An Overview of the Conventional and Novel Methods Employed for SARS-CoV-2 Neutralizing Antibody Measurement
Abstract
1. Introduction
2. Methods of Neutralizing Antibody Measurement
2.1. Plaque Reduction Neutralization Test—The Gold Standard Assay
2.2. Pseudovirus Neutralization Test
2.3. Foci Reduction Neutralization Test
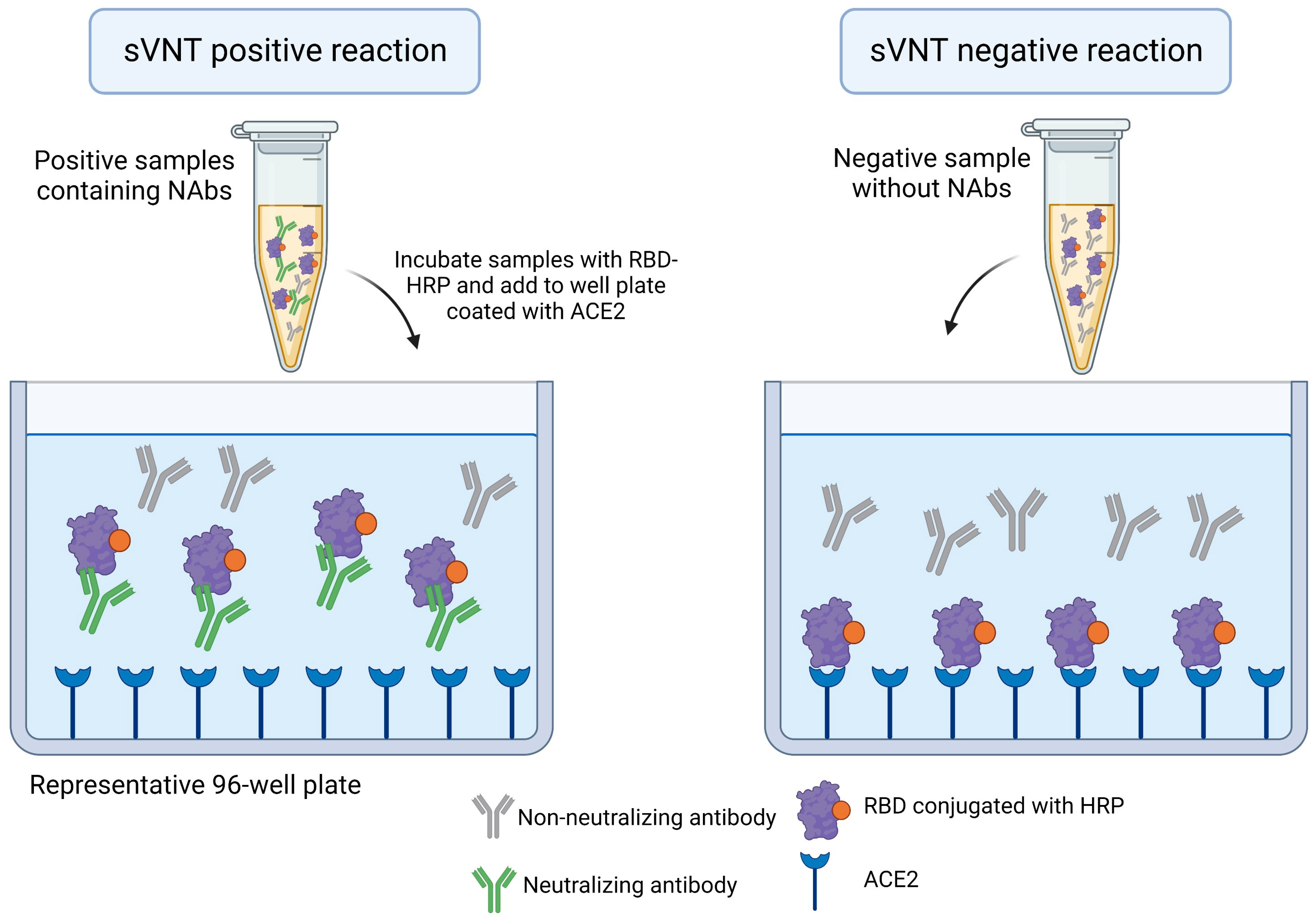
2.4. Surrogate Virus Neutralization Test—An ELISA-Based Approach
3. Innovative Strategies for the Detection and Quantification of SARS-CoV-2 Neutralizing Antibodies
4. Perspectives and Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard|WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. Available online: https://covid19.who.int/ (accessed on 17 April 2023).
- Chen, P.; Nirula, A.; Heller, B.; Gottlieb, R.L.; Boscia, J.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with COVID-19. N. Engl. J. Med. 2021, 384, 229–237. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Vanderheiden, A.; Edara, V.V.; Floyd, K.; Kauffman, R.C.; Mantus, G.; Anderson, E.; Rouphael, N.; Edupuganti, S.; Shi, P.Y.; Menachery, V.D.; et al. Development of a Rapid Focus Reduction Neutralization Test Assay for Measuring SARS-CoV-2 Neutralizing Antibodies. Curr. Protoc. Immunol. 2020, 131, e116. [Google Scholar] [CrossRef]
- Machado, B.A.S.; Hodel, K.V.S.; Fonseca, L.M.D.S.; Pires, V.C.; Mascarenhas, L.A.B.; da Silva Andrade, L.P.C.; Moret, M.A.; Badaró, R. The Importance of Vaccination in the Context of the COVID-19 Pandemic: A Brief Update Regarding the Use of Vaccines. Vaccines 2022, 10, 591. [Google Scholar] [CrossRef]
- Castro Dopico, X.; Ols, S.; Loré, K.; Karlsson Hedestam, G.B. Immunity to SARS-CoV-2 Induced by Infection or Vaccination. J. Intern. Med. 2022, 291, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing Antibody Levels Are Highly Predictive of Immune Protection from Symptomatic SARS-CoV-2 Infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Counoupas, C.; Pino, P.; Stella, A.O.; Ashley, C.; Lukeman, H.; Bhattacharyya, N.D.; Tada, T.; Anchisi, S.; Metayer, C.; Martinis, J.; et al. High-Titer Neutralizing Antibodies against the SARS-CoV-2 Delta Variant Induced by Alhydroxyquim-II-Adjuvanted Trimeric Spike Antigens. Microbiol. Spectr. 2022, 10, e01695-21. [Google Scholar] [CrossRef]
- Liu, K.T.; Han, Y.J.; Wu, G.H.; Huang, K.Y.A.; Huang, P.N. Overview of Neutralization Assays and International Standard for Detecting SARS-CoV-2 Neutralizing Antibody. Viruses 2022, 14, 1560. [Google Scholar] [CrossRef]
- Baum, A.; Ajithdoss, D.; Copin, R.; Zhou, A.; Lanza, K.; Negron, N.; Ni, M.; Wei, Y.; Mohammadi, K.; Musser, B.; et al. REGN-COV2 Antibodies Prevent and Treat SARS-CoV-2 Infection in Rhesus Macaques and Hamsters. Science 2020, 370, 1110–1115. [Google Scholar] [CrossRef] [PubMed]
- Morales-Núñez, J.J.; Muñoz-Valle, J.F.; Torres-Hernández, P.C.; Hernández-Bello, J. Overview of Neutralizing Antibodies and Their Potential in COVID-19. Vaccines 2021, 9, 1376. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Cromer, D.; Steain, M.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Kent, S.J.; Triccas, J.A.; Khoury, D.S.; Davenport, M.P. Neutralising Antibody Titres as Predictors of Protection against SARS-CoV-2 Variants and the Impact of Boosting: A Meta-Analysis. Lancet Microbe 2022, 3, e52–e61. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Pérez-Then, E.; Lucas, C.; Monteiro, V.S.; Miric, M.; Brache, V.; Cochon, L.; Vogels, C.B.F.; Malik, A.A.; De la Cruz, E.; Jorge, A.; et al. Neutralizing Antibodies against the SARS-CoV-2 Delta and Omicron Variants Following Heterologous CoronaVac plus BNT162b2 Booster Vaccination. Nat. Med. 2022, 28, 481–485. [Google Scholar] [CrossRef]
- Looi, M.K. What Do We Know about the Arcturus XBB.1.16 Subvariant? BMJ 2023, 381, p1074. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.C.; Lu, R.M.; Su, S.C.; Chiang, P.Y.; Ko, S.H.; Ke, F.Y.; Liang, K.H.; Hsieh, T.Y.; Wu, H.C. Monoclonal Antibodies for COVID-19 Therapy and SARS-CoV-2 Detection. J. Biomed. Sci. 2022, 29, 1. [Google Scholar] [CrossRef]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.J. A Human Monoclonal Antibody Blocking SARS-CoV-2 Infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Baral, P.K.; Yin, J.; James, M.N.G. Treatment and Prevention Strategies for the COVID 19 Pandemic: A Review of Immunotherapeutic Approaches for Neutralizing SARS-CoV-2. Int. J. Biol. Macromol. 2021, 186, 490–500. [Google Scholar] [CrossRef]
- Self, W.H.; Sandkovsky, U.; Reilly, C.S.; Vock, D.M.; Gottlieb, R.L.; Mack, M.; Golden, K.; Dishner, E.; Vekstein, A.; Ko, E.R.; et al. Efficacy and Safety of Two Neutralising Monoclonal Antibody Therapies, Sotrovimab and BRII-196 plus BRII-198, for Adults Hospitalised with COVID-19 (TICO): A Randomised Controlled Trial. Lancet Infect. Dis. 2022, 22, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.J.; Lennette, E.H. Virology on the Bookshelf. Application of Tissue Culture Technics to Diagnostic Virology in the Public Health Laboratory. Am. J. Public. Health Nations Health 1961, 51, 511–516. [Google Scholar] [CrossRef]
- Valcourt, E.J.; Manguiat, K.; Robinson, A.; Lin, Y.-C.; Abe, K.T.; Mubareka, S.; Shigayeva, A.; Zhong, Z.; Girardin, R.C.; DuPuis, A.; et al. Evaluating Humoral Immunity against SARS-CoV-2: Validation of a Plaque-Reduction Neutralization Test and a Multilaboratory Comparison of Conventional and Surrogate Neutralization Assays. Microbiol. Spectr. 2021, 9, e0088621. [Google Scholar] [CrossRef]
- Russell, P.K.; Nisalak, A.; Sukhavachana, P.; Vivona, S. A Plaque Reduction Test for Dengue Virus Neutralizing Antibodies. J. Immunol. 1967, 99, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.J.; Nisalak, A.; Anderson, K.B.; Libraty, D.H.; Kalayanarooj, S.; Vaughn, D.W.; Putnak, R.; Gibbons, R.V.; Jarman, R.; Endy, T.P. Dengue Plaque Reduction Neutralization Test (PRNT) in Primary and Secondary Dengue Virus Infections: How Alterations in Assay Conditions Impact Performance. Am. J. Trop. Med. Hyg. 2009, 81, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, E.J.; Manguiat, K.; Wood, H.; Drebot, M. Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020, 57, ecpmc105. [Google Scholar] [CrossRef]
- Maeda, A.; Maeda, J. Review of Diagnostic Plaque Reduction Neutralization Tests for Flavivirus Infection. Vet. J. 2013, 195, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Bewley, K.R.; Coombes, N.S.; Gagnon, L.; McInroy, L.; Baker, N.; Shaik, I.; St-Jean, J.R.; St-Amant, N.; Buttigieg, K.R.; Humphries, H.E.; et al. Quantification of SARS-CoV-2 Neutralizing Antibody by Wild-Type Plaque Reduction Neutralization, Microneutralization and Pseudotyped Virus Neutralization Assays. Nat. Protoc. 2021, 16, 3114–3140. [Google Scholar] [CrossRef]
- Shen, C.; Wang, Z.; Zhao, F.; Yang, Y.; Li, J.; Yuan, J.; Wang, F.; Li, D.; Yang, M.; Xing, L.; et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA 2020, 323, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.A.; Ganesan, S.; Naik, S.; Bissar, S.; Al Zamel, I.; Warren, K.; Zaher, W.A.; Khan, G. Serological Assays for Assessing Postvaccination SARS-CoV-2 Antibody Response. Microbiol. Spectr. 2021, 9, e00733-21. [Google Scholar] [CrossRef]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 MRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II Study of COVID-19 RNA Vaccine BNT162b1 in Adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and Immunogenicity of the ChAdOx1 NCoV-19 Vaccine against SARS-CoV-2: A Preliminary Report of a Phase 1/2, Single-Blind, Randomised Controlled Trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Leung, N.H.L.; Cheng, S.M.S.; Martín-Sánchez, M.; Au, N.Y.M.; Ng, Y.Y.; Luk, L.L.H.; Chan, K.C.K.; Li, J.K.C.; Leung, Y.W.Y.; Tsang, L.C.H.; et al. Immunogenicity of a Third Dose of BNT162b2 to Ancestral Severe Acute Respiratory Syndrome Coronavirus 2 and the Omicron Variant in Adults Who Received 2 Doses of Inactivated Vaccine. Clin. Infect. Dis. 2023, 76, e299–e307. [Google Scholar] [CrossRef]
- Sanders, J.-S.F.; Messchendorp, A.L.; de Vries, R.D.; Baan, C.C.; van Baarle, D.; van Binnendijk, R.; Diavatopoulos, D.A.; Geers, D.; Schmitz, K.S.; GeurtsvanKessel, C.H.; et al. Antibody and T-Cell Responses 6 Months after Coronavirus Disease 2019 Messenger RNA-1273 Vaccination in Patients with Chronic Kidney Disease, on Dialysis, or Living with a Kidney Transplant. Clin. Infect. Dis. 2023, 76, e188–e199. [Google Scholar] [CrossRef] [PubMed]
- Valim, V.; Martins-Filho, O.A.; Gouvea, M.d.P.G.; Camacho, L.A.B.; Villela, D.A.M.; de Lima, S.M.B.; Azevedo, A.S.; Neto, L.F.P.; Domingues, C.M.A.S.; de Medeiros Junior, N.F.; et al. Effectiveness, Safety, and Immunogenicity of Half Dose ChAdOx1 NCoV-19 COVID-19 Vaccine: Viana Project. Front. Immunol. 2022, 13, 966416. [Google Scholar] [CrossRef]
- Banga Ndzouboukou, J.-L.; Zhang, Y.-D.; Fan, X.-L. Recent Developments in SARS-CoV-2 Neutralizing Antibody Detection Methods. Curr. Med. Sci. 2021, 41, 1052–1064. [Google Scholar] [CrossRef]
- Crawford, K.H.D.; Eguia, R.; Dingens, A.S.; Loes, A.N.; Bloom, J.D.; Crawford, K. Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Protocols.io 2021. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell 2021, 184, 2372–2383.e9. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, H.; Wang, M.; et al. Establishment and Validation of a Pseudovirus Neutralization Assay for SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 680–686. [Google Scholar] [CrossRef]
- Case, J.B.; Rothlauf, P.W.; Chen, R.E.; Liu, Z.; Zhao, H.; Kim, A.S.; Bloyet, L.M.; Zeng, Q.; Tahan, S.; Droit, L.; et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe 2020, 28, 475–485.e5. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Tran, J.T.; Peng, L.; Yang, L.; Suhandynata, R.T.; Hoffman, M.A.; Zhao, F.; Song, G.; He, W.; Limbo, O.; et al. A Rapid Assay for SARS-CoV-2 Neutralizing Antibodies That Is Insensitive to Antiretroviral Drugs. J. Immunol. 2021, 207, 344–351. [Google Scholar] [CrossRef]
- Powell, S.K.; Artlip, M.; Kaloss, M.; Brazinski, S.; Lyons, R.; McGarrity, G.J.; Otto, E. Efficacy of Antiretroviral Agents against Murine Replication-Competent Retrovirus Infection in Human Cells. J. Virol. 1999, 73, 8813–8816. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; St Denis, K.J.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. MRNA-Based COVID-19 Vaccine Boosters Induce Neutralizing Immunity against SARS-CoV-2 Omicron Variant. Cell 2022, 185, 457–466.e4. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Lam, E.C.; Astudillo, M.G.; Yang, D.; Miller, T.E.; Feldman, J.; Hauser, B.M.; Caradonna, T.M.; Clayton, K.L.; Nitido, A.D.; et al. COVID-19-Neutralizing Antibodies Predict Disease Severity and Survival. Cell 2021, 184, 476–488.e11. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Xia, H.; Shi, P.Y.; Xie, X.; Ren, P. A Single-Round Infection Fluorescent SARS-CoV-2 Neutralization Test for COVID-19 Serological Testing at a Biosafety Level-2 Laboratory. Viruses 2022, 14, 1211. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Lui, B.G.; Wallisch, A.K.; Bacher, M.; Mühl, J.; Reinholz, J.; Ozhelvaci, O.; Beckmann, N.; Garcia, R.d.l.C.G.; Poran, A.; et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 MRNA Vaccine-Elicited Human Sera. Science 2022, 375, 678–680. [Google Scholar] [CrossRef]
- Barrett, J.R.; Belij-Rammerstorfer, S.; Dold, C.; Ewer, K.J.; Folegatti, P.M.; Gilbride, C.; Halkerston, R.; Hill, J.; Jenkin, D.; Stockdale, L.; et al. Phase 1/2 Trial of SARS-CoV-2 Vaccine ChAdOx1 NCoV-19 with a Booster Dose Induces Multifunctional Antibody Responses. Nat. Med. 2021, 27, 279–288. [Google Scholar] [CrossRef]
- Niyomnaitham, S.; Jongkaewwattana, A.; Meesing, A.; Pinpathomrat, N.; Nanthapisal, S.; Hirankarn, N.; Siwamogsatham, S.; Kirdlarp, S.; Chaiwarith, R.; Lawpoolsri, S.; et al. Immunogenicity of a Fractional or Full Third Dose of AZD1222 Vaccine or BNT162b2 Messenger RNA Vaccine after Two Doses of CoronaVac Vaccines against the Delta and Omicron Variants. Int. J. Infect. Dis. 2023, 129, 19–31. [Google Scholar] [CrossRef]
- Chantasrisawad, N.; Techasaensiri, C.; Kosalaraksa, P.; Phongsamart, W.; Tangsathapornpong, A.; Jaru-Ampornpan, P.; Sophonphan, J.; Suntarattiwong, P.; Puthanakit, T. The Immunogenicity of an Extended Dosing Interval of BNT162b2 against SARS-CoV-2 Omicron Variant among Healthy School-Aged Children, a Randomized Controlled Trial. Int. J. Infect. Dis. 2023, 130, 52–59. [Google Scholar] [CrossRef]
- Ali, K.; Berman, G.; Zhou, H.; Deng, W.; Faughnan, V.; Coronado-Voges, M.; Ding, B.; Dooley, J.; Girard, B.; Hillebrand, W.; et al. Evaluation of MRNA-1273 SARS-CoV-2 Vaccine in Adolescents. N. Engl. J. Med. 2021, 385, 2241–2251. [Google Scholar] [CrossRef]
- Borobia, A.M.; Carcas, A.J.; Pérez-Olmeda, M.; Castaño, L.; Bertran, M.J.; García-Pérez, J.; Campins, M.; Portolés, A.; González-Pérez, M.; García Morales, M.T.; et al. Immunogenicity and Reactogenicity of BNT162b2 Booster in ChAdOx1-S-Primed Participants (CombiVacS): A Multicentre, Open-Label, Randomised, Controlled, Phase 2 Trial. Lancet 2021, 398, 121–130. [Google Scholar] [CrossRef]
- Alter, G.; Yu, J.; Liu, J.; Chandrashekar, A.; Borducchi, E.N.; Tostanoski, L.H.; McMahan, K.; Jacob-Dolan, C.; Martinez, D.R.; Chang, A.; et al. Immunogenicity of Ad26.COV2.S Vaccine against SARS-CoV-2 Variants in Humans. Nature 2021, 596, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 NCoV-19 COVID-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Madhi, S.A.; Kwatra, G.; Richardson, S.I.; Koen, A.L.; Baillie, V.; Cutland, C.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; et al. Durability of ChAdOx1 NCoV-19 (AZD1222) Vaccine and Hybrid Humoral Immunity against Variants Including Omicron BA.1 and BA.4 6 Months after Vaccination (COV005): A Post-Hoc Analysis of a Randomised, Phase 1b-2a Trial. Lancet Infect. Dis. 2023, 23, 295–306. [Google Scholar] [CrossRef] [PubMed]
- El Sahly, H.M.; Baden, L.R.; Essink, B.; Montefiori, D.; McDermont, A.; Rupp, R.; Lewis, M.; Swaminathan, S.; Griffin, C.; Fragoso, V.; et al. Humoral Immunogenicity of the MRNA-1273 Vaccine in the Phase 3 Coronavirus Efficacy (COVE) Trial. J. Infect. Dis. 2022, 226, 1731–1742. [Google Scholar] [CrossRef] [PubMed]
- Benkeser, D.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Janes, H.E.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Comparing Antibody Assays as Correlates of Protection against COVID-19 in the COVE MRNA-1273 Vaccine Efficacy Trial. Sci. Transl. Med. 2023, 15, eade9078. [Google Scholar] [CrossRef] [PubMed]
- Fournier, C.; Duverlie, G.; François, C.; Schnuriger, A.; Dedeurwaerder, S.; Brochot, E.; Capron, D.; Wychowski, C.; Thibault, V.; Castelain, S. A Focus Reduction Neutralization Assay for Hepatitis C Virus Neutralizing Antibodies. Virol. J. 2007, 4, 35. [Google Scholar] [CrossRef]
- Suntronwong, N.; Assawakosri, S.; Kanokudom, S.; Yorsaeng, R.; Auphimai, C.; Thongmee, T.; Vichaiwattana, P.; Duangchinda, T.; Chantima, W.; Pakchotanon, P.; et al. Strong Correlations between the Binding Antibodies against Wild-Type and Neutralizing Antibodies against Omicron BA.1 and BA.2 Variants of SARS-CoV-2 in Individuals Following Booster (Third-Dose) Vaccination. Diagnostics 2022, 12, 1781. [Google Scholar] [CrossRef]
- Annen, K.; Morrison, T.E.; DomBourian, M.G.; McCarthy, M.K.; Huey, L.; Merkel, P.A.; Andersen, G.; Schwartz, E.; Knight, V. Presence and Short-Term Persistence of SARS-CoV-2 Neutralizing Antibodies in COVID-19 Convalescent Plasma Donors. Transfusion 2021, 61, 1148–1159. [Google Scholar] [CrossRef]
- Whiteman, M.C.; Bogardus, L.; Giacone, D.G.; Rubinstein, L.J.; Antonello, J.M.; Sun, D.; Daijogo, S.; Gurney, K.B. Virus Reduction Neutralization Test: A Single-Cell Imaging High-Throughput Virus Neutralization Assay for Dengue. Am. J. Trop. Med. Hyg. 2018, 99, 1430–1439. [Google Scholar] [CrossRef]
- Sancilio, A.E.; D’Aquila, R.T.; McNally, E.M.; Velez, M.P.; Ison, M.G.; Demonbreun, A.R.; McDade, T.W. A Surrogate Virus Neutralization Test to Quantify Antibody-Mediated Inhibition of SARS-CoV-2 in Finger Stick Dried Blood Spot Samples. Sci. Rep. 2021, 11, 15321. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.C.; Tiu, C.; Hu, Z.; Chen, V.C.W.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 Surrogate Virus Neutralization Test Based on Antibody-Mediated Blockage of ACE2–Spike Protein–Protein Interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Sholukh, A.M.; Fiore-Gartland, A.; Ford, E.S.; Miner, M.D.; Hou, Y.J.; Tse, L.V.; Kaiser, H.; Zhu, H.; Lu, J.; Madarampalli, B.; et al. Evaluation of Cell-Based and Surrogate SARS-CoV-2 Neutralization Assays. J. Clin. Microbiol. 2021, 59, 527–548. [Google Scholar] [CrossRef]
- Wisnewski, A.V.; Liu, J.; Lucas, C.; Klein, J.; Iwasaki, A.; Cantley, L.; Fazen, L.; Luna, J.C.; Slade, M.; Redlich, C.A. Development and Utilization of a Surrogate SARS-CoV-2 Viral Neutralization Assay to Assess MRNA Vaccine Responses. PLoS ONE 2022, 17, e0262657. [Google Scholar] [CrossRef]
- Abe, K.T.; Li, Z.; Samson, R.; Samavarchi-Tehrani, P.; Valcourt, E.J.; Wood, H.; Budylowski, P.; Dupuis, A.P.; Girardin, R.C.; Rathod, B.; et al. A Simple Protein-Based Surrogate Neutralization Assay for SARS-CoV-2. JCI Insight 2020, 5, e142362. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Purcell, L.A.; Snell, G.; Veesler, D. Tackling COVID-19 with Neutralizing Monoclonal Antibodies. Cell 2021, 184, 4593–4595. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Zhang, N.; Richardson, S.A.; Wu, J.V. Rapid Lateral Flow Tests for the Detection of SARS-CoV-2 Neutralizing Antibodies. Expert. Rev. Mol. Diagn. 2021, 21, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Li, Z.; He, A.; Wu, B.; Yang, H.; Wu, Y.; Hu, F.; Lin, G.; Zhang, D. Ultrabright Nanoparticle-Labeled Lateral Flow Immunoassay for Detection of Anti-SARS-CoV-2 Neutralizing Antibodies in Human Serum. Biomaterials 2022, 288, 121694. [Google Scholar] [CrossRef]
- Dong, T.; Han, C.; Jiang, M.; Zhang, T.; Kang, Q.; Wang, P.; Zhou, F. A Four-Channel Surface Plasmon Resonance Sensor Functionalized Online for Simultaneous Detections of Anti-SARS-CoV-2 Antibody, Free Viral Particles, and Neutralized Viral Particles. ACS Sens. 2022, 7, 3560–3570. [Google Scholar] [CrossRef]
- Batool, R.; Soler, M.; Colavita, F.; Fabeni, L.; Matusali, G.; Lechuga, L.M. Biomimetic Nanoplasmonic Sensor for Rapid Evaluation of Neutralizing SARS-CoV-2 Monoclonal Antibodies as Antiviral Therapy. Biosens. Bioelectron. 2023, 226, 115137. [Google Scholar] [CrossRef]
- Schultz, J.S.; McCarthy, M.K.; Rester, C.; Sabourin, K.R.; Annen, K.; DomBourian, M.; Eisenmesser, E.; Frazer-Abel, A.; Knight, V.; Jaenisch, T.; et al. Development and Validation of a Multiplex Microsphere Immunoassay Using Dried Blood Spots for SARS-CoV-2 Seroprevalence: Application in First Responders in Colorado, USA. J. Clin. Microbiol. 2021, 59, 1110–1128. [Google Scholar] [CrossRef]
- Liang, Z.; Peng, T.; Jiao, X.; Zhao, Y.; Xie, J.; Jiang, Y.; Meng, B.; Fang, X.; Yu, X.; Dai, X. Latex Microsphere-Based Bicolor Immunochromatography for Qualitative Detection of Neutralizing Antibody against SARS-CoV-2. Biosensors 2022, 12, 103. [Google Scholar] [CrossRef]
- Muruato, A.E.; Fontes-Garfias, C.R.; Ren, P.; Garcia-Blanco, M.A.; Menachery, V.D.; Xie, X.; Shi, P.Y. A High-Throughput Neutralizing Antibody Assay for COVID-19 Diagnosis and Vaccine Evaluation. Nat. Commun. 2020, 11, 4059. [Google Scholar] [CrossRef] [PubMed]
- Mravinacova, S.; Jönsson, M.; Christ, W.; Klingström, J.; Yousef, J.; Hellström, C.; Hedhammar, M.; Havervall, S.; Thålin, C.; Pin, E.; et al. A Cell-Free High Throughput Assay for Assessment of SARS-CoV-2 Neutralizing Antibodies. New Biotechnol. 2022, 66, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kon, E.; Han, X.; Zhang, X.; Kong, N.; Mitchell, M.J.; Peer, D.; Tao, W. Nanotechnology-Based Strategies against SARS-CoV-2 Variants. Nat. Nanotechnol. 2022, 17, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Danh, K.; Karp, D.G.; Singhal, M.; Tankasala, A.; Gebhart, D.; de Jesus Cortez, F.; Tandel, D.; Robinson, P.V.; Seftel, D.; Stone, M.; et al. Detection of Neutralizing Antibodies against Multiple SARS-CoV-2 Strains in Dried Blood Spots Using Cell-Free PCR. Nat. Commun. 2022, 13, 4212. [Google Scholar] [CrossRef]
- Nunez, F.A.; Castro, A.C.H.; de Oliveira, V.L.; Lima, A.C.; Oliveira, J.R.; de Medeiros, G.X.; Sasahara, G.L.; Santos, K.S.; Lanfredi, A.J.C.; Alves, W.A. Electrochemical Immunosensors Based on Zinc Oxide Nanorods for Detection of Antibodies Against SARS-CoV-2 Spike Protein in Convalescent and Vaccinated Individuals. ACS Biomater. Sci. Eng. 2023, 9, 458–473. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Kimura, M.; Tan, L.; Yoshida, Y.; Ozawa, T.; Kishi, H.; Fukushi, S.; Saijo, M.; Sano, K.; Suzuki, T.; et al. Evaluation of SARS-CoV-2 Neutralizing Antibodies Using a Vesicular Stomatitis Virus Possessing SARS-CoV-2 Spike Protein. Virol. J. 2021, 18, 16. [Google Scholar] [CrossRef]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef]
- Jeong, B.S.; Cha, J.S.; Hwang, I.; Kim, U.; Adolf-Bryfogle, J.; Coventry, B.; Cho, H.S.; Kim, K.D.; Oh, B.H. Computational Design of a Neutralizing Antibody with Picomolar Binding Affinity for All Concerning SARS-CoV-2 Variants. In Mabs; Taylor & Francis: Abingdon, UK, 2022; Volume 14. [Google Scholar] [CrossRef]
- Gattinger, P.; Niespodziana, K.; Stiasny, K.; Sahanic, S.; Tulaeva, I.; Borochova, K.; Dorofeeva, Y.; Schlederer, T.; Sonnweber, T.; Hofer, G.; et al. Neutralization of SARS-CoV-2 Requires Antibodies against Conformational Receptor-Binding Domain Epitopes. Allergy 2022, 77, 230–242. [Google Scholar] [CrossRef]




| Main Articles Reported | References |
|---|---|
| These works applied the PRNT 1 as a reference to compare the performance of other methods, such as the microneutralization assay, surrogate virus neutralization test and pseudotyped virus neutralization assay. The authors showed the main advantages and limitations of each method | [27,29] |
| This work applied PRNT to measure the beneficial effect of convalescent plasma treatment of critically ill patients. They used PRNT to determine viral neutralization | [28] |
| Safety and immunogenicity studies of COVID-19 vaccines which were subjected to the PRNT assay | [30,31,32,33,34,35] |
| Works that applied pVNT 2 to measure NAbs 3, including those in which the immunogenicity of COVID-19 vaccines was evaluated | [32,38,43,44,45,46,47,48,49,50,51,52,53,54,55,56,73] |
| Works that applied FRNT 4, including clinical trials | [4,32,48,53,58,59] |
| sVNT 5 reported, including a randomized clinical trial | [33,49,62] |
| SARS-CoV-2 neutralization assays based on LFIA 6 | [72] |
| SARS-CoV-2 neutralization assays based on SPR 7 | [69,70] |
| SARS-CoV-2 neutralization assays based on MIA 8 | [71,74] |
| Nanotechnology-based neutralization assays | [41,68,75,76,77] |
| SARS-CoV-2 neutralization assays based CRNT 9 | [78] |
| Assay | Principle | Advantages | Limitations |
|---|---|---|---|
| Plaque reduction neutralization test (PRNT) | Infection of Vero cells with live viruses. The measurement of NAbs’ potency is determined through the quantification of the plaque-forming units (PFU) in specimens containing the antibodies, such as plasma, serum, or blood. |
|
|
| Pseudovirus neutralization test (pVNT) | Production of lentivirus expressing spike in the surface and transduction of HEK cells overexpressing ACE2. HEK cells are transduced to express GFP or luciferase. The redout is based on the reduction of the percentage of GFP positive cells of the decrease in the luciferase bioluminescence. |
|
|
| Foci reduction neutralization test (FRNT) or microneutralization assay (MNA) | FRNT has a similar principle to the PRNT assay. However, the readout is based on a monoclonal antibody conjugated to horseradish peroxidase (HRP) directed against the spike protein. This method uses live viruses. The PFU is quantified by a microanalyzer. |
|
|
| Surrogate virus neutralization test (sVNT) | The method is based on an enzyme linked immunosorbent Assay (ELISA) and measures the RBD-ACE2 interaction. The RBD conjugates with HRP interacts with ACE2 in an ELISA plate in the absence of NAbs |
|
|
| Lateral flow immunoassay (LFIA) | The assay uses RBD or full-length spike conjugated to a colloidal gold, colorful microsphere or fluorescent beads. NAbs from the sample interact with RBD in the conjugation pad. NAbs-RBD binding will avoid RBD binding to ACE2 coated in the test line. In this case the test line will be less colorful, or less fluorescent depending on the NAbs titer. |
|
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocha, V.P.C.; Quadros, H.C.; Fernandes, A.M.S.; Gonçalves, L.P.; Badaró, R.J.d.S.; Soares, M.B.P.; Machado, B.A.S. An Overview of the Conventional and Novel Methods Employed for SARS-CoV-2 Neutralizing Antibody Measurement. Viruses 2023, 15, 1504. https://doi.org/10.3390/v15071504
Rocha VPC, Quadros HC, Fernandes AMS, Gonçalves LP, Badaró RJdS, Soares MBP, Machado BAS. An Overview of the Conventional and Novel Methods Employed for SARS-CoV-2 Neutralizing Antibody Measurement. Viruses. 2023; 15(7):1504. https://doi.org/10.3390/v15071504
Chicago/Turabian StyleRocha, Vinícius Pinto Costa, Helenita Costa Quadros, Antônio Márcio Santana Fernandes, Luana Pereira Gonçalves, Roberto José da Silva Badaró, Milena Botelho Pereira Soares, and Bruna Aparecida Souza Machado. 2023. "An Overview of the Conventional and Novel Methods Employed for SARS-CoV-2 Neutralizing Antibody Measurement" Viruses 15, no. 7: 1504. https://doi.org/10.3390/v15071504
APA StyleRocha, V. P. C., Quadros, H. C., Fernandes, A. M. S., Gonçalves, L. P., Badaró, R. J. d. S., Soares, M. B. P., & Machado, B. A. S. (2023). An Overview of the Conventional and Novel Methods Employed for SARS-CoV-2 Neutralizing Antibody Measurement. Viruses, 15(7), 1504. https://doi.org/10.3390/v15071504






