Broad-Spectrum Antivirals Derived from Natural Products
Abstract
:1. Introduction
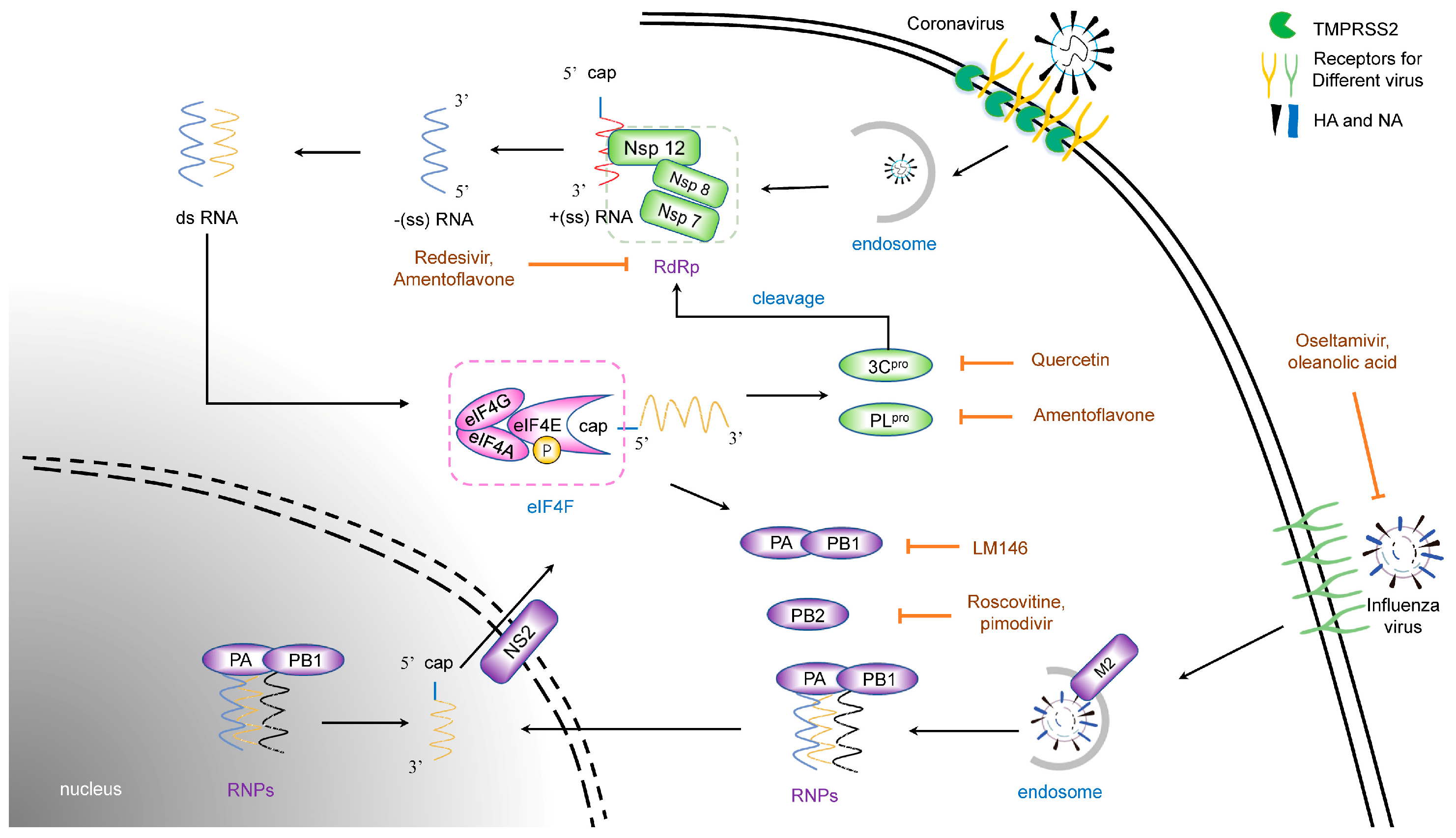
2. Life Cycles of Different Viruses
2.1. Life Cycles of Coronavirus
2.2. Life Cycles of Influenza Virus
2.3. Life Cycles of the Herpes Simplex Virus
3. Therapeutic Natural Antivirals for Virus Entry
3.1. Natural Products Targeting Coronavirus Entry
3.1.1. Receptors
3.1.2. Membrane Protease
3.2. Natural Products Targeting Membrane Fusion and Endocytosis
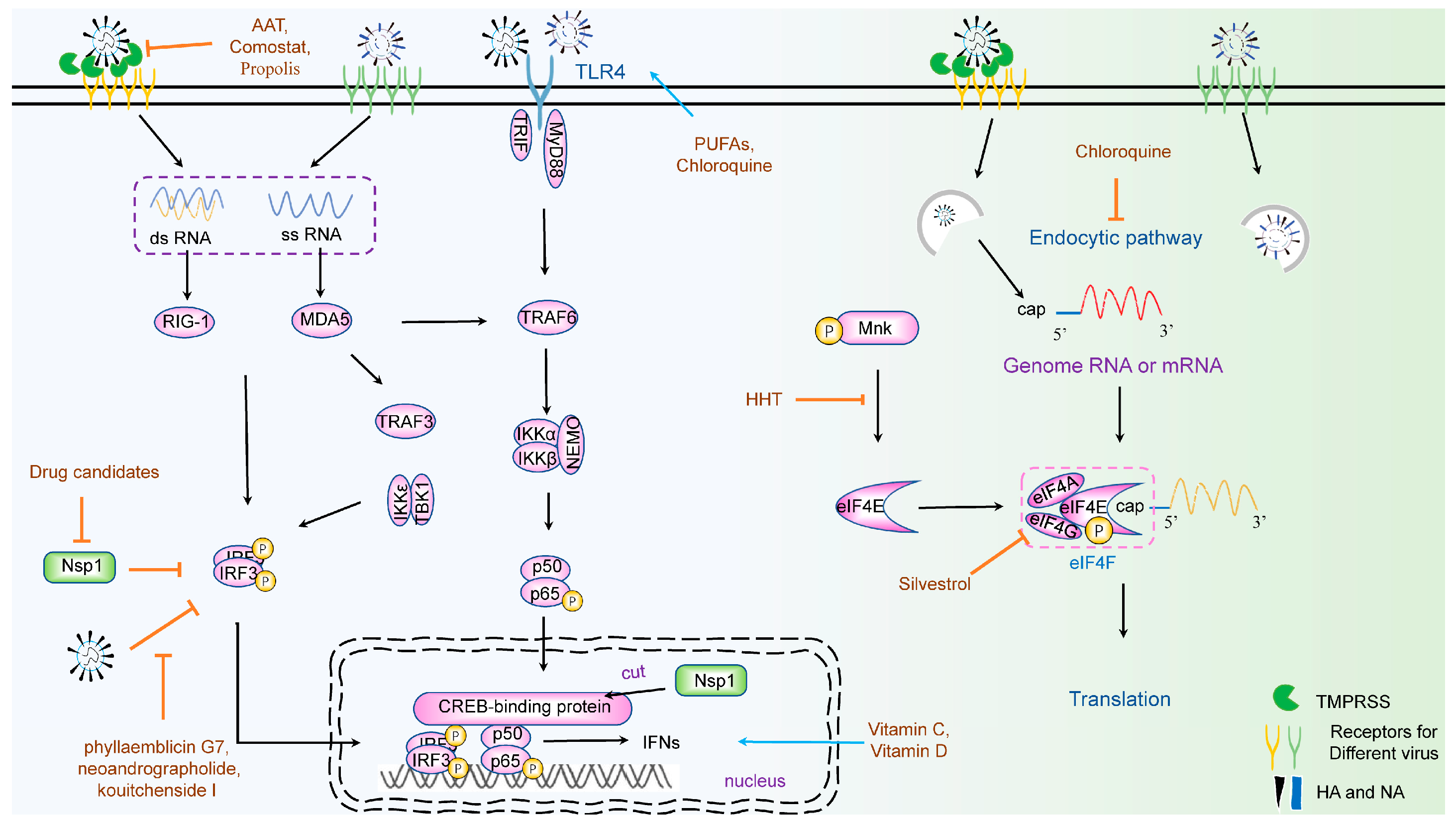
4. Natural Products Targeting Viral Translation
4.1. Targeting the Translation Initiation Factor
4.1.1. eIF4A
4.1.2. Phosphorylation of eIF4E (p-eIF4E) and eIF4E
5. Natural Products Targeting an Innate Immune Response
6. Virus-Targeting Antivirals
6.1. Targeting Coronavirus Protein
6.1.1. Nsp1
6.1.2. Nsp3 (PLpro) and Nsp5 (3CLpro)
6.1.3. RdRp
6.2. Targeting the Influenza Virus Protein
6.2.1. HA and NA
6.2.2. RNA Polymerase
6.3. Targeting the Herpes Simplex Virus Protein
6.3.1. Surface Glycoproteins
6.3.2. DNA Polymerase
7. Discussion
Funding
Conflicts of Interest
References
- Andronico, A.; Kiem, C.T.; Paireau, J.; Succo, T.; Bosetti, P.; Lefrancq, N.; Nacher, M.; Djossou, F.; Sanna, A.; Flamand, C.; et al. Evaluating the impact of curfews and other measures on SARS-CoV-2 transmission in French Guiana. Nat. Commun. 2021, 12, 1634. [Google Scholar] [CrossRef]
- Menéndez-Arias, L.; Delgado, R. Update and latest advances in antiretroviral therapy. Trends Pharmacol. Sci. 2022, 43, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.-D.; Rappuoli, R. SARS—Beginning to understand a new virus. Nat. Rev. Genet. 2003, 1, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Uribe, M.; Rodríguez-Posada, M.E.; Ramirez-Nieto, G.C. Molecular evidence of orthomyxovirus presence in colombian neotropical bats. Front. Microbiol. 2022, 13, 845546. [Google Scholar] [CrossRef]
- Luo, W.; Tian, L.; Gan, Y.; Chen, E.; Shen, X.; Pan, J.; Irwin, D.M.; Chen, R.-A.; Shen, Y. The fit of codon usage of human-isolated avian influenza A viruses to human. Infect. Genet. Evol. 2020, 81, 104181. [Google Scholar] [CrossRef]
- Shriner, S.A.; Root, J.J. A review of avian influenza a virus associations in synanthropic birds. Viruses 2020, 12, 1209. [Google Scholar] [CrossRef]
- Martini, M.; Gazzaniga, V.; Bragazzi, N.; Barberis, I. The Spanish influenza pandemic: A lesson from history 100 years after 1918. J. Prev. Med. Hyg. 2019, 60, E64–E67. [Google Scholar] [CrossRef]
- Fineberg, H.V. Pandemic Preparedness and Response—Lessons from the H1N1 Influenza of 2009. N. Engl. J. Med. 2014, 370, 1335–1342. [Google Scholar] [CrossRef]
- Crimi, S.; Fiorillo, L.; Bianchi, A.; D’amico, C.; Amoroso, G.; Gorassini, F.; Mastroieni, R.; Marino, S.; Scoglio, C.; Catalano, F.; et al. Herpes Virus, Oral clinical signs and QoL: Systematic review of recent data. Viruses 2019, 11, 463. [Google Scholar] [CrossRef]
- James, C.; Harfouche, M.; Welton, N.J.; Turner, K.M.E.; Abu-Raddad, L.J.; Gottlieb, S.L.; Looker, K.J. Herpes simplex virus: Global infection prevalence and incidence estimates, 2016. Bull. World Health Organ. 2020, 98, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Zmasek, C.M.; Lefkowitz, E.J.; Niewiadomska, A.; Scheuermann, R.H. Genomic evolution of the Coronaviridae family. Virology 2022, 570, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Author Correction: Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Genet. 2022, 20, 315. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Millet, J.K.; A Jaimes, J.; Whittaker, G.R. Molecular diversity of coronavirus host cell entry receptors. FEMS Microbiol. Rev. 2021, 45, fuaa057. [Google Scholar] [CrossRef]
- Liu, C.; Ma, Y.; Yang, Y.; Zheng, Y.; Shang, J.; Zhou, Y.; Jiang, S.; Du, L.; Li, J.; Li, F. Cell Entry of Porcine Epidemic Diarrhea Coronavirus Is Activated by Lysosomal Proteases. J. Biol. Chem. 2016, 291, 24779–24786. [Google Scholar] [CrossRef]
- Lapointe, C.P.; Grosely, R.; Johnson, A.G.; Wang, J.; Fernández, I.S.; Puglisi, J.D. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc. Natl. Acad. Sci. USA 2021, 118, e2017715118. [Google Scholar] [CrossRef]
- Glatthaar-Saalmüller, B.; Fal, A.; Schönknecht, K.; Conrad, F.; Sievers, H.; Saalmüller, A. Antiviral activity of an aqueous extract derived from Aloe arborescens Mill. against a broad panel of viruses causing infections of the upper respiratory tract. Phytomedicine 2015, 22, 911–920. [Google Scholar] [CrossRef]
- Sempere Borau, M.; Stertz, S. Entry of influenza A virus into host cells—Recent progress and remaining challenges. Curr. Opin. Virol. 2021, 48, 23–29. [Google Scholar] [CrossRef]
- Vanderlinden, E.; Naesens, L. Emerging Antiviral Strategies to Interfere with Influenza Virus Entry. Med. Res. Rev. 2014, 34, 301–339. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Ojha, C.R.; Russell, C.J. Relationship between hemagglutinin stability and influenza virus persistence after exposure to low pH or supraphysiological heating. PLoS Pathog. 2021, 17, e1009910. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Gui, M.; Xiang, Y. Structural intermediates in the low pH-induced transition of influenza hemagglutinin. PLoS Pathog. 2020, 16, e1009062. [Google Scholar] [CrossRef]
- Miyake, Y.; Keusch, J.J.; Decamps, L.; Ho-Xuan, H.; Iketani, S.; Gut, H.; Kutay, U.; Helenius, A.; Yamauchi, Y. Influenza virus uses transportin 1 for vRNP debundling during cell entry. Nat. Microbiol. 2019, 4, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the centre: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Te Velthuis, A.J.; Fodor, E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.C.; Charles, P.D.; Hester, S.S.; Thomas, B.; Trudgian, D.; Martínez-Alonso, M.; Fodor, E. Conserved and host-specific features of influenza virion architecture. Nat. Commun. 2014, 5, 4816. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Watanabe, S.; Kawaoka, Y. Cellular Networks Involved in the Influenza Virus Life Cycle. Cell Host Microbe 2010, 7, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Duarte, L.F.; Farías, M.A.; Álvarez, D.M.; Bueno, S.M.; Riedel, C.A.; González, P.A. Herpes Simplex Virus Type 1 Infection of the Central Nervous System: Insights into Proposed Interrelationships with Neurodegenerative Disorders. Front. Cell. Neurosci. 2019, 13, 46. [Google Scholar] [CrossRef]
- Wang, S.; Ljubimov, A.V.; Jin, L.; Pfeffer, K.; Kronenberg, M.; Ghiasi, H. Herpes Simplex Virus 1 Latency and the Kinetics of Reactivation Are Regulated by a Complex Network of Interactions between the Herpesvirus Entry Mediator, Its Ligands (gD, BTLA, LIGHT, and CD160), and the Latency-Associated Transcript. J. Virol. 2018, 92, e01451-18. [Google Scholar] [CrossRef]
- Steiner, I.; Benninger, F. Update on Herpes Virus Infections of the Nervous System. Curr. Neurol. Neurosci. Rep. 2013, 13, 414. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, H.; Shi, J.; Zhang, Z.; Gong, R. Identification of a Novel Inhibitor against Middle East Respiratory Syndrome Coronavirus. Viruses 2017, 9, 255. [Google Scholar] [CrossRef] [PubMed]
- Eggink, D.; Bontjer, I.; de Taeye, S.W.; Langedijk, J.P.M.; Berkhout, B.; Sanders, R.W. HIV-1 anchor inhibitors and membrane fusion inhibitors target distinct but overlapping steps in virus entry. J. Biol. Chem. 2019, 294, 5736–5746. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, K.; Farasat, A.; Rostamian, M.; Johari, B.; Madanchi, H. Enfuvirtide, an HIV-1 fusion inhibitor peptide, can act as a potent SARS-CoV-2 fusion inhibitor: An in silico drug repurposing study. J. Biomol. Struct. Dyn. 2022, 40, 5566–5576. [Google Scholar] [CrossRef]
- Bruno, B.J.; Miller, G.D.; Lim, C.S.; Harrington, A.; Tal-Gan, Y.; Richard, J.; Agrahari, V.; Agrahari, V.; Mitra, A.K.; Panicucci, R.; et al. Basics and recent advances in peptide and protein drug delivery. Ther. Deliv. 2013, 4, 1443–1467. [Google Scholar] [CrossRef] [PubMed]
- Arranz-Gibert, P.; Ciudad, S.; Seco, J.; García, J.; Giralt, E.; Teixidó, M. Immunosilencing peptides by stereochemical inversion and sequence reversal: Retro-D-peptides. Sci. Rep. 2018, 8, 6446. [Google Scholar] [CrossRef] [PubMed]
- Vagner, J.; Qu, H.; Hruby, V.J. Peptidomimetics, a synthetic tool of drug discovery. Curr. Opin. Chem. Biol. 2008, 12, 292–296. [Google Scholar] [CrossRef]
- Schütz, D.; Ruiz-Blanco, Y.B.; Münch, J.; Kirchhoff, F.; Sanchez-Garcia, E.; Müller, J.A. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv. Drug Deliv. Rev. 2020, 167, 47–65. [Google Scholar] [CrossRef]
- Bai, X.; Hippensteel, J.; Leavitt, A.; Maloney, J.P.; Beckham, D.; Garcia, C.; Li, Q.; Freed, B.M.; Ordway, D.; Sandhaus, R.A.; et al. Hypothesis: Alpha-1-antitrypsin is a promising treatment option for COVID-19. Med. Hypotheses 2021, 146, 110394. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Zhao, H.; Meng, X.; Peng, Z.; Lam, H.; Zhang, C.; Zhou, X.; Chan, J.F.-W.; Kao, R.Y.T.; To, K.K.-W.; Yuen, K.-Y. Fusion-inhibition peptide broadly inhibits influenza virus and SARS-CoV-2, including Delta and Omicron variants. Emerg. Microbes Infect. 2022, 11, 926–937. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Tsuda, M.; Nanbo, A.; Hattori, T.; Sasaki, J.; Sasaki, T.; Miyazaki, T.; Ohba, Y. A Ca2+-dependent signalling circuit regulates influenza A virus internalization and infection. Nat. Commun. 2013, 4, 2763. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Nishide, S.; Ose, T.; Suzuki, T.; Kato, I.; Fukuhara, H.; Fujioka, M.; Horiuchi, K.; Satoh, A.O.; Nepal, P.; et al. A Sialylated Voltage-Dependent Ca2+ Channel Binds Hemagglutinin and Mediates Influenza A Virus Entry into Mammalian Cells. Cell Host Microbe 2018, 23, 809–818.e5. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, Y.; Chen, Z.; Chen, L.; Liang, J.; Zhang, C.; Zhang, Z.; Yang, J. Nisoldipine Inhibits Influenza A Virus Infection by Interfering with Virus Internalization Process. Viruses 2022, 14, 2738. [Google Scholar] [CrossRef]
- Simmons, G.; Reeves, J.D.; Rennekamp, A.J.; Amberg, S.M.; Piefer, A.J.; Bates, P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 2004, 101, 4240–4245. [Google Scholar] [CrossRef]
- Yang, Z.-Y.; Huang, Y.; Ganesh, L.; Leung, K.; Kong, W.-P.; Schwartz, O.; Subbarao, K.; Nabel, G.J. pH-Dependent Entry of Severe Acute Respiratory Syndrome Coronavirus Is Mediated by the Spike Glycoprotein and Enhanced by Dendritic Cell Transfer through DC-SIGN. J. Virol. 2004, 78, 5642–5650. [Google Scholar] [CrossRef]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005, 2, 69. [Google Scholar] [CrossRef]
- Savarino, A.; Shytaj, I.L. Chloroquine and beyond: Exploring anti-rheumatic drugs to reduce immune hyperactivation in HIV/AIDS. Retrovirology 2015, 12, 51. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Pastenkos, G.; Lee, B.; Pritchard, S.M.; Nicola, A.V. Bovine Herpesvirus 1 Entry by a Low-pH Endosomal Pathway. J. Virol. 2018, 92, e00839-18. [Google Scholar] [CrossRef]
- Aljadeed, R. The Rise and Fall of Hydroxychloroquine and Chloroquine in COVID-19. J. Pharm. Pr. 2022, 35, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Xiang, Y.; Wang, X.; Wang, Q.; Zhong, M.; Wang, S.; Wang, X.; Fan, J.; Kitazato, K.; Wang, Y. Epidermal Growth Factor Receptor-PI3K Signaling Controls Cofilin Activity to Facilitate Herpes Simplex Virus 1 Entry into Neuronal Cells. mBio 2014, 5, e00958-13. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chi, X.; Wang, J.; Potter, A.L.; Wang, X.; Yang, C.J. Small molecule RAF265 as an antiviral therapy acts against HSV-1 by regulating cytoskeleton rearrangement and cellular translation machinery. J. Med. Virol. 2023, 95, e28226. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, Y.; Li, F.; Cao, W.; Zeng, Q.; Qin, S.; Wang, Z.; Jia, J.; Xiao, J.; Hu, X.; et al. Hsp90 Inhibitors Inhibit the Entry of Herpes Simplex Virus 1 Into Neuron Cells by Regulating Cofilin-Mediated F-Actin Reorganization. Front. Microbiol. 2021, 12, 799890. [Google Scholar] [CrossRef]
- Leppek, K.; Das, R.; Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- Walker, M.J.; Shortridge, M.D.; Albin, D.D.; Cominsky, L.Y.; Varani, G. Structure of the RNA Specialized Translation Initiation Element that Recruits eIF3 to the 5′-UTR of c-Jun. J. Mol. Biol. 2020, 432, 1841–1855. [Google Scholar] [CrossRef]
- Venezia, N.D.; Vincent, A.; Marcel, V.; Catez, F.; Diaz, J.-J. Emerging Role of Eukaryote Ribosomes in Translational Control. Int. J. Mol. Sci. 2019, 20, 1226. [Google Scholar] [CrossRef]
- Gebauer, F.; Hentze, M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004, 5, 827–835. [Google Scholar] [CrossRef]
- Gandin, V.; Miluzio, A.; Barbieri, A.M.; Beugnet, A.; Kiyokawa, H.; Marchisio, P.C.; Biffo, S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 2008, 455, 684–688. [Google Scholar] [CrossRef]
- Merrick, W.C.; Pavitt, G.D. Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harb. Perspect. Biol. 2018, 10, a033092. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Vogel, S.B.; Wolfe, M.M.; McGuigan, J.E.; Hawkins, I.F.; Howard, R.J.; Woodward, E.R. Localization and Resection of Gastrinomas in Zollinger-Ellison Syndrome. Ann. Surg. 1987, 205, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Parsyan, A.; Svitkin, Y.; Shahbazian, D.; Gkogkas, C.; Lasko, P.; Merrick, W.C.; Sonenberg, N. mRNA helicases: The tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011, 12, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Woodard, J.L.; Lucas, D.M.; Fuchs, J.R.; Kinghorn, A.D. Rocaglamide, silvestrol and structurally related bioactive compounds from Aglaia species. Nat. Prod. Rep. 2014, 31, 924–939. [Google Scholar] [CrossRef]
- Saradhi, U.V.R.V.; Gupta, S.V.; Chiu, M.; Wang, J.; Ling, Y.; Liu, Z.; Newman, D.J.; Covey, J.M.; Kinghorn, A.D.; Marcucci, G.; et al. Characterization of Silvestrol Pharmacokinetics in Mice Using Liquid Chromatography–Tandem Mass Spectrometry. AAPS J. 2011, 13, 347–356. [Google Scholar] [CrossRef]
- Dong, H.-J.; Wang, J.; Zhang, X.-Z.; Li, C.-C.; Liu, J.-F.; Wang, X.-J. Proteomic screening identifies RPLp2 as a specific regulator for the translation of coronavirus. Int. J. Biol. Macromol. 2023, 230, 123191. [Google Scholar] [CrossRef]
- Walsh, D.; Mohr, I. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 2004, 18, 660–672. [Google Scholar] [CrossRef]
- Furic, L.; Rong, L.; Larsson, O.; Koumakpayi, I.H.; Yoshida, K.; Brueschke, A.; Petroulakis, E.; Robichaud, N.; Pollak, M.; Gaboury, L.A.; et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. USA 2010, 107, 14134–14139. [Google Scholar] [CrossRef]
- Proud, C.G. Mnks, eIF4E phosphorylation and cancer. Biochim. Biophys. Acta 2015, 1849, 766–773. [Google Scholar] [CrossRef]
- Dong, H.-J.; Wang, Z.-H.; Meng, W.; Li, C.-C.; Hu, Y.-X.; Zhou, L.; Wang, X.-J. The Natural Compound Homoharringtonine Presents Broad Antiviral Activity In Vitro and In Vivo. Viruses 2018, 10, 601. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.; McWhirter, S.M.; Faia, K.L.; Rowe, D.C.; Latz, E.; Golenbock, D.T.; Coyle, A.J.; Liao, S.-M.; Maniatis, T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003, 4, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Ji, X.; Lv, Y.; Cui, D.; Xie, J. The Roles of TRAF3 in Immune Responses. Dis. Markers 2023, 2023, 7787803. [Google Scholar] [CrossRef] [PubMed]
- Taft, J.; Markson, M.; Legarda, D.; Patel, R.; Chan, M.; Malle, L.; Richardson, A.; Gruber, C.; Martín-Fernández, M.; Mancini, G.M.; et al. Human TBK1 deficiency leads to autoinflammation driven by TNF-induced cell death. Cell 2021, 184, 4447–4463.e20. [Google Scholar] [CrossRef]
- Thoresen, D.; Wang, W.; Galls, D.; Guo, R.; Xu, L.; Pyle, A.M. The molecular mechanism of RIG-I activation and signaling. Immunol. Rev. 2021, 304, 154–168. [Google Scholar] [CrossRef]
- Panne, D.; McWhirter, S.M.; Maniatis, T.; Harrison, S.C. Interferon Regulatory Factor 3 Is Regulated by a Dual Phosphorylation-dependent Switch. J. Biol. Chem. 2007, 282, 22816–22822. [Google Scholar] [CrossRef]
- Siriwardena, A.K.; Andrade, R.; Prasad, U.S.; Cameron, E.W. Oesophagogastrostomy in the management of benign oesophageal strictures. J. R. Coll. Surg. Edinb. 1991, 36, 183–184. [Google Scholar]
- Zhang, K.; Lin, S.; Li, J.; Deng, S.; Zhang, J.; Wang, S. Modulation of Innate Antiviral Immune Response by Porcine Enteric Coronavirus. Front. Microbiol. 2022, 13, 845137. [Google Scholar] [CrossRef]
- Luo, M.; Xu, X.; Liu, X.; Shen, W.; Yang, L.; Zhu, Z.; Weng, S.; He, J.; Zuo, H. The Non-Receptor Protein Tyrosine Phosphatase PTPN6 Mediates a Positive Regulatory Approach from the Interferon Regulatory Factor to the JAK/STAT Pathway in Litopenaeus vannamei. Front. Immunol. 2022, 13, 913955. [Google Scholar] [CrossRef]
- Stark, G.R.; Darnell, J.E., Jr. The JAK-STAT Pathway at Twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- Verweij, M.C.; Wellish, M.; Whitmer, T.; Malouli, D.; Lapel, M.; Jonjic, S.; Haas, J.G.; DeFilippis, V.R.; Mahalingam, R.; Früh, K. Varicella Viruses Inhibit Interferon-Stimulated JAK-STAT Signaling through Multiple Mechanisms. PLoS Pathog. 2015, 11, e1004901. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.C.; Maggini, S. Vitamin C and Immune Function. Nutrients 2017, 9, 1211. [Google Scholar] [CrossRef] [PubMed]
- Mey, J.T.; Kirwan, J.P.; Axelrod, C.L. The Role of Nutrition in Mitigating the Effects of COVID-19 from Infection through PASC. Nutrients 2023, 15, 866. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.; Khanna, D. The Role of Vitamin C in Human Immunity and Its Treatment Potential Against COVID-19: A Review Article. Cureus 2023, 15, e33740. [Google Scholar] [CrossRef] [PubMed]
- Ran, L.; Zhao, W.; Wang, J.; Wang, H.; Zhao, Y.; Tseng, Y.; Bu, H. Extra Dose of Vitamin C Based on a Daily Supplementation Shortens the Common Cold: A Meta-Analysis of 9 Randomized Controlled Trials. BioMed Res. Int. 2018, 2018, 1837634. [Google Scholar] [CrossRef]
- Bae, M.; Kim, H. Mini-Review on the Roles of Vitamin C, Vitamin D, and Selenium in the Immune System against COVID-19. Molecules 2020, 25, 5346. [Google Scholar] [CrossRef]
- Prietl, B.; Treiber, G.; Pieber, T.R.; Amrein, K. Vitamin D and Immune Function. Nutrients 2013, 5, 2502–2521. [Google Scholar] [CrossRef]
- Milani, G.P.; Macchi, M.; Guz-Mark, A. Vitamin C in the Treatment of COVID-19. Nutrients 2021, 13, 1172. [Google Scholar] [CrossRef]
- Kumar, R.; Rathi, H.; Haq, A.; Wimalawansa, S.J.; Sharma, A. Putative roles of vitamin D in modulating immune response and immunopathology associated with COVID-19. Virus Res. 2021, 292, 198235. [Google Scholar] [CrossRef]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of chloroquine on viral infections: An old drug against today’s diseases. Lancet Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef]
- Bakadia, B.M.; He, F.; Souho, T.; Lamboni, L.; Ullah, M.W.; Boni, B.O.; Ahmed, A.A.Q.; Mukole, B.M.; Yang, G. Prevention and treatment of COVID-19: Focus on interferons, chloroquine/hydroxychloroquine, azithromycin, and vaccine. Biomed. Pharmacother. 2021, 133, 111008. [Google Scholar] [CrossRef] [PubMed]
- Eskiler, G.G.; Özkan, A.D.; Kaleli, S.; Bilir, C. Inhibition of TLR4/TRIF/IRF3 Signaling Pathway by Curcumin in Breast Cancer Cells. J. Pharm. Pharm. Sci. 2019, 22, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yu, J. Modulation of Toll-like receptor signaling in innate immunity by natural products. Int. Immunopharmacol. 2016, 37, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Albutti, A. Rescuing the Host Immune System by Targeting the Immune Evasion Complex ORF8-IRF3 in SARS-CoV-2 Infection with Natural Products Using Molecular Modeling Approaches. Int. J. Environ. Res. Public Health 2021, 19, 112. [Google Scholar] [CrossRef]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef]
- Ricciardi, S.; Guarino, A.M.; Giaquinto, L.; Polishchuk, E.V.; Santoro, M.; Di Tullio, G.; Wilson, C.; Panariello, F.; Soares, V.C.; Dias, S.S.G.; et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature 2022, 606, 761–768. [Google Scholar] [CrossRef]
- Park, G.J.; Osinski, A.; Hernandez, G.; Eitson, J.L.; Majumdar, A.; Tonelli, M.; Henzler-Wildman, K.; Pawłowski, K.; Chen, Z.; Li, Y.; et al. The mechanism of RNA capping by SARS-CoV-2. Nature 2022, 609, 793–800. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, K.; Yoo, D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 2016, 489, 252–268. [Google Scholar] [CrossRef]
- Shen, Z.; Zhang, G.; Yang, Y.; Li, M.; Yang, S.; Peng, G. Lysine 164 is critical for SARS-CoV-2 Nsp1 inhibition of host gene expression. J. Gen. Virol. 2021, 102, 001513. [Google Scholar] [CrossRef]
- Afsar, M.; Narayan, R.; Akhtar, N.; Das, D.; Rahil, H.; Nagaraj, S.K.; Eswarappa, S.M.; Tripathi, S.; Hussain, T.; Department of Molecular Reproduction; et al. Drug targeting Nsp1-ribosomal complex shows antiviral activity against SARS-CoV-2. Elife 2022, 11, e74877. [Google Scholar] [CrossRef]
- Shen, M.; Ding, P.; Luan, G.; Du, T.; Deng, S. The antiviral activity of a small molecule drug targeting the NSP1-ribosome complex against Omicron, especially in elderly patients. Front. Cell Infect. Microbiol. 2023, 13, 1141274. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Moccia, S.; Spagnuolo, C.; Tedesco, I.; Russo, G.L. Roles of flavonoids against coronavirus infection. Chem. Interactions 2020, 328, 109211. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, A.; Mujawdiya, P.K.; Lysiuk, R.; Shanaida, M.; Peana, M.; Benahmed, A.G.; Beley, N.; Kovalska, N.; Bjørklund, G. Quercetin in the Prevention and Treatment of Coronavirus Infections: A Focus on SARS-CoV-2. Pharmaceuticals 2022, 15, 1049. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Chakraborty, A.; Biswas, A.; Chowdhuri, S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors-an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2021, 39, 4362–4374. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.; Perron, M.; Murakami, E.; Bauer, K.; Park, Y.; Eckstrand, C.; Liepnieks, M.; Pedersen, N. The nucleoside analog GS-441524 strongly inhibits feline infectious peritonitis (FIP) virus in tissue culture and experimental cat infection studies. Veter- Microbiol. 2018, 219, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Perron, M.; Bannasch, M.; Montgomery, E.; Murakami, E.; Liepnieks, M.; Liu, H. Efficacy and safety of the nucleoside analog GS-441524 for treatment of cats with naturally occurring feline infectious peritonitis. J. Feline Med. Surg. 2019, 21, 271–281. [Google Scholar] [CrossRef]
- Jones, S.; Novicoff, W.; Nadeau, J.; Evans, S. Unlicensed GS-441524-Like Antiviral Therapy Can Be Effective for at-Home Treatment of Feline Infectious Peritonitis. Animals 2021, 11, 2257. [Google Scholar] [CrossRef]
- Ng, T.I.; Correia, I.; Seagal, J.; DeGoey, D.A.; Schrimpf, M.R.; Hardee, D.J.; Noey, E.L.; Kati, W.M. Antiviral Drug Discovery for the Treatment of COVID-19 Infections. Viruses 2022, 14, 961. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q.; et al. Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020, 395, 1569–1578. [Google Scholar] [CrossRef]
- Gandhi, S.; Klein, J.; Robertson, A.J.; Peña-Hernández, M.A.; Lin, M.J.; Roychoudhury, P.; Lu, P.; Fournier, J.; Ferguson, D.; Bakhash, S.A.K.M.; et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: A case report. Nat. Commun. 2022, 13, 1547. [Google Scholar] [CrossRef]
- Stevens, L.J.; Pruijssers, A.J.; Lee, H.W.; Gordon, C.J.; Tchesnokov, E.P.; Gribble, J.; George, A.S.; Hughes, T.M.; Lu, X.; Li, J.; et al. Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci. Transl. Med. 2022, 14, eabo0718. [Google Scholar] [CrossRef] [PubMed]
- Portilla-Martínez, A.; Ortiz-Flores, M.; Hidalgo, I.; Gonzalez-Ruiz, C.; Meaney, E.; Ceballos, G.; Nájera, N. In silico evaluation of flavonoids as potential inhibitors of SARS-CoV-2 main nonstructural proteins (Nsps)—Amentoflavone as a multitarget candidate. J. Mol. Model. 2022, 28, 404. [Google Scholar] [CrossRef] [PubMed]
- Li, H.C.; Yang, C.-H.; Lo, S.-Y. Strategies of Influenza A Virus to Ensure the Translation of Viral mRNAs. Pathogens 2022, 11, 1521. [Google Scholar] [CrossRef] [PubMed]
- Gubareva, L.V. Molecular mechanisms of influenza virus resistance to neuraminidase inhibitors. Virus Res. 2004, 103, 199–203. [Google Scholar] [CrossRef]
- Han, J.; Perez, J.; Schafer, A.; Cheng, H.; Peet, N.; Rong, L.; Manicassamy, B. Influenza Virus: Small Molecule Therapeutics and Mechanisms of Antiviral Resistance. Curr. Med. Chem. 2018, 25, 5115–5127. [Google Scholar] [CrossRef]
- Xiao, S.; Si, L.; Tian, Z.; Jiao, P.; Fan, Z.; Meng, K.; Zhou, X.; Wang, H.; Xu, R.; Han, X.; et al. Pentacyclic triterpenes grafted on CD cores to interfere with influenza virus entry: A dramatic multivalent effect. Biomaterials 2016, 78, 74–85. [Google Scholar] [CrossRef]
- Shao, L.; Yang, F.; Su, Y.; Li, W.; Zhang, J.; Xu, H.; Huang, B.; Sun, M.; Mu, Y.; Zhang, Y.; et al. Design and Synthesis of Oleanolic Acid Trimers to Enhance Inhibition of Influenza Virus Entry. ACS Med. Chem. Lett. 2021, 12, 1759–1765. [Google Scholar] [CrossRef]
- Haidari, M.; Ali, M.; Ward Casscells, S., 3rd; Madjid, M. Pomegranate (Punica granatum) purified polyphenol extract inhibits influenza virus and has a synergistic effect with oseltamivir. Phytomedicine 2009, 16, 1127–1136. [Google Scholar] [CrossRef]
- Ou, J.-L.; Mizushina, Y.; Wang, S.-Y.; Chuang, D.-Y.; Nadar, M.; Hsu, W.-L. Structure-activity relationship analysis of curcumin analogues on anti-influenza virus activity. FEBS J. 2013, 280, 5829–5840. [Google Scholar] [CrossRef]
- Mélin, L.; Gesner, E.; Attwell, S.; Kharenko, O.A.; van der Horst, E.H.; Hansen, H.C.; Gagnon, A. Design and Synthesis of LM146, a Potent Inhibitor of PB1 with an Improved Selectivity Profile over SMARCA2. ACS Omega 2021, 6, 21327–21338. [Google Scholar] [CrossRef]
- Huang, Q.; Zhong, Y.; Li, J.; Ye, Y.; Wu, W.; Chen, L.; Feng, M.; Yang, J.; Liu, S. Kinase inhibitor roscovitine as a PB2 cap-binding inhibitor against influenza a virus replication. Biochem. Biophys. Res. Commun. 2020, 526, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Gregor, J.; Radilová, K.; Brynda, J.; Fanfrlík, J.; Konvalinka, J.; Kožíšek, M. Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase. Molecules 2021, 26, 1007. [Google Scholar] [CrossRef] [PubMed]
- Soh, Y.Q.S.; Malone, K.D.; Eguia, R.T.; Bloom, J.D. Comprehensive Profiling of Mutations to Influenza Virus PB2 That Confer Resistance to the Cap-Binding Inhibitor Pimodivir. Viruses 2021, 13, 1196. [Google Scholar] [CrossRef] [PubMed]
- Kuzuhara, T.; Iwai, Y.; Takahashi, H.; Hatakeyama, D.; Echigo, N. Green tea catechins inhibit the endonuclease activity of influenza A virus RNA polymerase. PLoS Curr. 2009, 1, RRN1052. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gu, S.; Zhu, X.; Liu, M.; Cao, Z.; Qiu, P.; Li, S.; Liu, S.; Song, G. Discovery and optimization of new 6, 7-dihydroxy-1, 2, 3, 4-tetrahydroisoquinoline derivatives as potent influenza virus PAN inhibitors. Eur. J. Med. Chem. 2022, 227, 113929. [Google Scholar] [CrossRef]
- James, S.H.; Larson, K.B.; Acosta, E.P.; Prichard, M.N. Helicase-Primase as a Target of New Therapies for Herpes Simplex Virus Infections. Clin. Pharmacol. Ther. 2015, 97, 66–78. [Google Scholar] [CrossRef]
- Rodríguez, M.C.; Dybas, J.M.; Hughes, J.; Weitzman, M.D.; Boutell, C. The HSV-1 ubiquitin ligase ICP0: Modifying the cellular proteome to promote infection. Virus Res. 2020, 285, 198015. [Google Scholar] [CrossRef]
- Majmudar, H.; Hao, M.; Sankaranarayanan, N.V.; Zanotti, B.; Volin, M.V.; Desai, U.R.; Tiwari, V. A synthetic glycosaminoglycan mimetic blocks HSV-1 infection in human iris stromal cells. Antivir. Res. 2019, 161, 154–162. [Google Scholar] [CrossRef]
- Wu, J.; Power, H.; Miranda-Saksena, M.; Valtchev, P.; Schindeler, A.; Cunningham, A.L.; Dehghani, F. Identifying HSV-1 Inhibitors from Natural Compounds via Virtual Screening Targeting Surface Glycoprotein D. Pharmaceuticals 2022, 15, 361. [Google Scholar] [CrossRef]
- Cetina-Corona, A.; López-Sánchez, U.; Salinas-Trujano, J.; Méndez-Tenorio, A.; Barrón, B.L.; Torres-Flores, J. Peptides Derived from Glycoproteins H and B of Herpes Simplex Virus Type 1 and Herpes Simplex Virus Type 2 Are Capable of Blocking Herpetic Infection in vitro. Intervirology 2016, 59, 235–242. [Google Scholar] [CrossRef]
- Hassan, S.T.S.; Šudomová, M.; Berchová-Bímová, K.; Šmejkal, K.; Echeverría, J. Psoromic Acid, a Lichen-Derived Molecule, Inhibits the Replication of HSV-1 and HSV-2, and Inactivates HSV-1 DNA Polymerase: Shedding Light on Antiherpetic Properties. Molecules 2019, 24, 2912. [Google Scholar] [CrossRef] [PubMed]
- Wald, A.; Corey, L.; Timmler, B.; Magaret, A.; Warren, T.; Tyring, S.; Johnston, C.; Kriesel, J.; Fife, K.; Galitz, L.; et al. Helicase–Primase Inhibitor Pritelivir for HSV-2 Infection. N. Engl. J. Med. 2014, 370, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Lokhande, K.B.; Doiphode, S.; Vyas, R.; Swamy, K.V. Molecular docking and simulation studies on SARS-CoV-2 Mpro reveals Mitoxantrone, Leucovorin, Birinapant, and Dynasore as potent drugs against COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 7294–7305. [Google Scholar] [CrossRef] [PubMed]
- Rai, H.; Barik, A.; Singh, Y.P.; Suresh, A.; Singh, L.; Singh, G.; Nayak, U.Y.; Dubey, V.K.; Modi, G. Molecular docking, binding mode analysis, molecular dynamics, and prediction of ADMET/toxicity properties of selective potential antiviral agents against SARS-CoV-2 main protease: An effort toward drug repurposing to combat COVID-19. Mol. Divers. 2021, 25, 1905–1927. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, W.-J.; Wang, X.-J. Broad-Spectrum Antivirals Derived from Natural Products. Viruses 2023, 15, 1100. https://doi.org/10.3390/v15051100
Tian W-J, Wang X-J. Broad-Spectrum Antivirals Derived from Natural Products. Viruses. 2023; 15(5):1100. https://doi.org/10.3390/v15051100
Chicago/Turabian StyleTian, Wen-Jun, and Xiao-Jia Wang. 2023. "Broad-Spectrum Antivirals Derived from Natural Products" Viruses 15, no. 5: 1100. https://doi.org/10.3390/v15051100
APA StyleTian, W.-J., & Wang, X.-J. (2023). Broad-Spectrum Antivirals Derived from Natural Products. Viruses, 15(5), 1100. https://doi.org/10.3390/v15051100





