Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Protein Expression and Protein Purification
2.3. NLS Peptides
2.4. Crystallization and Data Processing
2.5. Fluorescence Polarization
2.6. EMSA
2.7. Co-Immunoprecipitation (Co-IP) Assay
2.8. Western Blot
2.9. Immunofluorescence Assays
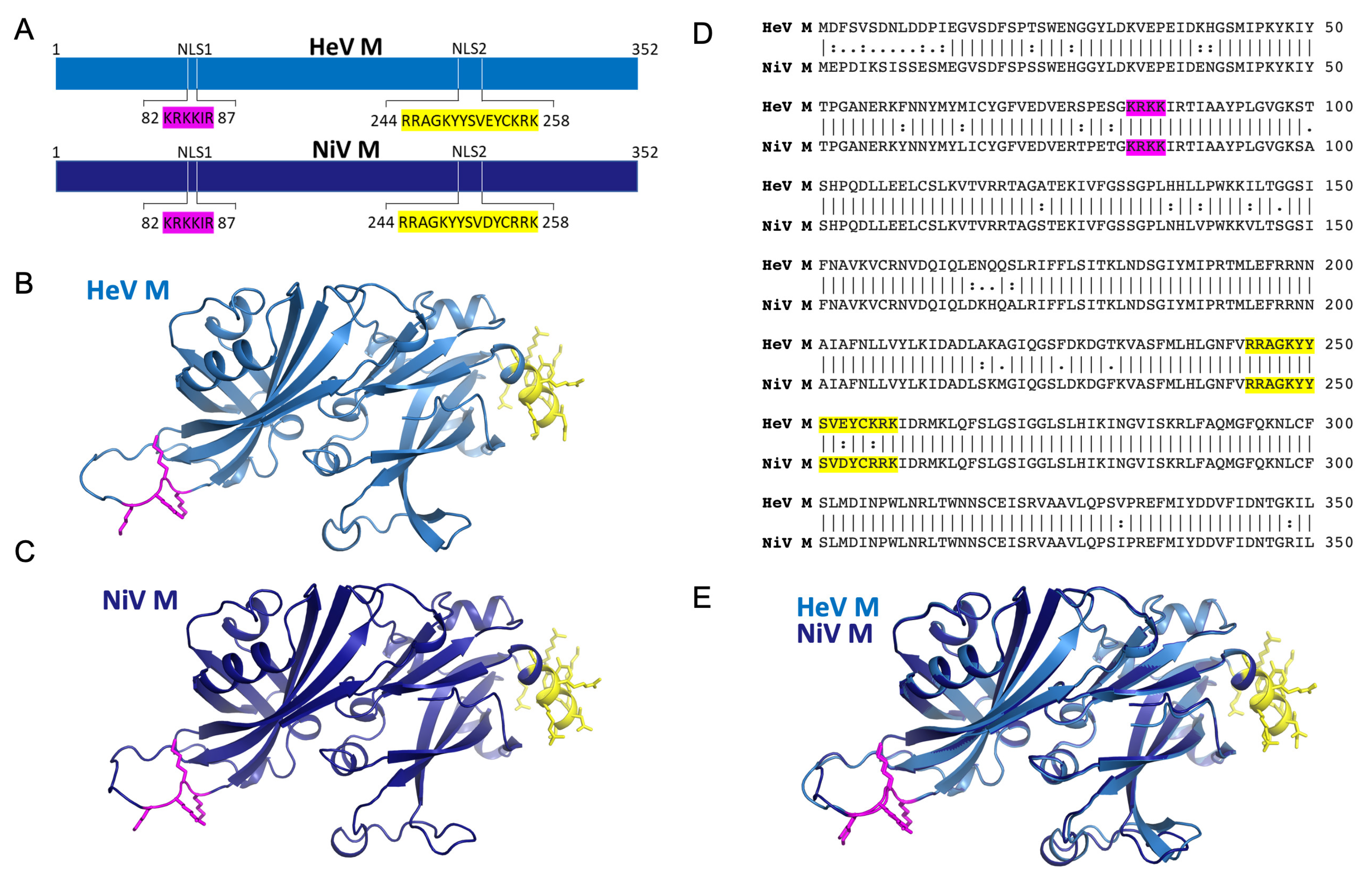
3. Results
3.1. Henipavirus M NLS1 Binds to IMPα at the Major Site
3.2. M NLS2 Binds to IMPα at the Minor Site
3.3. M NLS1 and NLS2 Bind IMPα Isoforms in Electromobility Shift Assays and Fluorescence Polarization Assays
3.4. The M NLS2 Is an Important Binding Interface in a Cellular Context
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, L.F.; Michalski, W.P.; Yu, M.; Pritchard, L.I.; Crameri, G.; Shiell, B.; Eaton, B.T. A novel P/V/C gene in a new member of the Paramyxoviridae family, which causes lethal infection in humans, horses, and other animals. J. Virol. 1998, 72, 1482–1490. [Google Scholar] [CrossRef]
- Wang, L.F.; Yu, M.; Hansson, E.; Pritchard, L.I.; Shiell, B.; Michalski, W.P.; Eaton, B.T. The exceptionally large genome of Hendra virus: Support for creation of a new genus within the family Paramyxoviridae. J. Virol. 2000, 74, 9972–9979. [Google Scholar] [CrossRef] [PubMed]
- Selvey, L.A.; Wells, R.M.; McCormack, J.G.; Ansford, A.J.; Murray, K.; Rogers, R.J.; Lavercombe, P.S.; Selleck, P.; Sheridan, J.W. Infection of humans and horses by a newly described morbillivirus. Med. J. Aust. 1995, 162, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Business Queensland. Summary of Hendra Virus Incidents in Horses. Available online: https://www.business.qld.gov.au/industries/service-industries-professionals/service-industries/veterinary-surgeons/guidelines-hendra/incident-summary (accessed on 29 September 2022).
- Broder, C.C. Henipavirus outbreaks to antivirals: The current status of potential therapeutics. Curr. Opin. Virol. 2012, 2, 176–187. [Google Scholar] [CrossRef]
- Wang, J.; Anderson, D.E.; Halpin, K.; Hong, X.; Chen, H.; Walker, S.; Valdeter, S.; van der Heide, B.; Neave, M.J.; Bingham, J.; et al. A new Hendra virus genotype found in Australian flying foxes. Virol. J. 2021, 18, 197. [Google Scholar] [CrossRef]
- Gurley, E.S.; Montgomery, J.M.; Hossain, M.J.; Bell, M.; Azad, A.K.; Islam, M.R.; Molla, M.A.; Carroll, D.S.; Ksiazek, T.G.; Rota, P.A.; et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis. 2007, 13, 1031–1037. [Google Scholar] [CrossRef]
- Ang, B.S.; Lim, T.C.; Wang, L. Nipah virus infection. J. Clin. Microbiol. 2018, 56, e01875-17. [Google Scholar] [CrossRef]
- Lo, M.K.; Rota, P.A. The emergence of Nipah virus, a highly pathogenic paramyxovirus. J. Clin. Virol. 2008, 43, 396–400. [Google Scholar] [CrossRef] [PubMed]
- AbuBakar, S.; Chang, L.-Y.; Ali, A.M.; Sharifah, S.; Yusoff, K.; Zamrod, Z. Isolation and molecular identification of Nipah virus from pigs. Emerg. Infect. Dis. 2004, 10, 2228. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, B.H.; Lowe, L.; Tamin, A.; Liu, X.; Bankamp, B.; Bowden, N.; Rollin, P.E.; Comer, J.A.; Ksiazek, T.G.; Hossain, M.J. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg. Infect. Dis. 2005, 11, 1594. [Google Scholar] [CrossRef] [PubMed]
- Arunkumar, G.; Chandni, R.; Mourya, D.T.; Singh, S.K.; Sadanandan, R.; Sudan, P.; Bhargava, B.; People, N.I.; Group, H.S. Outbreak Investigation of Nipah Virus Disease in Kerala, India, 2018. J. Infect. Dis. 2018, 219, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Arankalle, V.A.; Bandyopadhyay, B.T.; Ramdasi, A.Y.; Jadi, R.; Patil, D.R.; Rahman, M.; Majumdar, M.; Banerjee, P.S.; Hati, A.K.; Goswami, R.P.; et al. Genomic characterization of Nipah virus, West Bengal, India. Emerg. Infect. Dis. 2011, 17, 907–909. [Google Scholar] [CrossRef]
- Chadha, M.S.; Comer, J.A.; Lowe, L.; Rota, P.A.; Rollin, P.E.; Bellini, W.J.; Ksiazek, T.G.; Mishra, A. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 2006, 12, 235–240. [Google Scholar] [CrossRef]
- Madera, S.; Kistler, A.; Ranaivoson, H.C.; Ahyong, V.; Andrianiaina, A.; Andry, S.; Raharinosy, V.; Randriambolamanantsoa, T.H.; Fifi Ravelomanantsoa, N.A.; Tato, C.M.; et al. Discovery and Genomic Characterization of a Novel Henipavirus, Angavokely virus, from fruit bats in Madagascar. J. Virol. 2022, 96, e00921-22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.A.; Li, H.; Jiang, F.C.; Zhu, F.; Zhang, Y.F.; Chen, J.J.; Tan, C.W.; Anderson, D.E.; Fan, H.; Dong, L.Y.; et al. A Zoonotic Henipavirus in Febrile Patients in China. N. Engl. J. Med. 2022, 387, 470–472. [Google Scholar] [CrossRef] [PubMed]
- Hernández, L.H.A.; da Paz, T.Y.B.; Silva, S.P.d.; Silva, F.S.d.; Barros, B.C.V.d.; Nunes, B.T.D.; Casseb, L.M.N.; Medeiros, D.B.A.; Vasconcelos, P.F.d.C.; Cruz, A.C.R. First Genomic Evidence of a Henipa-like Virus in Brazil. Viruses 2022, 14, 2167. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Ciancanelli, M.J.; Basler, C.F. Mutation of YMYL in the Nipah Virus Matrix Protein Abrogates Budding and Alters Subcellular Localization. J. Virol. 2006, 80, 12070–12078. [Google Scholar] [CrossRef]
- Dietzel, E.; Kolesnikova, L.; Sawatsky, B.; Heiner, A.; Weis, M.; Kobinger, G.P.; Becker, S.; von Messling, V.; Maisner, A. Nipah virus matrix protein influences fusogenicity and is essential for particle infectivity and stability. J. Virol. 2016, 90, 2514–2522. [Google Scholar] [CrossRef]
- Norris, M.J.; Husby, M.L.; Kiosses, W.B.; Yin, J.; Saxena, R.; Rennick, L.J.; Heiner, A.; Harkins, S.S.; Pokhrel, R.; Schendel, S.L.; et al. Measles and Nipah virus assembly: Specific lipid binding drives matrix polymerization. Sci. Adv. 2022, 8, eabn1440. [Google Scholar] [CrossRef]
- Monaghan, P.; Green, D.; Pallister, J.; Klein, R.; White, J.; Williams, C.; McMillan, P.; Tilley, L.; Lampe, M.; Hawes, P.; et al. Detailed morphological characterisation of Hendra virus infection of different cell types using super-resolution and conventional imaging. Virol. J. 2014, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.E.; Park, A.; Lake, M.; Pentecost, M.; Torres, B.; Yun, T.E.; Wolf, M.C.; Holbrook, M.R.; Freiberg, A.N.; Lee, B. Ubiquitin-Regulated Nuclear-Cytoplasmic Trafficking of the Nipah Virus Matrix Protein Is Important for Viral Budding. PLoS Pathog. 2010, 6, e1001186. [Google Scholar] [CrossRef] [PubMed]
- McLinton, E.C.; Wagstaff, K.M.; Lee, A.; Moseley, G.W.; Marsh, G.A.; Wang, L.-F.; Jans, D.A.; Lieu, K.G.; Netter, H.J. Nuclear localization and secretion competence are conserved among henipavirus matrix proteins. J. Gen. Virol. 2017, 98, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Pentecost, M.; Vashisht, A.A.; Lester, T.; Voros, T.; Beaty, S.M.; Park, A.; Wang, Y.E.; Yun, T.E.; Freiberg, A.N.; Wohlschlegel, J.A.; et al. Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Paramyxovirinae Matrix Proteins. PLoS Pathog. 2015, 11, e1004739. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, S.; Zhao, T.; Rozario, A.; Rootes, C.; McMillan, P.; Purcell, A.; Woon, A.; Marsh, G.; Lieu, K.; Wang, L. Viral regulation of host cell biology by hijacking of the nucleolar DNA-damage response. Nat. Commun. 2018, 9, 3057. [Google Scholar] [CrossRef]
- Görlich, D.; Prehn, S.; Laskey, R.A.; Hartmann, E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 1994, 79, 767–778. [Google Scholar] [CrossRef]
- Moroianu, J.; Blobel, G.; Radu, A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc. Natl. Acad. Sci. USA 1995, 92, 2008–2011. [Google Scholar] [CrossRef]
- Weis, K.; Mattaj, L.W.; Lamond, A. Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science 1995, 268, 1049–1053. [Google Scholar] [CrossRef]
- Chi, N.C.; Adam, E.J.; Adam, S.A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell. Biol. 1995, 130, 265–274. [Google Scholar] [CrossRef]
- Fontes, M.R.M.; Teh, T.; Kobe, B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α11Edited by K. Nagai. J. Mol. Biol. 2000, 297, 1183–1194. [Google Scholar] [CrossRef]
- Graham, S.C.; Assenberg, R.; Delmas, O.; Verma, A.; Gholami, A.; Talbi, C.; Owens, R.J.; Stuart, D.I.; Grimes, J.M.; Bourhy, H. Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association. PLoS Pathog. 2008, 4, e1000251. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sankhala, R.S.; Florio, T.J.; Zhou, L.; Nguyen, N.L.T.; Lokareddy, R.K.; Cingolani, G.; Panté, N. Synergy of two low-affinity NLSs determines the high avidity of influenza A virus nucleoprotein NP for human importin α isoforms. Sci. Rep. 2017, 7, 11381. [Google Scholar] [CrossRef] [PubMed]
- Pumroy, R.A.; Ke, S.; Hart, D.J.; Zachariae, U.; Cingolani, G. Molecular Determinants for Nuclear Import of Influenza A PB2 by Importin α Isoforms 3 and 7. Structure 2015, 23, 374–384. [Google Scholar] [CrossRef]
- Nakada, R.; Matsuura, Y. Crystal structure of importin-α bound to the nuclear localization signal of Epstein-Barr virus EBNA-LP protein. Protein Sci. 2017, 26, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Ng, I.H.W.; Chan, K.W.-K.; Tan, M.J.A.; Gwee, C.P.; Smith, K.M.; Jeffress, S.J.; Saw, W.-G.; Swarbrick, C.M.D.; Watanabe, S.; Jans, D.A.; et al. Zika Virus NS5 Forms Supramolecular Nuclear Bodies That Sequester Importin-α and Modulate the Host Immune and Pro-Inflammatory Response in Neuronal Cells. ACS Infect. Dis. 2019, 5, 932–948. [Google Scholar] [CrossRef]
- Tay, M.Y.F.; Smith, K.; Ng, I.H.W.; Chan, K.W.K.; Zhao, Y.; Ooi, E.E.; Lescar, J.; Luo, D.; Jans, D.A.; Forwood, J.K.; et al. The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production. PLoS Pathog. 2016, 12, e1005886. [Google Scholar] [CrossRef]
- Munasinghe, T.S.; Edwards, M.R.; Tsimbalyuk, S.; Vogel, O.A.; Smith, K.M.; Stewart, M.; Foster, J.K.; Bosence, L.A.; Aragão, D.; Roby, J.A.; et al. MERS-CoV ORF4b employs an unusual binding mechanism to target IMPα and block innate immunity. Nat. Commun. 2022, 13, 1604. [Google Scholar] [CrossRef]
- Liu, Y.C.; Grusovin, J.; Adams, T.E. Electrostatic interactions between hendra virus matrix proteins are required for efficient virus-like-particle assembly. J. Virol. 2018, 92, e00143-18. [Google Scholar] [CrossRef]
- Reid, S.P.; Valmas, C.; Martinez, O.; Sanchez, F.M.; Basler, C.F. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin α proteins with activated STAT1. J. Virol. 2007, 81, 13469–13477. [Google Scholar] [CrossRef]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Aragão, D.; Aishima, J.; Cherukuvada, H.; Clarken, R.; Clift, M.; Cowieson, N.P.; Ericsson, D.J.; Gee, C.L.; Macedo, S.; Mudie, N.; et al. MX2: A high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 2018, 25, 885–891. [Google Scholar] [CrossRef]
- Battye, T.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Cryst. D Biol. Cryst. 2011, 67 Pt 4, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Evans, P. Scaling and assessment of data quality. Acta Cryst. D Biol. Cryst. 2006, 62 Pt 1, 72–82. [Google Scholar] [CrossRef]
- McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Cryst. D Biol. Cryst. 2007, 63 Pt 1, 32–41. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D Biol. Cryst. 2010, 66 Pt 2, 213–221. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst. D Biol. Cryst. 2012, 68 Pt 4, 352–367. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Cryst. D Biol. Cryst. 2004, 60 Pt 12, 2126–2132. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Alvisi, G.; Manaresi, E.; Cross, E.M.; Hoad, M.; Akbari, N.; Pavan, S.; Ariawan, D.; Bua, G.; Petersen, G.F.; Forwood, J.; et al. Importin alpha/beta-dependent nuclear transport of human parvovirus B19 nonstructural protein 1 is essential for viral replication. Antivir. Res. 2023, 213, 105588. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Tsimbalyuk, S.; Edwards, M.R.; Cross, E.M.; Batra, J.; Soares da Costa, T.P.; Aragão, D.; Basler, C.F.; Forwood, J.K. Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses. Nat. Commun. 2018, 9, 3703. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.L.; Cardenas, W.B.; Zamarin, D.; Palese, P.; Basler, C.F. Nuclear Localization of the Nipah Virus W Protein Allows for Inhibition of both Virus- and Toll-Like Receptor 3-Triggered Signaling Pathways. J. Virol. 2005, 79, 6078–6088. [Google Scholar] [CrossRef] [PubMed]
- Jagga, B.; Edwards, M.; Pagin, M.; Wagstaff, K.M.; Aragão, D.; Roman, N.; Nanson, J.D.; Raidal, S.R.; Dominado, N.; Stewart, M.; et al. Structural basis for nuclear import selectivity of pioneer transcription factor SOX2. Nat. Commun. 2021, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Deffrasnes, C.; Marsh, G.A.; Foo, C.H.; Rootes, C.L.; Gould, C.M.; Grusovin, J.; Monaghan, P.; Lo, M.K.; Tompkins, S.M.; Adams, T.E. Genome-wide siRNA screening at biosafety level 4 reveals a crucial role for fibrillarin in henipavirus infection. PLoS Pathog. 2016, 12, e1005478. [Google Scholar] [CrossRef]
- Bauer, A.; Neumann, S.; Karger, A.; Henning, A.-K.; Maisner, A.; Lamp, B.; Dietzel, E.; Kwasnitschka, L.; Balkema-Buschmann, A.; Keil, G.M. ANP32B is a nuclear target of henipavirus M proteins. PLoS ONE 2014, 9, e97233. [Google Scholar] [CrossRef]
- Patch, J.R.; Han, Z.; McCarthy, S.E.; Yan, L.; Wang, L.F.; Harty, R.N.; Broder, C.C. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol. J. 2008, 5, 137. [Google Scholar] [CrossRef]
- Rawlinson, S.M.; Zhao, T.; Ardipradja, K.; Zhang, Y.; Veugelers, P.F.; Harper, J.A.; David, C.T.; Sundaramoorthy, V.; Moseley, G.W. Henipaviruses and lyssaviruses target nucleolar treacle protein and regulate ribosomal RNA synthesis. Traffic 2023, 24, 146–157. [Google Scholar] [CrossRef]
- Chang, C.-W.; Couñago, R.M.; Williams, S.J.; Bodén, M.; Kobe, B. Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-α. Traffic 2013, 14, 1144–1154. [Google Scholar] [CrossRef]
- Marfori, M.; Mynott, A.; Ellis, J.J.; Mehdi, A.M.; Saunders, N.F.W.; Curmi, P.M.; Forwood, J.K.; Bodén, M.; Kobe, B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2011, 1813, 1562–1577. [Google Scholar] [CrossRef]
- Lott, K.; Bhardwaj, A.; Sims, P.J.; Cingolani, G. A Minimal Nuclear Localization Signal (NLS) in Human Phospholipid Scramblase 4 That Binds Only the Minor NLS-binding Site of Importin α1*. J. Biol. Chem. 2011, 286, 28160–28169. [Google Scholar] [CrossRef]
- Nakada, R.; Hirano, H.; Matsuura, Y. Structure of importin-α bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci. Rep. 2015, 5, 15055. [Google Scholar] [CrossRef] [PubMed]
- Mynott, A.V.; Harrop, S.J.; Brown, L.J.; Breit, S.N.; Kobe, B.; Curmi, P.M.G. Crystal structure of importin-α bound to a peptide bearing the nuclear localisation signal from chloride intracellular channel protein 4. FEBS J. 2011, 278, 1662–1675. [Google Scholar] [CrossRef] [PubMed]
- Selinger, M.; Novotný, R.; Sýs, J.; Roby, J.A.; Tykalová, H.; Ranjani, G.S.; Vancová, M.; Jaklová, K.; Kaufman, F.; Bloom, M.E.; et al. Tick-borne encephalitis virus capsid protein induces translational shutoff as revealed by its structural-biological analysis. J. Biol. Chem. 2022, 298, 102585. [Google Scholar] [CrossRef] [PubMed]
- Bharaj, P.; Wang, Y.E.; Dawes, B.E.; Yun, T.E.; Park, A.; Yen, B.; Basler, C.F.; Freiberg, A.N.; Lee, B.; Rajsbaum, R. The Matrix Protein of Nipah Virus Targets the E3-Ubiquitin Ligase TRIM6 to Inhibit the IKKε Kinase-Mediated Type-I IFN Antiviral Response. PLoS Pathog. 2016, 12, e1005880. [Google Scholar] [CrossRef]
- Smith, K.M.; Himiari, Z.; Tsimbalyuk, S.; Forwood, J.K. Structural Basis for Importin-α Binding of the Human Immunodeficiency Virus Tat. Sci. Rep. 2017, 7, 1650. [Google Scholar] [CrossRef]
- Róna, G.; Marfori, M.; Borsos, M.; Scheer, I.; Takács, E.; Tóth, J.; Babos, F.; Magyar, A.; Erdei, A.; Bozóky, Z.; et al. Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: Structural and mechanistic insights. Acta Cryst. D Biol. Cryst. 2013, 69 Pt 12, 2495–2505. [Google Scholar] [CrossRef]
- Battisti, A.J.; Meng, G.; Winkler, D.C.; McGinnes, L.W.; Plevka, P.; Steven, A.C.; Morrison, T.G.; Rossmann, M.G. Structure and assembly of a paramyxovirus matrix protein. Proc. Natl. Acad. Sci. USA 2012, 109, 13996–14000. [Google Scholar] [CrossRef]
- Duan, Z.; Xu, H.; Ji, X.; Zhao, J.; Xu, H.; Hu, Y.; Deng, S.; Hu, S.; Liu, X. Importin α5 negatively regulates importin β1-mediated nuclear import of Newcastle disease virus matrix protein and viral replication and pathogenicity in chicken fibroblasts. Virulence 2018, 9, 783–803. [Google Scholar] [CrossRef]



| Data Collection and Processing | IMPα2:HeV M NLS1 | IMPα2:HeV M NLS2 | IMPα3:HeV M NLS1 |
|---|---|---|---|
| Wavelength (Å) | 0.9537 | 0.9537 | 0.9537 |
| Resolution range (Å) | 24.42–1.9 (1.94–1.9) | 19.78–2.10 (2.16–2.10) | 29.78–2.75 (2.9–2.75) |
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Unit cell (Å, o) | 78.49 89.86 99.83 90 90 90 | 78.08 89.50 97.06, 90 90 90 | 49.06 64.27 158.58 90 90 90 |
| Total reflections | 405,486 (26,486) | 167,351 (12,946) | 154,985 (23,139) |
| Unique reflections | 56,298 (3749) | 40,357 (3253) | 13,681 (1966) |
| Multiplicity | 7.2 (7.1) | 4.1 (4.0) | (11.8) |
| Completeness (%) | 99.9 (100) | 99.9 (99.9) | 99.9 (100) |
| Mean I/sigma (I) | 11.9 (1.5) | 11.0 (1.9) | 7.8 (2.3) |
| Wilson B-factor Å2 | 29.65 | 35.12 | 55.58 |
| R-merge | 0.085 (1.376) | 0.063 (0.696) | 0.181 (1.032) |
| R-pim | 0.050 (0.820) | 0.035 (0.400) | 0.078 (0.446) |
| CC1/2 | 0.998 (0.602 | 0.998 (0.734) | 0.995 (0.887) |
| Refinement | |||
| Number of reflections | 56,237 (5567) | 40,289 (3941) | 13,635 (1323) |
| Number of R-free reflections | 2864 (322)) | 1981(180) | 671 (80) |
| R-work % | 0.1728 (0.2853) | 0.1791 (0.2520) | 0.2411 (0.3185) |
| R-free % | 0.1900 (0.3147) | 0.2053 (0.3058) | 0.2811 (0.3553) |
| RMS (bonds) | 0.015 | 0.006 | 0.003 |
| RMS (angles) | 1.18 | 0.77 | 0.58 |
| Ramachandran | |||
| favored (%) | 98.6 | 98.14 | 97.85 |
| allowed (%) | 1.4 | 1.86 | 2.15 |
| outliers (%) | 0.00 | 0.00 | 0.00 |
| Average B-factor Å2 | 43.57 | 50.33 | 72.53 |
| Clash score | 1.54 | 4.07 | 4.92 |
| PDB code | 8FUA | 8FUC | 8FUB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donnelly, C.M.; Vogel, O.A.; Edwards, M.R.; Taylor, P.E.; Roby, J.A.; Forwood, J.K.; Basler, C.F. Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism. Viruses 2023, 15, 1302. https://doi.org/10.3390/v15061302
Donnelly CM, Vogel OA, Edwards MR, Taylor PE, Roby JA, Forwood JK, Basler CF. Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism. Viruses. 2023; 15(6):1302. https://doi.org/10.3390/v15061302
Chicago/Turabian StyleDonnelly, Camilla M., Olivia A. Vogel, Megan R. Edwards, Paige E. Taylor, Justin A. Roby, Jade K. Forwood, and Christopher F. Basler. 2023. "Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism" Viruses 15, no. 6: 1302. https://doi.org/10.3390/v15061302
APA StyleDonnelly, C. M., Vogel, O. A., Edwards, M. R., Taylor, P. E., Roby, J. A., Forwood, J. K., & Basler, C. F. (2023). Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism. Viruses, 15(6), 1302. https://doi.org/10.3390/v15061302







