA | 380–400 | |
| RGNSP2.R1 | CAGCGCTCAATGAATGGATTT | 978–998 | |
| RGNSP2.R3 | GTACTGTTCTAACATGTCCGT | 750–770 | |
| RGNSP4.F3 | GATKCATTTAAGATTCTTT | NSP4 | 105–123 |
| RGNSP4.F4 | CARAATTCGAAGGAAATGGT | 384–403 | |
| RGNPS4.R3 | AACCCATCGGCTCTGCACC | 740–758 | |
| RGNSP4.R4 | TCCATYGATTCTTCAATTCC | 534–553 | |
| RGNSP5.F1 | GGAATATTAAAGTGTCGCTTGGTG | NSP5 | 1–24 |
| RGNSP5.F2 | GGTGGCTGGAAACACTGAGTGG | 21–42 | |
| RGNSP5.R1 | CCCAGAAATAATCAGTGGAGTC | 657–678 | |
| RGNSP5.R2 | TCAACTTCATCHGCCCAGTT | 315–334 | |
| RGNSP5.R3 | CTAATTTTCGAWATTTCAT | 445–463 |
| Viral Strain | Origin | Gene | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP1 | VP2 | VP3 | VP4 | VP6 | VP7 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | ||
| RVF | ||||||||||||
| Ch-03V0568 | Germany | JN596591 | JQ919995 | JQ919997 | HQ403603 | JQ919998 | JQ919999 | JQ920000 | JQ920002 | JQ920003 | ||
| Ch-361bR-k141_94516 | China | MZ218346 | MZ218350 | MZ218351 | ||||||||
| Ch-D62 | South Korea | KM254212 | KM254213 | KM254215 | KM254216 | KM254217 | KM254218 | KM254219 | KM254221 | KM254222 | ||
| Ch-D11 | South Korea | KM254224 | ||||||||||
| Ch-PB56-SII36 | Switzerland | OM469208 | ||||||||||
| Ch-ITA-956-1 | Italy | KT073228 | ||||||||||
| Ch-BR-15-4S-8 | Brazil | MG846380 | ||||||||||
| Ch-RS-BR-15-4S-7 | Brazil | MG846381 | ||||||||||
| Ch-RS-BR-15-5R | Brazil | MG846379 | ||||||||||
| Ch-AVRVFBR01 | Brazil | KF926653 | ||||||||||
| Ch-AVRVFBR02 | Brazil | KF926654 | ||||||||||
| Ch-AVRVFBR03 | Brazil | KF926655 | ||||||||||
| Ch-AVRVFBR04 | Brazil | KF926656 | ||||||||||
| Ch-AVRVFBR05 | Brazil | KF926657 | ||||||||||
| Ch-BRA27 | Brazil | KP824796 | ||||||||||
| Ch-BRA54 | Brazil | KP824800 | ||||||||||
| Ch-BRA57 | Brazil | KP824801 | ||||||||||
| Ch-BRA71 | Brazil | KP824803 | ||||||||||
| Ch-BRA81 | Brazil | KP824808 | ||||||||||
| RVG | ||||||||||||
| Ch-03V0567 | Germany | HQ403604 | JQ920012 | |||||||||
| Ch-MRC-DPRU1679 | South Africa | KJ752088 | KJ752083 | |||||||||
| Ty-Minnesota-1 | USA | KY689681 | ||||||||||
| Ty-Minnesota-2 | USA | MF120219 | ||||||||||
| Pg-HK18 | Hong Kong | KC876014 | ||||||||||
| Hg-H01-10385 | Netherlands | KP057507 | ||||||||||
| Pr-956-2 | Italy | KT073229 | ||||||||||
| Ch-BRA41 | Brazil | KP824797 | ||||||||||
| Ch-BRA43 | Brazil | KP824798 | ||||||||||
| Ch-BRA49 | Brazil | KP824799 | ||||||||||
| Ch-BRA68 | Brazil | KP824802 | ||||||||||
| Ch-BRA75 | Brazil | KP824804 | ||||||||||
| Ch-BRA76 | Brazil | KP824805 | ||||||||||
| Ch-BRA77 | Brazil | KP824806 | ||||||||||
| Ch-BRA78 | Brazil | KP824807 | ||||||||||
| Gene Segment | Virus Strains | |
|---|---|---|
| RVF | RVG | |
| VP1 | Complete ORF (3258 bp): BJ12 Partial ORF (1115 bp): BJ1, BJ7 | - |
| VP2 | Partial ORF (1200 bp): BJ1, BJ7, BJ12 | - |
| VP3 | - | - |
| VP4 | Complete ORF (2204 bp): BJ1, BJ7 Partial ORF (1130 bp): BJ12 | - |
| VP6 | Complete ORF (1188 bp): BJ1, BJ7, BJ12 Partial ORF (280 bp): BJ10, BJ11, BJ14, BJ22, BJ30, BJ31, BJ32, BJ37, BJ39, BJ41, BJ51, BJ53, MF06, MF53, MF54, MF67, MF69, MF70 | Complete ORF: MF48 (1173 bp) Partial ORF (642 bp): MF40, MF41 |
| VP7 | Partial ORF (398 bp): BJ1, BJ7, BJ12 | - |
| NSP1 | Complete ORF (1641 bp): BJ12 Partial ORF (997 bp): BJ1 | - |
| NSP2 | Complete ORF (954 bp): BJ1 Partial ORF (594 bp): BJ7, BJ12 | Partial ORF (953 bp): MF48 |
| NSP3 | - | - |
| NSP4 | Complete ORF (507 bp): BJ1, BJ7, BJ12, BJ37, MF104, MF110 | - |
| NSP5 | Complete ORF (654 bp): BJ1, BJ7, BJ10, BJ11, BJ12 Partial ORF (400 bp): BJ14, BJ30, BJ31, BJ32, BJ37, BJ39, BJ41, BJ51, MFO6 | Complete ORF (543 bp): MF40, MF41, MF48 |
References
- Matthijnssens, J.; Otto, P.H.; Ciarlet, M.; Desselberger, U.; Van Ranst, M.; Johne, R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 2012, 157, 1177–1182. [Google Scholar] [CrossRef]
- Mihalov-Kovács, E.; Gellért, Á.; Marton, S.; Farkas, S.L.; Fehér, E.; Oldal, M.; Jakab, F.; Martella, V.; Bányai, K. Candidate new rotavirus species in sheltered dogs, Hungary. Emerg. Infect. Dis. 2015, 21, 660–663. [Google Scholar] [CrossRef]
- Bányai, K.; Kemenesi, G.; Budinski, I.; Földes, F.; Zana, B.; Marton, S.; Varga-Kugler, R.; Oldal, M.; Kurucz, K.; Jakab, F. Candidate new rotavirus species in Schreiber’s bats, Serbia. Infect. Genet. Evol. 2017, 48, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Tausch, S.H.; Grützke, J.; Falkenhagen, A.; Patzina-Mehling, C.; Beer, M.; Höper, D.; Ulrich, R.G. Distantly Related Rotaviruses in Common Shrews, Germany, 2004–2014. Emerg. Infect. Dis. 2019, 25, 2310–2314. [Google Scholar] [CrossRef]
- Johne, R.; Schilling-Loeffler, K.; Ulrich, R.G.; Tausch, S.H. Whole Genome Sequence Analysis of a Prototype Strain of the Novel Putative Rotavirus Species L. Viruses 2022, 14, 462. [Google Scholar] [CrossRef]
- Estes, M.K.; Greenberg, H.B. Rotaviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Cohen, J.I., Griffin, D.E., Lamb, R.A., Martin, M.A., Racaniello, V.R., Rizman, B., Eds.; Wolters Kluver Health/Lippincott, Williams e Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 1347–1401. [Google Scholar]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef] [PubMed]
- Barro, M.; Patton, J.T. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J. Virol. 2007, 81, 4473–4481. [Google Scholar] [CrossRef]
- Graff, J.W.; Ettayebi, K.; Hardy, M.E. Rotavirus NSP1 inhibits NFkappaB activation by inducing proteasome-dependent degradation of beta-TrCP: A novel mechanism of IFN antagonism. PLoS Pathog. 2009, 5, e1000280. [Google Scholar] [CrossRef] [PubMed]
- Criglar, J.M.; Hu, L.; Crawford, S.E.; Hyser, J.M.; Broughman, J.R.; Prasad, B.V.; Estes, M.K. A novel form of rotavirus NSP2 and phosphorylation-dependent NSP2-NSP5 interactions are associated with viroplasm assembly. J. Virol. 2014, 88, 786–798. [Google Scholar] [CrossRef]
- Geiger, F.; Acker, J.; Papa, G.; Wang, X.; Arter, W.E.; Saar, K.L.; Erkamp, N.A.; Qi, R.; Bravo, J.P.; Strauss, S.; et al. Liquid-liquid phase separation underpins the formation of replication factories in rotaviruses. EMBO J. 2021, 40, e107711. [Google Scholar] [CrossRef]
- Vende, P.; Piron, M.; Castagné, N.; Poncet, D. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3’ end. J. Virol. 2000, 74, 7064–7071. [Google Scholar] [CrossRef] [PubMed]
- Ball, J.M.; Mitchell, D.M.; Gibbons, T.F.; Parr, R.D. Rotavirus NSP4: A multifunctional viral enterotoxin. Viral Immunol. 2005, 18, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Hyser, J.M.; Collinson-Pautz, M.R.; Utama, B.; Estes, M.K. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio 2010, 1, e00265-10. [Google Scholar] [CrossRef]
- Chang-Graham, A.L.; Perry, J.L.; Strtak, A.C.; Ramachandran, N.K.; Criglar, J.M.; Philip, A.A.; Patton, J.T.; Estes, M.K.; Hyser, J.M. Rotavirus Calcium Dysregulation Manifests as Dynamic Calcium Signaling in the Cytoplasm and Endoplasmic Reticulum. Sci. Rep. 2019, 9, 10822. [Google Scholar] [CrossRef]
- Bergeland, M.E.; McAdaragh, J.P.; Stotz, I. Rotaviral enteritis in turkey poults. In Proceedings of the 26th Western Poultry Diseases Conference, Davis, CA, USA, 21–24 March 1977; University of California: Davis, CA, USA, 1977; pp. 129–130. [Google Scholar]
- McNulty, M.S.; Allan, G.M.; Stuart, J.C. Rotavirus infection in avian species. Vet. Rec. 1978, 103, 319–320. [Google Scholar] [CrossRef]
- Dhama, K.; Saminathan, M.; Karthik, K.; Tiwari, R.; Shabbir, M.Z.; Kumar, N.; Malik, Y.S.; Singh, R.K. Avian rotavirus enteritis —An updated review. Vet. Q. 2015, 35, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Otto, P.; Liebler-Tenorio, E.M.; Elschner, M.; Reetz, J.; Löhren, U.; Diller, R. Detection of rotaviruses and intestinal lesions in broiler chicks from flocks with runting and stunting syndrome (RSS). Avian Dis. 2006, 50, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Otto, P.H.; Ahmed, M.U.; Hotzel, H.; Machnowska, P.; Reetz, J.; Roth, B.; Trojnar, E.; Johne, R. Detection of avian rotaviruses of groups A, D, F and G in diseased chickens and turkeys from Europe and Bangladesh. Vet. Microbiol. 2012, 156, 8–15. [Google Scholar] [CrossRef]
- Kindler, E.; Trojnar, E.; Heckel, G.; Otto, P.H.; Johne, R. Analysis of rotavirus species diversity and evolution including the newly determined full-length genome sequences of rotavirus F and G. Infect. Genet. Evol. 2013, 14, 58–67. [Google Scholar] [CrossRef]
- Phan, T.G.; Vo, N.P.; Boros, Á.; Pankovics, P.; Reuter, G.; Li, O.T.; Wang, C.; Deng, X.; Poon, L.L.; Delwart, E. The viruses of wild pigeon droppings. PLoS ONE 2013, 8, e72787. [Google Scholar] [CrossRef]
- Beserra, L.A.; Gregori, F. Description of rotavirus F in broilers from Brazilian poultry farms. Avian Dis. 2014, 58, 458–461. [Google Scholar] [CrossRef]
- Bodewes, R.; van Run, P.R.; Schürch, A.C.; Koopmans, M.P.; Osterhaus, A.D.; Baumgärtner, W.; Kuiken, T.; Smits, S.L. Virus characterization and discovery in formalin-fixed paraffin-embedded tissues. J. Virol. Methods. 2015, 214, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Stucker, K.M.; Stockwell, T.B.; Nyaga, M.M.; Halpin, R.A.; Fedorova, N.; Akopov, A.; Ngoveni, H.; Peenze, I.; Seheri, M.L.; Mphahlele, M.J.; et al. Complete genomic sequence for an avian group G rotavirus from South Africa. Genome Announc. 2015, 3, e00107–e00115. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.D.; Bezerra, D.A.; Silva, R.R.; Silva, M.J.; Júnior, E.C.; Soares, L.S. Detection of the VP6 gene of group F and G rotaviruses in broiler chicken fecal samples from the Amazon region of Brazil. Arch. Virol. 2016, 161, 2263–2268. [Google Scholar] [CrossRef]
- Ramig, R.F. Genetics of the rotaviruses. Annu. Rev. Microbiol. 1997, 51, 225–255. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Vashistt, J.; Changotra, H. Rotaviruses: Is their surveillance needed? Vaccine 2014, 32, 3367–3378. [Google Scholar] [CrossRef]
- Iturriza-Gómara, M.; Green, J.; Brown, D.W.; Ramsay, M.; Desselberger, U.; Gray, J.J. Molecular epidemiology of human group A rotavirus infections in the United Kingdom between 1995 and 1998. J. Clin. Microbiol. 2000, 38, 4394–4401. [Google Scholar] [CrossRef]
- Dóró, R.; Farkas, S.L.; Martella, V.; Bányai, K. Zoonotic transmission of rotavirus: Surveillance and control. Expert. Rev. Anti. Infect. Ther. 2015, 13, 1337–1350. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Rahman, M.; Attoui, H.; Bányai, K.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gómara, M.; Kirkwood, C.D.; Martella, V.; et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008, 153, 1621–1629. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Iturriza-Gómara, M.; Maes, P.; Patton, J.T.; et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008, 82, 3204–3219. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Bányai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Kuga, K.; Miyazaki, A.; Suzuki, T.; Takagi, M.; Hattori, N.; Katsuda, K.; Mase, M.; Sugiyama, M.; Tsunemitsu, H. Genetic diversity and classification of the outer capsid glycoprotein VP7 of porcine group B rotaviruses. Arch. Virol. 2009, 154, 1785–1795. [Google Scholar] [CrossRef] [PubMed]
- Marthaler, D.; Rossow, K.; Gramer, M.; Collins, J.; Goyal, S.; Tsunemitsu, H.; Kuga, K.; Suzuki, T.; Ciarlet, M.; Matthijnssens, J. Detection of substantial porcine group B rotavirus genetic diversity in the United States, resulting in a modified classification proposal for G genotypes. Virology 2012, 433, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Hasebe, A. A provisional complete genome-based genotyping system for rotavirus species C from terrestrial mammals. J. Gen. Virol. 2017, 98, 2647–2662. [Google Scholar] [CrossRef]
- Suzuki, T.; Inoue, D. Full genome-based genotyping system for rotavirus H and detection of potential gene recombination in nonstructural protein 3 between porcine rotavirus H and rotavirus C. J. Gen. Virol. 2018, 99, 1582–1589. [Google Scholar] [CrossRef]
- McNulty, M.S.; Todd, D.; Allan, G.M.; McFerran, J.B.; Greene, J.A. Epidemiology of rotavirus infection in broiler chickens: Recognition of four serogroups. Arch. Virol. 1984, 81, 113–121. [Google Scholar] [CrossRef]
- Kang, S.Y.; Nagaraja, K.V.; Newman, J.A. Electropherotypic analysis of rotaviruses isolated from turkeys. Avian Dis. 1986, 30, 794–801. [Google Scholar] [CrossRef]
- Theil, K.W.; Reynolds, D.L.; Saif, Y.M. Genomic variation among avian rotavirus-like viruses detected by polyacrylamide gel electrophoresis. Avian Dis. 1986, 30, 829–834. [Google Scholar] [CrossRef]
- Falcone, E.; Busi, C.; Lavazza, A.; Monini, M.; Bertoletti, M.; Canelli, E.; Vignolo, E.; Ruggeri, F.M.; Boniotti, M.B. Molecular characterization of avian rotaviruses circulating in Italian poultry flocks. Avian Pathol. 2015, 44, 509–515. [Google Scholar] [CrossRef]
- Duarte Júnior, J.W.B.; Chagas, E.H.N.; Serra, A.C.S.; Souto, L.C.D.S.; da Penha Júnior, E.T.; Bandeira, R.D.S.E.; Guimarães, R.J.P.S.; Oliveira, H.G.D.S.; Sousa, T.K.S.; Lopes, C.T.A.; et al. Ocurrence of rotavirus and picobirnavirus in wild and exotic avian from amazon forest. PLoS Negl. Trop. Dis. 2021, 15, e0008792. [Google Scholar] [CrossRef]
- Pinheiro, M.S.; Dias, J.B.L.; Cunha, B.R.A.V.; Petrucci, M.P.; Travassos, C.E.P.F.; Mendes, G.S.; Santos, N. Rotavirus F and G circulating in chickens in Southeastern Brazil. Trop. Anim. Health Prod. 2022, 54, 113. [Google Scholar] [CrossRef] [PubMed]
- Scorsato, A.P.; Telles, J.E.Q. Factors that affect the quality of DNA extracted from biological samples stored in paraffin blocks. J. Bras. Patol. Med. Lab. 2011, 47, 541–548. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Gouvea, V.; Brantly, M. Is rotavirus a population of reassortants? Trends Microbiol. 1995, 3, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Bányai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Vet Microbiol. 2010, 140, 246–255. [Google Scholar] [CrossRef]
- Patton, J.T. Rotavirus diversity and evolution in the post-vaccine world. Discov. Med. 2012, 13, 85–97. [Google Scholar] [PubMed]
- Degiuseppe, J.I.; Stupka, J.A. Genotype distribution of Group A rotavirus in children before and after massive vaccination in Latin America and the Caribbean: Systematic review. Vaccine 2020, 38, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Capua, I.; Alexander, D. Perspectives on the global threat: The challenge of avian influenza viruses for the world’s veterinary community. Avian Dis. 2010, 54 (Suppl. 1), 176–178. [Google Scholar] [CrossRef]
- Trovão, N.S.; Shepherd, F.K.; Herzberg, K.; Jarvis, M.C.; Lam, H.C.; Rovira, A.; Culhane, M.R.; Nelson, M.I.; Marthaler, D.G. Evolution of rotavirus C in humans and several domestic animal species. Zoonoses Public Health 2019, 66, 546–557. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinheiro, M.S.; Dias, J.B.L.; Petrucci, M.P.; Travassos, C.E.P.F.; Mendes, G.S.; Santos, N. Molecular Characterization of Avian Rotaviruses F and G Detected in Brazilian Poultry Flocks. Viruses 2023, 15, 1089. https://doi.org/10.3390/v15051089
Pinheiro MS, Dias JBL, Petrucci MP, Travassos CEPF, Mendes GS, Santos N. Molecular Characterization of Avian Rotaviruses F and G Detected in Brazilian Poultry Flocks. Viruses. 2023; 15(5):1089. https://doi.org/10.3390/v15051089
Chicago/Turabian StylePinheiro, Mariana S., Juliana B. L. Dias, Melissa P. Petrucci, Carlos E. P. F. Travassos, Gabriella S. Mendes, and Norma Santos. 2023. "Molecular Characterization of Avian Rotaviruses F and G Detected in Brazilian Poultry Flocks" Viruses 15, no. 5: 1089. https://doi.org/10.3390/v15051089
APA StylePinheiro, M. S., Dias, J. B. L., Petrucci, M. P., Travassos, C. E. P. F., Mendes, G. S., & Santos, N. (2023). Molecular Characterization of Avian Rotaviruses F and G Detected in Brazilian Poultry Flocks. Viruses, 15(5), 1089. https://doi.org/10.3390/v15051089





