The Re-Emergence of Hepatitis E Virus in Europe and Vaccine Development
Abstract
:1. Introduction
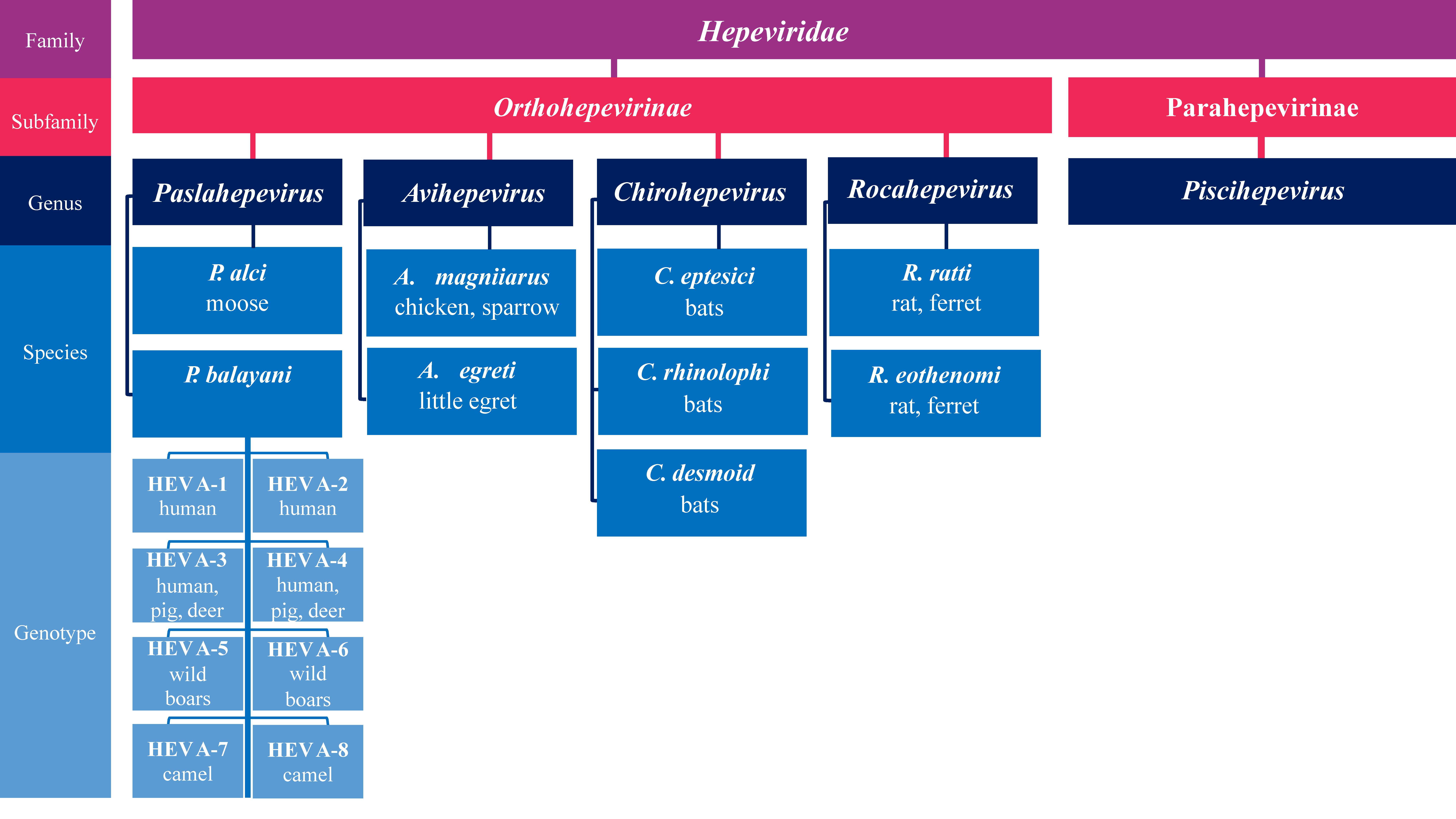
2. HEV Molecular Organization and Taxonomy
2.1. Virion Structure of HEV
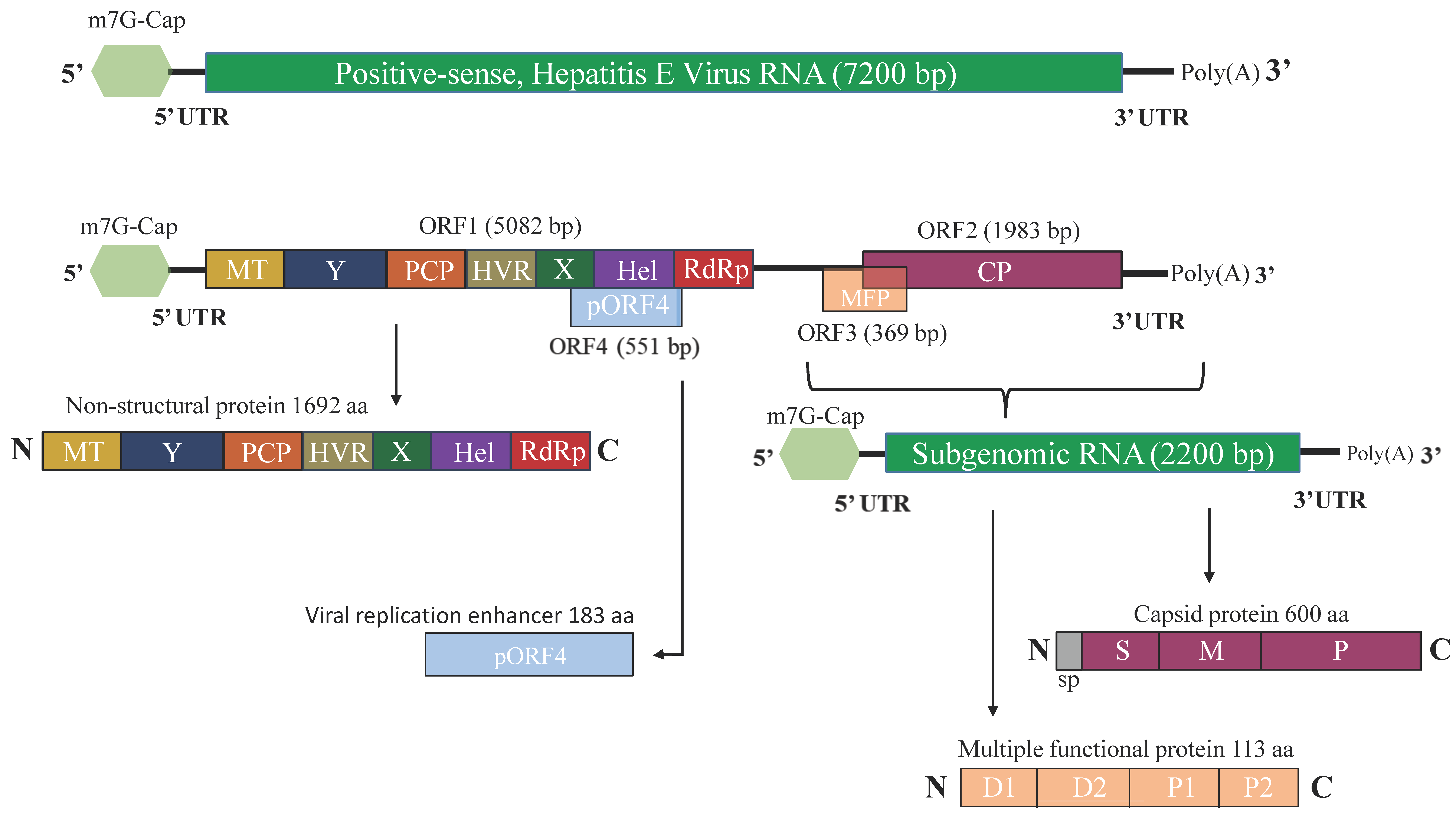
2.2. Genome Organization of HEV
2.3. HEV Proteins and Their Function
2.3.1. ORF1-Encoded Polyprotein
2.3.2. ORF2-Encoded Capsid Protein
2.3.3. ORF3-Encoded Multifunctional Protein
2.3.4. ORF4-Encoded Protein
3. HEV Transmission and Animal Reservoirs
3.1. HEV as a Zoonosis
3.2. Waterborne HEV in Developed Countries
3.3. HEV Transmission through Blood and Transplanted Organs
4. HEV Distribution in Europe
4.1. HEV in Humans
4.2. HEV in Animals
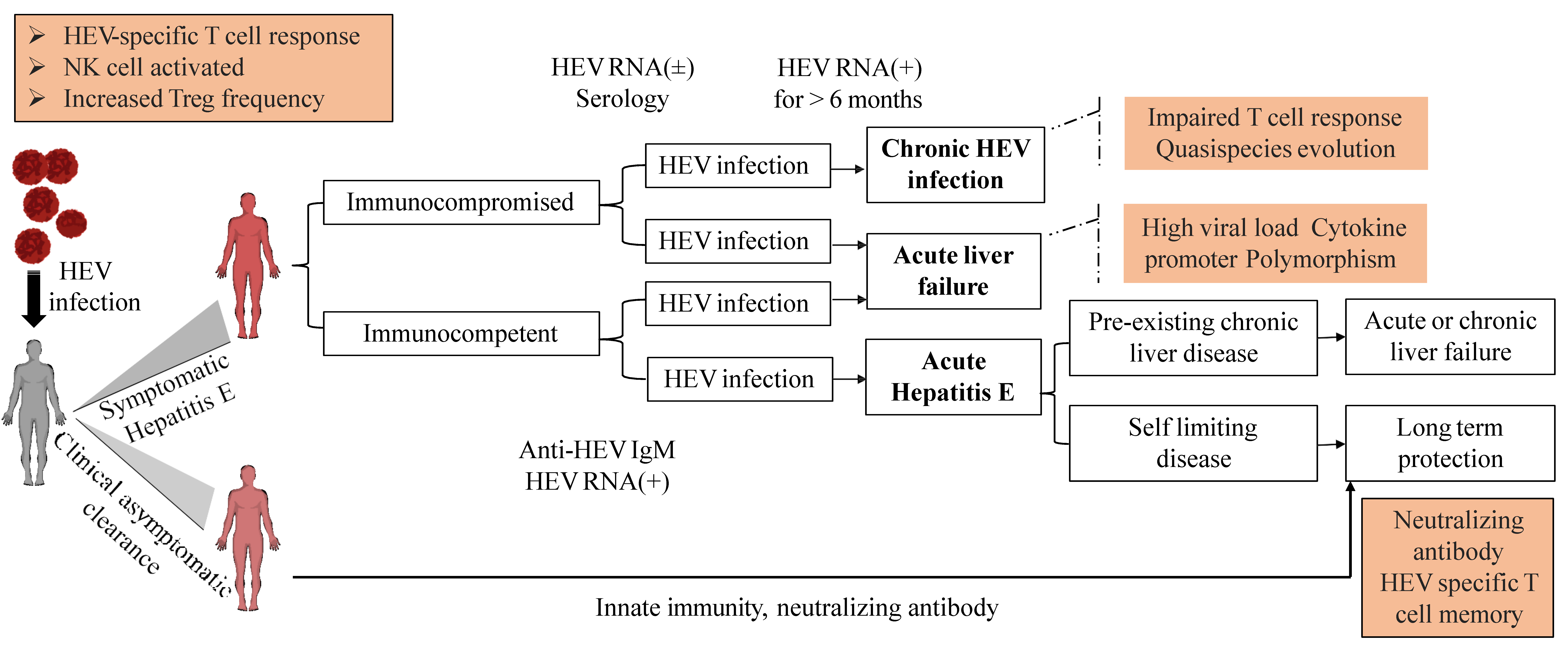
5. HEV Clinical Manifestations
5.1. Acute Hepatitis
5.2. Chronic HEV Infection. Reinfection
5.3. Complication of HEV Infection
5.3.1. Hepatitis E in Pregnant Women
5.3.2. Hepatitis E in Newborn
5.3.3. Hepatitis E in Individuals with Pre-Existing Liver Disease
5.3.4. Hepatitis E in Immunocompromised Individuals
6. HEV Diagnosis Establishment
7. HEV Therapy
8. Development of HEV Vaccine
8.1. Inactivated or Modified Live Virus Vaccine
8.2. Recombinant HEV Vaccines Production
8.2.1. E. coli Expression System
8.2.2. Insect Cell-Baculovirus Expression System
8.2.3. Yeast Expression System
8.2.4. Plant Expression System
8.3. Chimeric Vaccines
8.4. Vectored DNA Vaccines
9. Conclusions and Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Hepatitis E. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-e (accessed on 14 March 2023).
- ECDC Report: 10-Fold Increase of Hepatitis E Cases in the EU/EEA between 2005 and 2015. Available online: https://www.ecdc.europa.eu/en/news-events/ecdc-report-10-fold-increase-hepatitis-e-cases-eueea-between-2005-and-2015 (accessed on 23 February 2023).
- Bura, M.; Łagiedo-Żelazowska, M.; Michalak, M.; Sikora, J.; Mozer-Lisewska, I. Comparative Seroprevalence of Hepatitis A and E Viruses in Blood Donors from Wielkopolska Region, West-Central Poland. Pol. J. Microbiol. 2018, 67, 113–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalton, H.R.; Stableforth, W.; Hazeldine, S.; Thurairajah, P.; Ramnarace, R.; Warshow, U.; Ijaz, S.; Ellis, V.; Bendall, R. Autochthonous Hepatitis E in Southwest England: A Comparison with Hepatitis A. Eur. J. Clin. Microbiol. Infect. Dis. Off. Public Eur. Soc. Clin. Microbiol. 2008, 27, 579–585. [Google Scholar] [CrossRef]
- Hartl, J.; Otto, B.; Madden, R.G.; Webb, G.; Woolson, K.L.; Kriston, L.; Vettorazzi, E.; Lohse, A.W.; Dalton, H.R.; Pischke, S. Hepatitis E Seroprevalence in Europe: A Meta-Analysis. Viruses 2016, 8, 211. [Google Scholar] [CrossRef] [Green Version]
- Mansuy, J.-M.; Bendall, R.; Legrand-Abravanel, F.; Sauné, K.; Miédouge, M.; Ellis, V.; Rech, H.; Destruel, F.; Kamar, N.; Dalton, H.R.; et al. Hepatitis E Virus Antibodies in Blood Donors, France. Emerg. Infect. Dis. 2011, 17, 2309. [Google Scholar] [CrossRef] [PubMed]
- Mrzljak, A.; Balen, I.; Barbic, L.; Ilic, M.; Vilibic-Cavlek, T. Hepatitis E Virus in Professionally Exposed: A Reason for Concern? World J. Hepatol. 2021, 13, 723–730. [Google Scholar] [CrossRef]
- Di Profio, F.; Sarchese, V.; Palombieri, A.; Fruci, P.; Lanave, G.; Robetto, S.; Martella, V.; Di Martino, B. Current Knowledge of Hepatitis E Virus (HEV) Epidemiology in Ruminants. Pathogens 2022, 11, 1124. [Google Scholar] [CrossRef]
- Priemer, G.; Cierniak, F.; Wolf, C.; Ulrich, R.G.; Groschup, M.H.; Eiden, M. Co-Circulation of Different Hepatitis E Virus Genotype 3 Subtypes in Pigs and Wild Boar in North-East Germany, 2019. Pathog. Basel Switz. 2022, 11, 773. [Google Scholar] [CrossRef] [PubMed]
- Doceul, V.; Bagdassarian, E.; Demange, A.; Pavio, N. Zoonotic Hepatitis E Virus: Classification, Animal Reservoirs and Transmission Routes. Viruses 2016, 8, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izopet, J.; Tremeaux, P.; Marion, O.; Migueres, M.; Capelli, N.; Chapuy-Regaud, S.; Mansuy, J.-M.; Abravanel, F.; Kamar, N.; Lhomme, S. Hepatitis E Virus Infections in Europe. J. Clin. Virol. Off. Public Pan Am. Soc. Clin. Virol. 2019, 120, 20–26. [Google Scholar] [CrossRef]
- Boxall, E.; Herborn, A.; Kochethu, G.; Pratt, G.; Adams, D.; Ijaz, S.; Teo, C.-G. Transfusion-Transmitted Hepatitis E in a ‘Nonhyperendemic’ Country. Transfus. Med. 2006, 16, 79–83. [Google Scholar] [CrossRef]
- Kamar, N.; Selves, J.; Mansuy, J.-M.; Ouezzani, L.; Péron, J.-M.; Guitard, J.; Cointault, O.; Esposito, L.; Abravanel, F.; Danjoux, M.; et al. Hepatitis E Virus and Chronic Hepatitis in Organ-Transplant Recipients. N. Engl. J. Med. 2008, 358, 811–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and Regional Mortality from 235 Causes of Death for 20 Age Groups in 1990 and 2010: A Systematic Analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Faber, M.; Willrich, N.; Schemmerer, M.; Rauh, C.; Kuhnert, R.; Stark, K.; Wenzel, J.J. Hepatitis E Virus Seroprevalence, Seroincidence and Seroreversion in the German Adult Population. J. Viral Hepat. 2018, 25, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Guillois, Y.; Abravanel, F.; Miura, T.; Pavio, N.; Vaillant, V.; Lhomme, S.; Le Guyader, F.S.; Rose, N.; Le Saux, J.-C.; King, L.A.; et al. High Proportion of Asymptomatic Infections in an Outbreak of Hepatitis E Associated with a Spit-Roasted Piglet, France, 2013. Clin. Infect. Dis. 2016, 62, 351–357. [Google Scholar] [CrossRef] [Green Version]
- Dalton, H.R.; Bendall, R.P.; Keane, F.E.; Tedder, R.S.; Ijaz, S. Persistent Carriage of Hepatitis E Virus in Patients with HIV Infection. N. Engl. J. Med. 2009, 361, 1025–1027. [Google Scholar] [CrossRef]
- Kamar, N.; Pischke, S. Acute and Persistent Hepatitis E Virus Genotype 3 and 4 Infection: Clinical Features, Pathogenesis, and Treatment. Cold Spring Harb. Perspect. Med. 2019, 9, a031872. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.P.; Jain, A.K.; Naik, G.; Soni, N.; Chitnis, D.S. Viral Hepatitis during Pregnancy. Int. J. Gynaecol. Obstet. Off. Organ. Int. Fed. Gynaecol. Obstet. 2001, 72, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Navaneethan, U.; Al Mohajer, M.; Shata, M.T. Hepatitis E and Pregnancy: Understanding the Pathogenesis. Liver Int. Off. J. Int. Assoc. Study Liver 2008, 28, 1190–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rein, D.B.; Stevens, G.A.; Theaker, J.; Wittenborn, J.S.; Wiersma, S.T. The Global Burden of Hepatitis E Virus Genotypes 1 and 2 in 2005. Hepatol. Baltim. Md 2012, 55, 988–997. [Google Scholar] [CrossRef]
- Khuroo, M.S.; Kamili, S.; Khuroo, M.S. Clinical Course and Duration of Viremia in Vertically Transmitted Hepatitis E Virus (HEV) Infection in Babies Born to HEV-Infected Mothers. J. Viral Hepat. 2009, 16, 519–523. [Google Scholar] [CrossRef]
- Pischke, S.; Hartl, J.; Pas, S.D.; Lohse, A.W.; Jacobs, B.C.; Van der Eijk, A.A. Hepatitis E Virus: Infection beyond the Liver? J. Hepatol. 2017, 66, 1082–1095. [Google Scholar] [CrossRef] [Green Version]
- Khuroo, M.S. Study of an Epidemic of Non-A, Non-B Hepatitis: Possibility of Another Human Hepatitis Virus Distinct from Post-Transfusion Non-A, Non-B Type. Am. J. Med. 1980, 68, 818–824. [Google Scholar] [CrossRef]
- Purdy, M.A.; Drexler, J.F.; Meng, X.-J.; Norder, H.; Okamoto, H.; Van der Poel, W.H.M.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 001778. [Google Scholar] [CrossRef]
- Smith, D.B.; Simmonds, P.; Jameel, S.; Emerson, S.U.; Harrison, T.J.; Meng, X.-J.; Okamoto, H.; Van der Poel, W.H.M.; Purdy, M.A. Consensus Proposals for Classification of the Family Hepeviridae. J. Gen. Virol. 2014, 95, 2223–2232. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Cao, K.-Y.; Wernery, U.; Schountz, T.; Chiu, T.H.; Tsang, A.K.L.; Wong, P.-C.; Wong, E.Y.M.; et al. New Hepatitis E Virus Genotype in Bactrian Camels, Xinjiang, China, 2013. Emerg. Infect. Dis. 2016, 22, 2219–2221. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Tsang, A.K.L.; Joseph, M.; Wong, E.Y.M.; Tang, Y.; Sivakumar, S.; Xie, J.; Bai, R.; et al. New Hepatitis E Virus Genotype in Camels, the Middle East. Emerg. Infect. Dis. 2014, 20, 1044–1048. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Simmonds, P.; Izopet, J.; Oliveira-Filho, E.F.; Ulrich, R.G.; Johne, R.; Koenig, M.; Jameel, S.; Harrison, T.J.; Meng, X.-J.; et al. Proposed Reference Sequences for Hepatitis E Virus Subtypes. J. Gen. Virol. 2016, 97, 537–542. [Google Scholar] [CrossRef]
- Wang, B.; Meng, X.-J. Hepatitis E Virus: Host Tropism and Zoonotic Infection. Curr. Opin. Microbiol. 2021, 59, 8–15. [Google Scholar] [CrossRef]
- Wang, B.; Harms, D.; Yang, X.-L.; Bock, C.-T. Orthohepevirus C: An Expanding Species of Emerging Hepatitis E Virus Variants. Pathogens 2020, 9, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batts, W.; Yun, S.; Hedrick, R.; Winton, J. A Novel Member of the Family Hepeviridae from Cutthroat Trout (Oncorhynchus Clarkii). Virus Res. 2011, 158, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Nagashima, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Nishiyama, T.; Primadharsini, P.P.; Okamoto, H. Characterization of the Quasi-Enveloped Hepatitis E Virus Particles Released by the Cellular Exosomal Pathway. J. Virol. 2017, 91, e00822-17. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Matsuura, Y. Structure of Hepatitis E Viral Particle. Virus Res. 2011, 161, 59–64. [Google Scholar] [CrossRef]
- Xing, L.; Li, T.-C.; Mayazaki, N.; Simon, M.N.; Wall, J.S.; Moore, M.; Wang, C.-Y.; Takeda, N.; Wakita, T.; Miyamura, T.; et al. Structure of Hepatitis E Virion-Sized Particle Reveals an RNA-Dependent Viral Assembly Pathway. J. Biol. Chem. 2010, 285, 33175–33183. [Google Scholar] [CrossRef] [Green Version]
- Li, T.C.; Yamakawa, Y.; Suzuki, K.; Tatsumi, M.; Razak, M.A.; Uchida, T.; Takeda, N.; Miyamura, T. Expression and Self-Assembly of Empty Virus-like Particles of Hepatitis E Virus. J. Virol. 1997, 71, 7207–7213. [Google Scholar] [CrossRef]
- Purdy, M.A.; Harrison, T.J.; Jameel, S.; Meng, X.-J.; Okamoto, H.; Van der Poel, W.H.M.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae. J. Gen. Virol. 2017, 98, 2645–2646. [Google Scholar] [CrossRef] [PubMed]
- Emerson, S.U.; Zhang, M.; Meng, X.J.; Nguyen, H.; St Claire, M.; Govindarajan, S.; Huang, Y.K.; Purcell, R.H. Recombinant Hepatitis E Virus Genomes Infectious for Primates: Importance of Capping and Discovery of a Cis-Reactive Element. Proc. Natl. Acad. Sci. USA 2001, 98, 15270–15275. [Google Scholar] [CrossRef]
- Reyes, G.R.; Huang, C.-C.; Tam, A.W.; Purdy, M.A. Molecular Organization and Replication of Hepatitis E Virus (HEV). In Proceedings of the Unconventional Agents and Unclassified Viruses; Kaaden, O.-R., Eichhorn, W., Czerny, C.-P., Eds.; Springer: Vienna, Austria, 1993; pp. 15–25. [Google Scholar]
- Huang, Y.W.; Opriessnig, T.; Halbur, P.G.; Meng, X.J. Initiation at the Third In-Frame AUG Codon of Open Reading Frame 3 of the Hepatitis E Virus Is Essential for Viral Infectivity In Vivo. J. Virol. 2007, 81, 3018–3026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, V.P.; Anang, S.; Subramani, C.; Madhvi, A.; Bakshi, K.; Srivastava, A.; Shalimar; Nayak, B.; Ct, R.K.; Surjit, M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus. PLoS Pathog. 2016, 12, e1005521. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Haqshenas, G.; Kasorndorkbua, C.; Halbur, P.G.; Emerson, S.U.; Meng, X.J. Capped RNA Transcripts of Full-Length CDNA Clones of Swine Hepatitis E Virus Are Replication Competent When Transfected into Huh7 Cells and Infectious When Intrahepatically Inoculated into Pigs. J. Virol. 2005, 79, 1552–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sooryanarain, H.; Heffron, C.L.; Meng, X.-J. The U-Rich Untranslated Region of the Hepatitis E Virus Induces Differential Type I and Type III Interferon Responses in a Host Cell-Dependent Manner. mBio 2020, 11, e03103-19. [Google Scholar] [CrossRef] [Green Version]
- Magden, J.; Takeda, N.; Li, T.; Auvinen, P.; Ahola, T.; Miyamura, T.; Merits, A.; Kääriäinen, L. Virus-Specific MRNA Capping Enzyme Encoded by Hepatitis E Virus. J. Virol. 2001, 75, 6249–6255. [Google Scholar] [CrossRef] [Green Version]
- Ahola, T.; Karlin, D.G. Sequence Analysis Reveals a Conserved Extension in the Capping Enzyme of the Alphavirus Supergroup, and a Homologous Domain in Nodaviruses. Biol. Direct 2015, 10, 16. [Google Scholar] [CrossRef] [Green Version]
- Purdy, M.A. Evolution of the Hepatitis E Virus Polyproline Region: Order from Disorder. J. Virol. 2012, 86, 10186–10193. [Google Scholar] [CrossRef] [Green Version]
- Nan, Y.; Yu, Y.; Ma, Z.; Khattar, S.K.; Fredericksen, B.; Zhang, Y.-J. Hepatitis E Virus Inhibits Type I Interferon Induction by ORF1 Products. J. Virol. 2014, 88, 11924–11932. [Google Scholar] [CrossRef] [Green Version]
- Nan, Y.; Zhang, Y.-J. Molecular Biology and Infection of Hepatitis E Virus. Front. Microbiol. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montpellier, C.; Wychowski, C.; Sayed, I.M.; Meunier, J.-C.; Saliou, J.-M.; Ankavay, M.; Bull, A.; Pillez, A.; Abravanel, F.; Helle, F.; et al. Hepatitis E Virus Lifecycle and Identification of 3 Forms of the ORF2 Capsid Protein. Gastroenterology 2018, 154, 211–223.e8. [Google Scholar] [CrossRef] [PubMed]
- Ankavay, M.; Montpellier, C.; Sayed, I.M.; Saliou, J.-M.; Wychowski, C.; Saas, L.; Duvet, S.; Aliouat-Denis, C.-M.; Farhat, R.; de Masson d’Autume, V.; et al. New Insights into the ORF2 Capsid Protein, a Key Player of the Hepatitis E Virus Lifecycle. Sci. Rep. 2019, 9, 6243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Ying, D.; Lhomme, S.; Tang, Z.; Walker, C.M.; Xia, N.; Zheng, Z.; Feng, Z. Origin, Antigenicity, and Function of a Secreted Form of ORF2 in Hepatitis E Virus Infection. Proc. Natl. Acad. Sci. USA 2018, 115, 4773–4778. [Google Scholar] [CrossRef] [Green Version]
- Ding, Q.; Heller, B.; Capuccino, J.M.V.; Song, B.; Nimgaonkar, I.; Hrebikova, G.; Contreras, J.E.; Ploss, A. Hepatitis E Virus ORF3 Is a Functional Ion Channel Required for Release of Infectious Particles. Proc. Natl. Acad. Sci. USA 2017, 114, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Nan, Y.-C. Open Reading Frame 3 Protein of Hepatitis E Virus: Multi-Function Protein with Endless Potential. World J. Gastroenterol. 2021, 27, 2458–2473. [Google Scholar] [CrossRef] [PubMed]
- Subramani, C.; Nair, V.P.; Anang, S.; Mandal, S.D.; Pareek, M.; Kaushik, N.; Srivastava, A.; Saha, S.; Shalimar; Nayak, B.; et al. Host-Virus Protein Interaction Network Reveals the Involvement of Multiple Host Processes in the Life Cycle of Hepatitis E Virus. mSystems 2018, 3, e00135-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavio, N.; Meng, X.-J.; Doceul, V. Zoonotic Origin of Hepatitis E. Curr. Opin. Virol. 2015, 10, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Denner, J.; Pischke, S.; Steinmann, E.; Blümel, J.; Glebe, D. Why All Blood Donations Should Be Tested for Hepatitis E Virus (HEV). BMC Infect. Dis. 2019, 19, 541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenney, S.P. The Current Host Range of Hepatitis E Viruses. Viruses 2019, 11, 452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavio, N.; Doceul, V.; Bagdassarian, E.; Johne, R. Recent Knowledge on Hepatitis E Virus in Suidae Reservoirs and Transmission Routes to Human. Vet. Res. 2017, 48, 78. [Google Scholar] [CrossRef] [Green Version]
- Baymakova, M.; Terzieva, K.; Popov, R.; Grancharova, E.; Kundurzhiev, T.; Pepovich, R.; Tsachev, I. Seroprevalence of Hepatitis E Virus Infection among Blood Donors in Bulgaria. Viruses 2021, 13, 492. [Google Scholar] [CrossRef]
- Dremsek, P.; Wenzel, J.J.; Johne, R.; Ziller, M.; Hofmann, J.; Groschup, M.H.; Werdermann, S.; Mohn, U.; Dorn, S.; Motz, M.; et al. Seroprevalence Study in Forestry Workers from Eastern Germany Using Novel Genotype 3- and Rat Hepatitis E Virus-Specific Immunoglobulin G ELISAs. Med. Microbiol. Immunol. 2012, 201, 189–200. [Google Scholar] [CrossRef]
- Gérolami, R.; Moal, V.; Colson, P. Chronic Hepatitis E with Cirrhosis in a Kidney-Transplant Recipient. N. Engl. J. Med. 2008, 358, 859–860. [Google Scholar] [CrossRef] [Green Version]
- Berto, A.; Grierson, S.; Hakze-van der Honing, R.; Martelli, F.; Johne, R.; Reetz, J.; Ulrich, R.G.; Pavio, N.; Van der Poel, W.H.M.; Banks, M. Hepatitis E Virus in Pork Liver Sausage, France. Emerg. Infect. Dis. 2013, 19, 264–266. [Google Scholar] [CrossRef]
- Anheyer-Behmenburg, H.E.; Szabo, K.; Schotte, U.; Binder, A.; Klein, G.; Johne, R. Hepatitis E Virus in Wild Boars and Spillover Infection in Red and Roe Deer, Germany, 2013–2015. Emerg. Infect. Dis. 2017, 23, 130–133. [Google Scholar] [CrossRef] [Green Version]
- Boadella, M.; Casas, M.; Martín, M.; Vicente, J.; Segalés, J.; de la Fuente, J.; Gortázar, C. Increasing Contact with Hepatitis E Virus in Red Deer, Spain. Emerg. Infect. Dis. 2010, 16, 1994–1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutjes, S.A.; Lodder-Verschoor, F.; Lodder, W.J.; van der Giessen, J.; Reesink, H.; Bouwknegt, M.; de Roda Husman, A.M. Seroprevalence and Molecular Detection of Hepatitis E Virus in Wild Boar and Red Deer in The Netherlands. J. Virol. Methods 2010, 168, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Trojnar, E.; Kästner, B.; Johne, R. No Evidence of Hepatitis E Virus Infection in Farmed Deer in Germany. Food Environ. Virol. 2020, 12, 81–83. [Google Scholar] [CrossRef]
- Liang, H.; Chen, J.; Xie, J.; Sun, L.; Ji, F.; He, S.; Zheng, Y.; Liang, C.; Zhang, G.; Su, S.; et al. Hepatitis E Virus Serosurvey among Pet Dogs and Cats in Several Developed Cities in China. PLoS ONE 2014, 9, e98068. [Google Scholar] [CrossRef]
- Schlosser, J.; Dähnert, L.; Dremsek, P.; Tauscher, K.; Fast, C.; Ziegler, U.; Gröner, A.; Ulrich, R.G.; Groschup, M.H.; Eiden, M. Different Outcomes of Experimental Hepatitis E Virus Infection in Diverse Mouse Strains, Wistar Rats, and Rabbits. Viruses 2019, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Shukla, P.; Nguyen, H.T.; Torian, U.; Engle, R.E.; Faulk, K.; Dalton, H.R.; Bendall, R.P.; Keane, F.E.; Purcell, R.H.; Emerson, S.U. Cross-Species Infections of Cultured Cells by Hepatitis E Virus and Discovery of an Infectious Virus–Host Recombinant. Proc. Natl. Acad. Sci. USA 2011, 108, 2438–2443. [Google Scholar] [CrossRef]
- Ryll, R.; Bernstein, S.; Heuser, E.; Schlegel, M.; Dremsek, P.; Zumpe, M.; Wolf, S.; Pépin, M.; Bajomi, D.; Müller, G.; et al. Detection of Rat Hepatitis E Virus in Wild Norway Rats (Rattus norvegicus) and Black Rats (Rattus rattus) from 11 European Countries. Vet. Microbiol. 2017, 208, 58–68. [Google Scholar] [CrossRef]
- De Sabato, L.; Ianiro, G.; Monini, M.; De Lucia, A.; Ostanello, F.; Di Bartolo, I. Detection of Hepatitis E Virus RNA in Rats Caught in Pig Farms from Northern Italy. Zoonoses Public Health 2020, 67, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Ryll, R.; Eiden, M.; Heuser, E.; Weinhardt, M.; Ziege, M.; Höper, D.; Groschup, M.H.; Heckel, G.; Johne, R.; Ulrich, R.G. Hepatitis E Virus in Feral Rabbits along a Rural-Urban Transect in Central Germany. Infect. Genet. Evol. 2018, 61, 155–159. [Google Scholar] [CrossRef]
- Wang, L.; Liu, L.; Wang, L. An Overview: Rabbit Hepatitis E Virus (HEV) and Rabbit Providing an Animal Model for HEV Study. Rev. Med. Virol. 2018, 28, e1961. [Google Scholar] [CrossRef]
- Abravanel, F.; Lhomme, S.; El Costa, H.; Schvartz, B.; Peron, J.-M.; Kamar, N.; Izopet, J. Rabbit Hepatitis E Virus Infections in Humans, France. Emerg. Infect. Dis. 2017, 23, 1191–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahli, R.; Fraga, M.; Semela, D.; Moradpour, D.; Gouttenoire, J. Rabbit HEV in Immunosuppressed Patients with Hepatitis E Acquired in Switzerland. J. Hepatol. 2019, 70, 1023–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivero-Juarez, A.; Frias, M.; Perez, A.B.; Pineda, J.A.; Reina, G.; Fuentes-Lopez, A.; Freyre-Carrillo, C.; Ramirez-Arellano, E.; Alados, J.C.; Rivero, A.; et al. Orthohepevirus C Infection as an Emerging Cause of Acute Hepatitis in Spain: First Report in Europe. J. Hepatol. 2022, 77, 326–331. [Google Scholar] [CrossRef]
- Rodriguez, C.; Marchand, S.; Sessa, A.; Cappy, P.; Pawlotsky, J.-M. Orthohepevirus C Hepatitis, an Underdiagnosed Disease? J. Hepatol. 2023, 79, e39–e41. [Google Scholar] [CrossRef]
- Sridhar, S.; Yip, C.C.Y.; Wu, S.; Cai, J.; Zhang, A.J.-X.; Leung, K.-H.; Chung, T.W.H.; Chan, J.F.W.; Chan, W.-M.; Teng, J.L.L.; et al. Rat Hepatitis E Virus as Cause of Persistent Hepatitis after Liver Transplant. Emerg. Infect. Dis. 2018, 24, 2241–2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andonov, A.; Robbins, M.; Borlang, J.; Cao, J.; Hatchette, T.; Stueck, A.; Deschambault, Y.; Murnaghan, K.; Varga, J.; Johnston, L. Rat Hepatitis E Virus Linked to Severe Acute Hepatitis in an Immunocompetent Patient. J. Infect. Dis. 2019, 220, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, Y.; Li, Y.; Jin, W.; Duan, S.; Xu, H.; Zhao, Y.; He, Z.; Ami, Y.; Suzaki, Y.; et al. Experimental Cross-Species Transmission of Rat Hepatitis E Virus to Rhesus and Cynomolgus Monkeys. Viruses 2022, 14, 293. [Google Scholar] [CrossRef]
- Qian, Z.; Hao, X.; Xia, Y.; Yu, W.; Huang, F. Rat Hepatitis E Virus Is a Potential Zoonotic Pathogen to Humans. J. Hepatol. 2022, 77, 868–870. [Google Scholar] [CrossRef]
- Lee, G.-H.; Tan, B.-H.; Teo, E.C.-Y.; Lim, S.-G.; Dan, Y.-Y.; Wee, A.; Aw, P.P.K.; Zhu, Y.; Hibberd, M.L.; Tan, C.-K.; et al. Chronic Infection with Camelid Hepatitis E Virus in a Liver Transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology 2016, 150, 355–357.e3. [Google Scholar] [CrossRef] [Green Version]
- Capai, L.; Charrel, R.; Falchi, A. Hepatitis E in High-Income Countries: What Do We Know? And What Are the Knowledge Gaps? Viruses 2018, 10, 285. [Google Scholar] [CrossRef] [Green Version]
- Yugo, D.M.; Meng, X.-J. Hepatitis E Virus: Foodborne, Waterborne and Zoonotic Transmission. Int. J. Environ. Res. Public. Health 2013, 10, 4507–4533. [Google Scholar] [CrossRef] [Green Version]
- Fogeda, M.; Avellón, A.; Cilla, C.G.; Echevarría, J.M. Imported and Autochthonous Hepatitis E Virus Strains in Spain. J. Med. Virol. 2009, 81, 1743–1749. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, J.R.; Almeida-Santos, M.; Fernandes, M.; Maltez, F.; Lino, S.; Alves, L.; Abreu-Silva, J.; Ricardo, M.O.; Curran, M.D.; Maria, S.N. Hepatitis E Virus Genotype 1 Cases Imported to Portugal from India, 2016. Ann. Hepatol. 2018, 17, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, H.; Chassaing, M.; Berger, S.; Gantzer, C.; Bertrand, I.; Schvoerer, E. Transmission of Hepatitis E Virus by Water: An Issue Still Pending in Industrialized Countries. Water Res. 2019, 151, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Givens, C.E.; Kolpin, D.W.; Borchardt, M.A.; Duris, J.W.; Moorman, T.B.; Spencer, S.K. Detection of Hepatitis E Virus and Other Livestock-Related Pathogens in Iowa Streams. Sci. Total Environ. 2016, 566–567, 1042–1051. [Google Scholar] [CrossRef] [Green Version]
- Kokkinos, P.; Kozyra, I.; Lazic, S.; Bouwknegt, M.; Rutjes, S.; Willems, K.; Moloney, R.; de Roda Husman, A.M.; Kaupke, A.; Legaki, E.; et al. Harmonised Investigation of the Occurrence of Human Enteric Viruses in the Leafy Green Vegetable Supply Chain in Three European Countries. Food Environ. Virol. 2012, 4, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Maunula, L.; Kaupke, A.; Vasickova, P.; Söderberg, K.; Kozyra, I.; Lazic, S.; van der Poel, W.H.M.; Bouwknegt, M.; Rutjes, S.; Willems, K.A.; et al. Tracing Enteric Viruses in the European Berry Fruit Supply Chain. Int. J. Food Microbiol. 2013, 167, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Terio, V.; Bottaro, M.; Pavoni, E.; Losio, M.N.; Serraino, A.; Giacometti, F.; Martella, V.; Mottola, A.; Di Pinto, A.; Tantillo, G. Occurrence of Hepatitis A and E and Norovirus GI and GII in Ready-to-Eat Vegetables in Italy. Int. J. Food Microbiol. 2017, 249, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Lhomme, S.; Le Saux, J.-C.; Le Mehaute, P.; Guillois, Y.; Couturier, E.; Izopet, J.; Abranavel, F.; Le Guyader, F.S. Detection of Hepatitis E Virus in Sewage After an Outbreak on a French Island. Food Environ. Virol. 2016, 8, 194–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iaconelli, M.; Purpari, G.; Libera, S.D.; Petricca, S.; Guercio, A.; Ciccaglione, A.R.; Bruni, R.; Taffon, S.; Equestre, M.; Fratini, M.; et al. Hepatitis A and E Viruses in Wastewaters, in River Waters, and in Bivalve Molluscs in Italy. Food Environ. Virol. 2015, 7, 316–324. [Google Scholar] [CrossRef]
- Matos, A.M.; Mesquita, J.R.; Gonçalves, D.; Abreu-Silva, J.; Luxo, C.; Nascimento, M.S.J. Hepatitis E Virus Subgenotypes 3i and 3f in Wastewater of Treatment Plants of Portugal. J. Clin. Virol. 2016, 82, S71. [Google Scholar] [CrossRef]
- Myrmel, M.; Lange, H.; Rimstad, E. A 1-Year Quantitative Survey of Noro-, Adeno-, Human Boca-, and Hepatitis E Viruses in Raw and Secondarily Treated Sewage from Two Plants in Norway. Food Environ. Virol. 2015, 7, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Mansuy, J.M.; Gallian, P.; Dimeglio, C.; Saune, K.; Arnaud, C.; Pelletier, B.; Morel, P.; Legrand, D.; Tiberghien, P.; Izopet, J. A Nationwide Survey of Hepatitis E Viral Infection in French Blood Donors. Hepatology 2016, 63, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Crossan, C.; Baker, P.J.; Craft, J.; Takeuchi, Y.; Dalton, H.R.; Scobie, L. Hepatitis E Virus Genotype 3 in Shellfish, United Kingdom. Emerg. Infect. Dis. J. 2012, 18, 2085–2087. [Google Scholar] [CrossRef] [PubMed]
- Said, B.; Ijaz, S.; Kafatos, G.; Booth, L.; Thomas, H.L.; Walsh, A.; Ramsay, M.; Morgan, D. Hepatitis E Outbreak on Cruise Ship. Emerg. Infect. Dis. J. 2009, 15, 1738–1744. [Google Scholar] [CrossRef]
- Gallian, P.; Piquet, Y.; Assal, A.; Djoudi, R.; Chiaroni, J.; Izopet, J.; Tiberghien, P. Hepatitis E virus: Blood transfusion implications. Transfus. Clin. Biol. 2014, 21, 173–177. [Google Scholar] [CrossRef]
- Harvala, H.; Hewitt, P.E.; Reynolds, C.; Pearson, C.; Haywood, B.; Tettmar, K.I.; Ushiro-Lumb, I.; Brailsford, S.R.; Tedder, R.; Ijaz, S. Hepatitis E Virus in Blood Donors in England, 2016 to 2017: From Selective to Universal Screening. Eurosurveillance 2019, 24, 1800386. [Google Scholar] [CrossRef] [Green Version]
- Boland, F.; Martinez, A.; Pomeroy, L.; O’Flaherty, N. Blood Donor Screening for Hepatitis E Virus in the European Union. Transfus. Med. Hemother. 2019, 46, 95–103. [Google Scholar] [CrossRef]
- Riveiro-Barciela, M.; Sauleda, S.; Quer, J.; Salvador, F.; Gregori, J.; Pirón, M.; Rodríguez-Frías, F.; Buti, M. Red Blood Cell Transfusion-Transmitted Acute Hepatitis E in an Immunocompetent Subject in Europe: A Case Report. Transfusion 2017, 57, 244–247. [Google Scholar] [CrossRef]
- KYODO. In World First, Woman’s Death Linked to Hepatitis E Infection after Blood Transfusion at Japanese Facility. Available online: https://www.japantimes.co.jp/news/2018/02/01/national/world-first-womans-death-linked-hepatitis-e-infection-blood-transfusion-japanese-facility/ (accessed on 7 February 2023).
- Dreier, J.; Knabbe, C.; Vollmer, T. Transfusion-Transmitted Hepatitis E: NAT Screening of Blood Donations and Infectious Dose. Front. Med. 2018, 5, 5. [Google Scholar] [CrossRef]
- Yin, X.; Ambardekar, C.; Lu, Y.; Feng, Z. Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses. J. Virol. 2016, 90, 4232–4242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Vos, A.S.; Janssen, M.P.; Zaaijer, H.L.; Hogema, B.M. Cost-Effectiveness of the Screening of Blood Donations for Hepatitis E Virus in the Netherlands. Transfusion 2017, 57, 258–266. [Google Scholar] [CrossRef]
- Kamp, C.; Blümel, J.; Baylis, S.A.; Bekeredjian-Ding, I.; Chudy, M.; Heiden, M.; Henseler, O.; Keller-Stanislawski, B.; de Vos, A.S.; Funk, M.B. Impact of Hepatitis E Virus Testing on the Safety of Blood Components in Germany—Results of a Simulation Study. Vox Sang. 2018, 113, 811–813. [Google Scholar] [CrossRef]
- Schlosser, B.; Stein, A.; Neuhaus, R.; Pahl, S.; Ramez, B.; Krüger, D.H.; Berg, T.; Hofmann, J. Liver Transplant from a Donor with Occult HEV Infection Induced Chronic Hepatitis and Cirrhosis in the Recipient. J. Hepatol. 2012, 56, 500–502. [Google Scholar] [CrossRef] [Green Version]
- Koenecke, C.; Pischke, S.; Beutel, G.; Ritter, U.; Ganser, A.; Wedemeyer, H.; Eder, M. Hepatitis E Virus Infection in a Hematopoietic Stem Cell Donor. Bone Marrow Transplant. 2014, 49, 159–160. [Google Scholar] [CrossRef]
- Denner, J. Hepatitis E Virus (HEV)—The Future. Viruses 2019, 11, 251. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Liu, J.; Li, Y.; Su, J.; Ma, Z.; Bramer, W.M.; Cao, W.; de Man, R.A.; Peppelenbosch, M.P.; Pan, Q. The Global Epidemiology of Hepatitis E Virus Infection: A Systematic Review and Meta-Analysis. Liver Int. Off. J. Int. Assoc. Study Liver 2020, 40, 1516–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melgaço, J.G.; Gardinali, N.R.; Mello, V.d.M.d.; Leal, M.; Lewis-Ximenez, L.L.; Pinto, M.A. Hepatitis E: Update on Prevention and Control. BioMed Res. Int. 2018, 2018, e5769201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aspinall, E.J.; Couturier, E.; Faber, M.; Said, B.; Ijaz, S.; Tavoschi, L.; Takkinen, J.; Adlhoch, C.; on behalf of the country experts. Hepatitis E Virus Infection in Europe: Surveillance and Descriptive Epidemiology of Confirmed Cases, 2005 to 2015. Eurosurveillance 2017, 22, 30561. [Google Scholar] [CrossRef] [Green Version]
- Mrzljak, A.; Dinjar-Kujundzic, P.; Jemersic, L.; Prpic, J.; Barbic, L.; Savic, V.; Stevanovic, V.; Vilibic-Cavlek, T. Epidemiology of Hepatitis e in South-East Europe in the “One Health” Concept. World J. Gastroenterol. 2019, 25, 3168–3182. [Google Scholar] [CrossRef]
- Wilhelm, B.J.; Rajić, A.; Greig, J.; Waddell, L.; Trottier, G.; Houde, A.; Harris, J.; Borden, L.N.; Price, C. A Systematic Review/Meta-Analysis of Primary Research Investigating Swine, Pork or Pork Products as a Source of Zoonotic Hepatitis E Virus. Epidemiol. Infect. 2011, 139, 1127–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breum, S.Ø.; Hjulsager, C.K.; de Deus, N.; Segalés, J.; Larsen, L.E. Hepatitis E Virus Is Highly Prevalent in the Danish Pig Population. Vet. Microbiol. 2010, 146, 144–149. [Google Scholar] [CrossRef]
- Ivanova, A.; Tefanova, V.; Reshetnjak, I.; Kuznetsova, T.; Geller, J.; Lundkvist, Å.; Janson, M.; Neare, K.; Velström, K.; Jokelainen, P.; et al. Hepatitis E Virus in Domestic Pigs, Wild Boars, Pig Farm Workers, and Hunters in Estonia. Food Environ. Virol. 2015, 7, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Lange, H.; Øverbø, J.; Borgen, K.; Dudman, S.; Hoddevik, G.; Urdahl, A.M.; Vold, L.; Sjurseth, S.K. Hepatitis E in Norway: Seroprevalence in Humans and Swine. Epidemiol. Infect. 2017, 145, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Salines, M.; Andraud, M.; Rose, N. From the Epidemiology of Hepatitis E Virus (HEV) within the Swine Reservoir to Public Health Risk Mitigation Strategies: A Comprehensive Review. Vet. Res. 2017, 48, 31. [Google Scholar] [CrossRef] [Green Version]
- Martinelli, N.; Luppi, A.; Cordioli, P.; Lombardi, G.; Lavazza, A. Prevalence of Hepatitis E Virus Antibodies in Pigs in Northern Italy. Infect. Ecol. Epidemiol. 2011, 1, 7331. [Google Scholar] [CrossRef]
- Seminati, C.; Mateu, E.; Peralta, B.; de Deus, N.; Martin, M. Distribution of Hepatitis E Virus Infection and Its Prevalence in Pigs on Commercial Farms in Spain. Vet. J. 2008, 175, 130–132. [Google Scholar] [CrossRef]
- Takova, K.; Koynarski, T.; Minkov, I.; Ivanova, Z.; Toneva, V.; Zahmanova, G. Increasing Hepatitis E Virus Seroprevalence in Domestic Pigs and Wild Boar in Bulgaria. Animals 2020, 10, 1521. [Google Scholar] [CrossRef]
- Tsachev, I.; Baymakova, M.; Marutsov, P.; Gospodinova, K.; Kundurzhiev, T.; Petrov, V.; Pepovich, R. Seroprevalence of Hepatitis E Virus Infection Among Wild Boars in Western Bulgaria. Vector-Borne Zoonotic Dis. 2021, 21, 441–445. [Google Scholar] [CrossRef]
- Pugliese, M.; La Maestra, R.; Guercio, A.; Purpari, G.; Di Bella, S.; Vullo, S.; Niutta, P.P. Hepatitis E Virus Seroprevalence among Cows in a Rural Area of Southern Italy. Vet. Arh. 2021, 91, 333–338. [Google Scholar] [CrossRef]
- Vercouter, A.-S.; Sayed, I.M.; Lipkens, Z.; De Bleecker, K.; De Vliegher, S.; Colman, R.; Koppelman, M.; Supré, K.; Meuleman, P. Absence of Zoonotic Hepatitis E Virus Infection in Flemish Dairy Cows. Int. J. Food Microbiol. 2018, 281, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Baechlein, C.; Becher, P. No Evidence for Zoonotic Hepatitis E Virus Infection through Dairy Milk in Germany. Hepatology 2017, 65, 394. [Google Scholar] [CrossRef]
- Tialla, D.; Cissé, A.; Ouédraogo, G.A.; Hübschen, J.M.; Tarnagda, Z.; Snoeck, C.J. Prevalence of Hepatitis E Virus Antibodies in Cattle in Burkina Faso Associated with Swine Mixed Farming. J. Vet. Sci. 2022, 23, e33. [Google Scholar] [CrossRef]
- Hammerschmidt, F.; Schwaiger, K.; Dähnert, L.; Vina-Rodriguez, A.; Höper, D.; Gareis, M.; Groschup, M.H.; Eiden, M. Hepatitis E Virus in Wild Rabbits and European Brown Hares in Germany. Zoonoses Public Health 2017, 64, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolo, I.; De Sabato, L.; Marata, A.; Martinelli, N.; Magistrali, C.F.; Monini, M.; Ponterio, E.; Ostanello, F.; Ruggeri, F.M. Serological Survey of Hepatitis E Virus Infection in Farmed and Pet Rabbits in Italy. Arch. Virol. 2016, 161, 1343–1346. [Google Scholar] [CrossRef] [PubMed]
- Simanavicius, M.; Juskaite, K.; Verbickaite, A.; Jasiulionis, M.; Tamosiunas, P.L.; Petraityte-Burneikiene, R.; Zvirbliene, A.; Ulrich, R.G.; Kucinskaite-Kodze, I. Detection of Rat Hepatitis E Virus, but Not Human Pathogenic Hepatitis E Virus Genotype 1–4 Infections in Wild Rats from Lithuania. Vet. Microbiol. 2018, 221, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Lack, J.B.; Volk, K.; Bussche, R.A.V.D. Hepatitis E Virus Genotype 3 in Wild Rats, United States. Emerg. Infect. Dis. 2012, 18, 1268. [Google Scholar] [CrossRef]
- Mulder, A.C.; Kroneman, A.; Franz, E.; Vennema, H.; Tulen, A.D.; Takkinen, J.; Hofhuis, A.; Adlhoch, C. HEVnet: A One Health, Collaborative, Interdisciplinary Network and Sequence Data Repository for Enhanced Hepatitis E Virus Molecular Typing, Characterisation and Epidemiological Investigations. Eurosurveillance 2019, 24, 1800407. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Ling, B.; Guo, N.; Zhai, G.; Li, M.; Guo, Y. Immunological Manifestations of Hepatitis E-Associated Acute and Chronic Liver Failure and Its Regulatory Mechanisms. Front. Med. 2021, 8, 725993. [Google Scholar] [CrossRef]
- Klöhn, M.; Schrader, J.A.; Brüggemann, Y.; Todt, D.; Steinmann, E. Beyond the Usual Suspects: Hepatitis E Virus and Its Implications in Hepatocellular Carcinoma. Cancers 2021, 13, 5867. [Google Scholar] [CrossRef]
- Horvatits, T.; Schulze zur Wiesch, J.; Lütgehetmann, M.; Lohse, A.W.; Pischke, S. The Clinical Perspective on Hepatitis E. Viruses 2019, 11, 617. [Google Scholar] [CrossRef] [Green Version]
- Damiris, K.; Meybodi, M.A.; Niazi, M.; Pyrsopoulos, N. Hepatitis E in Immunocompromised Individuals. World J. Hepatol. 2022, 14, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Aslan, A.T.; Balaban, H.Y. Hepatitis E Virus: Epidemiology, Diagnosis, Clinical Manifestations, and Treatment. World J. Gastroenterol. 2020, 26, 5543–5560. [Google Scholar] [CrossRef] [PubMed]
- Fousekis, F.S.; Mitselos, I.V.; Christodoulou, D.K. Extrahepatic Manifestations of Hepatitis E Virus: An Overview. Clin. Mol. Hepatol. 2019, 26, 16–23. [Google Scholar] [CrossRef]
- Wu, J.; Xiang, Z.; Zhu, C.; Yao, Y.; Bortolanza, M.; Cao, H.; Li, L. Extrahepatic Manifestations Related to Hepatitis E Virus Infection and Their Triggering Mechanisms. J. Infect. 2021, 83, 298–305. [Google Scholar] [CrossRef]
- Rawla, P.; Raj, J.P.; Kannemkuzhiyil, A.J.; Aluru, J.S.; Thandra, K.C.; Gajendran, M. A Systematic Review of the Extra-Hepatic Manifestations of Hepatitis E Virus Infection. Med. Sci. 2020, 8, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pischke, S.; Wedemeyer, H. Hepatitis E Virus Infection: Multiple Faces of an Underestimated Problem. J. Hepatol. 2013, 58, 1045–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arends, J.E.; Ghisetti, V.; Irving, W.; Dalton, H.R.; Izopet, J.; Hoepelman, A.I.M.; Salmon, D. Hepatitis E: An Emerging Infection in High Income Countries. J. Clin. Virol. 2014, 59, 81–88. [Google Scholar] [CrossRef]
- Anderson, D.A. Hepatitis E. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Mandell, G.L., Bennett, J.E., Dolin, R., Eds.; Elsevier: Philadelphia, PA, USA, 2010; Volume 1, pp. 2411–2421. ISBN 978-0-4430-6839-3. [Google Scholar]
- Kamar, N.; Dalton, H.R.; Abravanel, F.; Izopet, J. Hepatitis E Virus Infection. Clin. Microbiol. Rev. 2014, 27, 116–138. [Google Scholar] [CrossRef] [Green Version]
- Abravanel, F.; Barragué, H.; Dörr, G.; Sauné, K.; Péron, J.-M.; Alric, L.; Kamar, N.; Izopet, J.; Champagne, E. Conventional and Innate Lymphocytes Response at the Acute Phase of HEV Infection in Transplanted Patients. J. Infect. 2016, 72, 723–730. [Google Scholar] [CrossRef]
- Fujiwara, S.; Yokokawa, Y.; Morino, K.; Hayasaka, K.; Kawabata, M.; Shimizu, T. Chronic Hepatitis E: A Review of the Literature. J. Viral Hepat. 2014, 21, 78–89. [Google Scholar] [CrossRef]
- de Niet, A.; Zaaijer, H.L.; ten Berge, I.; Weegink, C.J.; Reesink, H.W.; Beuers, U. Chronic Hepatitis E after Solid Organ Transplantation. Neth. J. Med. 2012, 70, 261–266. [Google Scholar] [PubMed]
- Haagsma, E.B.; Niesters, H.G.M.; van den Berg, A.P.; Riezebos-Brilman, A.; Porte, R.J.; Vennema, H.; Reimerink, J.H.J.; Koopmans, M.P.G. Prevalence of Hepatitis E Virus Infection in Liver Transplant Recipients. Liver Transpl. 2009, 15, 1225–1228. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Garrouste, C.; Haagsma, E.B.; Garrigue, V.; Pischke, S.; Chauvet, C.; Dumortier, J.; Cannesson, A.; Cassuto–Viguier, E.; Thervet, E.; et al. Factors Associated with Chronic Hepatitis in Patients With Hepatitis E Virus Infection Who Have Received Solid Organ Transplants. Gastroenterology 2011, 140, 1481–1489. [Google Scholar] [CrossRef]
- Gérolami, R.; Moal, V.; Picard, C.; Colson, P. Hepatitis E Virus as an Emerging Cause of Chronic Liver Disease in Organ Transplant Recipients. J. Hepatol. 2009, 50, 622–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khuroo, M.S. Chronic Liver Disease after Non-A, Non-B Hepatitis. Lancet Lond. Engl. 1980, 2, 860–861. [Google Scholar] [CrossRef]
- Kamar, N.; Rostaing, L.; Abravanel, F.; Garrouste, C.; Lhomme, S.; Esposito, L.; Basse, G.; Cointault, O.; Ribes, D.; Nogier, M.B.; et al. Ribavirin Therapy Inhibits Viral Replication on Patients with Chronic Hepatitis e Virus Infection. Gastroenterology 2010, 139, 1612–1618. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, G.; Pan, Q.; Zhao, J. Chronic Hepatitis E in a Renal Transplant Recipient: The First Report of Genotype 4 Hepatitis E Virus Caused Chronic Infection in Organ Recipient. Gastroenterology 2018, 154, 1199–1201. [Google Scholar] [CrossRef] [Green Version]
- Sridhar, S.; Chan, J.F.W.; Yap, D.Y.H.; Teng, J.L.L.; Huang, C.; Yip, C.C.Y.; Hung, I.F.N.; Tang, S.C.W.; Lau, S.K.P.; Woo, P.C.Y.; et al. Genotype 4 Hepatitis E Virus Is a Cause of Chronic Hepatitis in Renal Transplant Recipients in Hong Kong. J. Viral Hepat. 2018, 25, 209–213. [Google Scholar] [CrossRef]
- Ma, Z.; de Man, R.A.; Kamar, N.; Pan, Q. Chronic Hepatitis E: Advancing Research and Patient Care. J. Hepatol. 2022, 77, 1109–1123. [Google Scholar] [CrossRef]
- Whitsett, M.; Feldman, D.M.; Jacobson, I. Hepatitis E Virus Infection in the United States: Current Understanding of the Prevalence and Significance in the Liver Transplant Patient Population and Proposed Diagnostic and Treatment Strategies. Liver Transplant. 2020, 26, 709–717. [Google Scholar] [CrossRef]
- Abravanel, F.; Lhomme, S.; Chapuy-Regaud, S.; Mansuy, J.-M.; Muscari, F.; Sallusto, F.; Rostaing, L.; Kamar, N.; Izopet, J. Hepatitis E Virus Reinfections in Solid-Organ-Transplant Recipients Can Evolve into Chronic Infections. J. Infect. Dis. 2014, 209, 1900–1906. [Google Scholar] [CrossRef]
- Dalton, H.R.; Hazeldine, S.; Banks, M.; Ijaz, S.; Bendall, R. Locally Acquired Hepatitis E in Chronic Liver Disease. Lancet Lond. Engl. 2007, 369, 1260. [Google Scholar] [CrossRef]
- Pradhan, M.; Anand, B.; Singh, A. Hepatitis E Virus Infection Causing Isolated Fetal Ascites: A Case Report. Fetal Diagn. Ther. 2012, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Krain, L.J.; Atwell, J.E.; Nelson, K.E.; Labrique, A.B. Fetal and Neonatal Health Consequences of Vertically Transmitted Hepatitis E Virus Infection. Am. J. Trop. Med. Hyg. 2014, 90, 365–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beniwal, M.; Kumar, A.; Kar, P.; Jilani, N.; Sharma, J.B. Prevalence and Severity of Acute Viral Hepatitis and Fulminant Hepatitis during Pregnancy: A Prospective Study from North India. Indian J. Med. Microbiol. 2003, 21, 184–185. [Google Scholar] [CrossRef]
- Kumar, A.; Beniwal, M.; Kar, P.; Sharma, J.B.; Murthy, N.S. Hepatitis E in Pregnancy. Int. J. Gynecol. Obstet. 2004, 85, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Nanji, A.A.; French, S.W. Relationship between Pork Consumption and Cirrhosis. Lancet Lond. Engl. 1985, 1, 681–683. [Google Scholar] [CrossRef]
- Cheng, S.-H.; Mai, L.; Zhu, F.-Q.; Pan, X.-F.; Sun, H.-X.; Cao, H.; Shu, X.; Ke, W.-M.; Li, G.; Xu, Q.-H. Influence of Chronic HBV Infection on Superimposed Acute Hepatitis E. World J. Gastroenterol. WJG 2013, 19, 5904–5909. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, S.-Y.; Zhang, D.-D.; Li, X.-Y.; Zhang, Y.-L.; Li, W.-X.; Yan, J.-J.; Wang, M.; Xun, J.-N.; Lu, C.; et al. Clinical Features of Acute Hepatitis E Super-Infections on Chronic Hepatitis B. World J. Gastroenterol. 2016, 22, 10388–10397. [Google Scholar] [CrossRef]
- Kumar, M.; Sharma, B.C.; Sarin, S.K. Hepatitis E Virus as an Etiology of Acute Exacerbation of Previously Unrecognized Asymptomatic Patients with Hepatitis B Virus-Related Chronic Liver Disease. J. Gastroenterol. Hepatol. 2008, 23, 883–887. [Google Scholar] [CrossRef]
- Aggarwal, A.; Perumpail, R.B.; Tummala, S.; Ahmed, A. Hepatitis E Virus Infection in the Liver Transplant Recipients: Clinical Presentation and Management. World J. Hepatol. 2016, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Khuroo, M.S.; Kamili, S. Aetiology, Clinical Course and Outcome of Sporadic Acute Viral Hepatitis in Pregnancy. J. Viral Hepat. 2003, 10, 61–69. [Google Scholar] [CrossRef]
- Li, M.; Bu, Q.; Gong, W.; Li, H.; Wang, L.; Li, S.; Sridhar, S.; CY Woo, P.; Wang, L. Hepatitis E Virus Infection and Its Associated Adverse Feto-Maternal Outcomes among Pregnant Women in Qinhuangdao, China. J. Matern. Fetal Neonatal Med. 2020, 33, 3647–3651. [Google Scholar] [CrossRef]
- Khuroo, M. Association of Severity of Hepatitis e Virus Infection in Mother and Vertically Transmitted Infection in Feuts. JK Pract. 2006, 13, 70–74. [Google Scholar]
- Dalton, H.R.; Bendall, R.P.; Pritchard, C.; Henley, W.; Melzer, D. National Mortality Rates from Chronic Liver Disease and Consumption of Alcohol and Pig Meat. Epidemiol. Infect. 2010, 138, 174–182. [Google Scholar] [CrossRef]
- Kumar, A.; Aggarwal, R.; Naik, S.R.; Saraswat, V.; Ghoshal, U.C.; Naik, S. Hepatitis E Virus Is Responsible for Decompensation of Chronic Liver Disease in an Endemic Region. Indian J. Gastroenterol. Off. J. Indian Soc. Gastroenterol. 2004, 23, 59–62. [Google Scholar]
- Blasco-Perrin, H.; Madden, R.G.; Stanley, A.; Crossan, C.; Hunter, J.G.; Vine, L.; Lane, K.; Devooght-Johnson, N.; Mclaughlin, C.; Petrik, J.; et al. Hepatitis E Virus in Patients with Decompensated Chronic Liver Disease: A Prospective UK/French Study. Aliment. Pharmacol. Ther. 2015, 42, 574–581. [Google Scholar] [CrossRef]
- Hamid, S.S.; Atiq, M.; Shehzad, F.; Yasmeen, A.; Nissa, T.; Salam, A.; Siddiqui, A.; Jafri, W. Hepatitis E Virus Superinfection in Patients with Chronic Liver Disease. Hepatol. Baltim. Md. 2002, 36, 474–478. [Google Scholar] [CrossRef]
- Ramachandran, J.; Eapen, C.E.; Kang, G.; Abraham, P.; Hubert, D.D.J.; Kurian, G.; Hephzibah, J.; Mukhopadhya, A.; Chandy, G.M. Hepatitis E Superinfection Produces Severe Decompensation in Patients with Chronic Liver Disease. J. Gastroenterol. Hepatol. 2004, 19, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Te, H.S.; Drobeniuc, J.; Kamili, S.; Dong, C.; Hart, J.; Sharapov, U.M. Hepatitis E Virus Infection in a Liver Transplant Recipient in the United States: A Case Report. Transplant. Proc. 2013, 45, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Tavitian, S.; Péron, J.-M.; Huynh, A.; Mansuy, J.-M.; Ysebaert, L.; Huguet, F.; Vinel, J.-P.; Attal, M.; Izopet, J.; Récher, C. Hepatitis E Virus Excretion Can Be Prolonged in Patients with Hematological Malignancies. J. Clin. Virol. Off. Public Pan Am. Soc. Clin. Virol. 2010, 49, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Weclawiak, H.; Guilbeau-Frugier, C.; Legrand-Abravanel, F.; Cointault, O.; Ribes, D.; Esposito, L.; Cardeau-Desangles, I.; Guitard, J.; Sallusto, F.; et al. Hepatitis E Virus and the Kidney in Solid-Organ Transplant Patients. Transplantation 2012, 93, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Höner zu Siederdissen, C.; Pischke, S.; Schlue, J.; Deterding, K.; Hellms, T.; Schuler-Lüttmann, S.; Schwarz, A.; Manns, M.P.; Cornberg, M.; Wedemeyer, H. Chronic Hepatitis E Virus Infection beyond Transplantation or Human Immunodeficiency Virus Infection. Hepatology 2014, 60, 1112. [Google Scholar] [CrossRef]
- Pishmisheva, M.; Shikov, P.; Golkocheva-Markova, E.; Kotsev, S.; Naseva, E.; Vatev, N.; Argirova, R. Spread of Hepatitis E Viral Infection among Hemodialysis Patients in Pazardzhik District, Bulgaria. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 1086–1092. [Google Scholar] [CrossRef]
- Hosseini-Moghaddam, S.M.; Zarei, A.; Alavian, S.M.; Mansouri, M. Hepatitis E Virus Infection: A General Review with a Focus on Hemodialysis and Kidney Transplant Patients. Am. J. Nephrol. 2010, 31, 398–407. [Google Scholar] [CrossRef]
- Sylvan, S.P.E.; Jacobson, S.H.; Christenson, B. Prevalence of Antibodies to Hepatitis E Virus among Hemodialysis Patients in Sweden. J. Med. Virol. 1998, 54, 38–43. [Google Scholar] [CrossRef]
- Li, F.; Zhuang, H.; Kolivas, S.; Locarnini, S.A.; Anderson, D.A. Persistent and Transient Antibody Responses to Hepatitis E Virus Detected by Western Immunoblot Using Open Reading Frame 2 and 3 and Glutathione S-Transferase Fusion Proteins. J. Clin. Microbiol. 1994, 32, 2060–2066. [Google Scholar] [CrossRef] [Green Version]
- Hoofnagle, J.H.; Nelson, K.E.; Purcell, R.H. Hepatitis E. N. Engl. J. Med. 2012, 367, 1237–1244. [Google Scholar] [CrossRef]
- Takahashi, M.; Kusakai, S.; Mizuo, H.; Suzuki, K.; Fujimura, K.; Masuko, K.; Sugai, Y.; Aikawa, T.; Nishizawa, T.; Okamoto, H. Simultaneous Detection of Immunoglobulin A (IgA) and IgM Antibodies against Hepatitis E Virus (HEV) Is Highly Specific for Diagnosis of Acute HEV Infection. J. Clin. Microbiol. 2005, 43, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Dawson, G.J.; Chau, K.H.; Cabal, C.M.; Yarbough, P.O.; Reyes, G.R.; Mushahwar, I.K. Solid-Phase Enzyme-Linked Immunosorbent Assay for Hepatitis E Virus IgG and IgM Antibodies Utilizing Recombinant Antigens and Synthetic Peptides. J. Virol. Methods 1992, 38, 175–186. [Google Scholar] [CrossRef]
- Tsarev, S.A.; Tsareva, T.S.; Emerson, S.U.; Kapikian, A.Z.; Ticehurst, J.; London, W.; Purcell, R.H. ELISA for Antibody to Hepatitis E Virus (HEV) Based on Complete Open-Reading Frame-2 Protein Expressed in Insect Cells: Identification of HEV Infection in Primates. J. Infect. Dis. 1993, 168, 369–378. [Google Scholar] [CrossRef]
- Norder, H.; Karlsson, M.; Mellgren, Å.; Konar, J.; Sandberg, E.; Lasson, A.; Castedal, M.; Magnius, L.; Lagging, M. Diagnostic Performance of Five Assays for Anti-Hepatitis E Virus IgG and IgM in a Large Cohort Study. J. Clin. Microbiol. 2016, 54, 549–555. [Google Scholar] [CrossRef] [Green Version]
- Wedemeyer, H.; Pischke, S.; Manns, M.P. Pathogenesis and Treatment of Hepatitis E Virus Infection. Gastroenterology 2012, 142, 1388–1397.e1. [Google Scholar] [CrossRef]
- Zhang, S.; Tian, D.; Zhang, Z.; Xiong, J.; Yuan, Q.; Ge, S.; Zhang, J.; Xia, N. Clinical Significance of Anti-HEV IgA in Diagnosis of Acute Genotype 4 Hepatitis E Virus Infection Negative for Anti-HEV IgM. Dig. Dis. Sci. 2009, 54, 2512–2518. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Subhadra, S.; Singh, B.; Panda, B.K. Hepatitis E Virus: The Current Scenario. Int. J. Infect. Dis. 2013, 17, e228–e233. [Google Scholar] [CrossRef] [Green Version]
- Baylis, S.A.; Hanschmann, K.-M.; Blümel, J.; Nübling, C.M. HEV Collaborative Study Group Standardization of Hepatitis E Virus (HEV) Nucleic Acid Amplification Technique-Based Assays: An Initial Study to Evaluate a Panel of HEV Strains and Investigate Laboratory Performance. J. Clin. Microbiol. 2011, 49, 1234–1239. [Google Scholar] [CrossRef] [Green Version]
- Mallet, V.; Nicand, E.; Sultanik, P.; Chakvetadze, C.; Tessé, S.; Thervet, E.; Mouthon, L.; Sogni, P.; Pol, S. Brief Communication: Case Reports of Ribavirin Treatment for Chronic Hepatitis E. Ann. Intern. Med. 2010, 153, 85–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhou, X.; Debing, Y.; Chen, K.; Van Der Laan, L.J.W.; Neyts, J.; Janssen, H.L.A.; Metselaar, H.J.; Peppelenbosch, M.P.; Pan, Q. Calcineurin Inhibitors Stimulate and Mycophenolic Acid Inhibits Replication of Hepatitis E Virus. Gastroenterology 2014, 146, 1775–1783. [Google Scholar] [CrossRef]
- Huang, Y.-W.; Qin, A.; Tsai, C.-Y.; Chen, P.-J. Novel Pegylated Interferon for the Treatment of Chronic Viral Hepatitis. Viruses 2022, 14, 1128. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Rostaing, L.; Abravanel, F.; Garrouste, C.; Esposito, L.; Cardeau-Desangles, I.; Mansuy, J.M.; Selves, J.; Peron, J.M.; Otal, P.; et al. Pegylated Interferon-Alpha for Treating Chronic Hepatitis E Virus Infection after Liver Transplantation. Clin. Infect. Dis. Off. Public Infect. Dis. Soc. Am. 2010, 50, e30–e33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamar, N.; Abravanel, F.; Garrouste, C.; Cardeau-Desangles, I.; Mansuy, J.M.; Weclawiak, H.; Izopet, J.; Rostaing, L. Three-Month Pegylated Interferon-Alpha-2a Therapy for Chronic Hepatitis E Virus Infection in a Haemodialysis Patient. Nephrol. Dial. Transplant. 2010, 25, 2792–2795. [Google Scholar] [CrossRef] [Green Version]
- Dao Thi, V.L.; Debing, Y.; Wu, X.; Rice, C.M.; Neyts, J.; Moradpour, D.; Gouttenoire, J. Sofosbuvir Inhibits Hepatitis E Virus Replication In Vitro and Results in an Additive Effect When Combined with Ribavirin. Gastroenterology 2016, 150, 82–85.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drinane, M.; Jing Wang, X.; Watt, K. Sofosbuvir and Ribavirin Eradication of Refractory Hepatitis E in an Immunosuppressed Kidney Transplant Recipient. Hepatol. Baltim. Md. 2019, 69, 2297–2299. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, N.; Subramani, C.; Anang, S.; Muthumohan, R.; Shalimar; Nayak, B.; Ranjith-Kumar, C.T.; Surjit, M. Zinc Salts Block Hepatitis E Virus Replication by Inhibiting the Activity of Viral RNA-Dependent RNA Polymerase. J. Virol. 2017, 91, e00754-17. [Google Scholar] [CrossRef] [Green Version]
- Todt, D.; Moeller, N.; Praditya, D.; Kinast, V.; Friesland, M.; Engelmann, M.; Verhoye, L.; Sayed, I.M.; Behrendt, P.; Dao Thi, V.L.; et al. The Natural Compound Silvestrol Inhibits Hepatitis E Virus (HEV) Replication in Vitro and in Vivo. Antiviral Res. 2018, 157, 151–158. [Google Scholar] [CrossRef]
- Netzler, N.E.; Enosi Tuipulotu, D.; Vasudevan, S.G.; Mackenzie, J.M.; White, P.A. Antiviral Candidates for Treating Hepatitis E Virus Infection. Antimicrob. Agents Chemother. 2019, 63, e00003-19. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, T.; Kobayashi, T.; Jirintai, S.; Kii, I.; Nagashima, S.; Prathiwi Primadharsini, P.; Nishizawa, T.; Okamoto, H. Screening of Novel Drugs for Inhibiting Hepatitis E Virus Replication. J. Virol. Methods 2019, 270, 1–11. [Google Scholar] [CrossRef]
- Soon, C.F.; Zhang, S.; Suneetha, P.V.; Antunes, D.A.; Manns, M.P.; Raha, S.; Schultze-Florey, C.; Prinz, I.; Wedemeyer, H.; Sällberg Chen, M.; et al. Hepatitis E Virus (HEV)-Specific T Cell Receptor Cross-Recognition: Implications for Immunotherapy. Front. Immunol. 2019, 10, 2076. [Google Scholar] [CrossRef] [Green Version]
- Johne, R.; Trojnar, E.; Filter, M.; Hofmann, J. Thermal Stability of Hepatitis E Virus as Estimated by a Cell Culture Method. Appl. Environ. Microbiol. 2016, 82, 4225–4231. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Takahashi, M.; Takahashi, H.; Ichiyama, K.; Hoshino, Y.; Nagashima, S.; Mizuo, H.; Okamoto, H. Development and Characterization of a Genotype 4 Hepatitis E Virus Cell Culture System Using a HE-JF5/15F Strain Recovered from a Fulminant Hepatitis Patient. J. Clin. Microbiol. 2009, 47, 1906–1910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lhomme, S.; Abravanel, F.; Dubois, M.; Sandres-Saune, K.; Mansuy, J.-M.; Rostaing, L.; Kamar, N.; Izopet, J. Characterization of the Polyproline Region of the Hepatitis E Virus in Immunocompromised Patients. J. Virol. 2014, 88, 12017–12025. [Google Scholar] [CrossRef] [Green Version]
- Tam, A.W.; Smith, M.M.; Guerra, M.E.; Huang, C.-C.; Bradley, D.W.; Fry, K.E.; Reyes, G.R. Hepatitis E Virus (HEV): Molecular Cloning and Sequencing of the Full-Length Viral Genome. Virology 1991, 185, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.J. Hepatitis E Virus: Animal Reservoirs and Zoonotic Risk. Vet. Microbiol. 2010, 140, 256–265. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Dai, X.; Chang, J.C.; Lopareva, E.; Pillot, J.; Fields, H.A.; Khudyakov, Y.E. Identification and Characterization of the Neutralization Epitope(s) of the Hepatitis E Virus. Virology 2001, 288, 203–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, T.; Mori, Y.; Miyazaki, N.; Cheng, R.H.; Yoshimura, M.; Unno, H.; Shima, R.; Moriishi, K.; Tsukihara, T.; Li, T.C.; et al. Biological and Immunological Characteristics of Hepatitis E Virus-like Particles Based on the Crystal Structure. Proc. Natl. Acad. Sci. USA 2009, 106, 12986–12991. [Google Scholar] [CrossRef]
- Nagashima, S.; Takahashi, M.; Jirintai, S.; Tanggis; Kobayashi, T.; Nishizawa, T.; Okamoto, H. The Membrane on the Surface of Hepatitis E Virus Particles Is Derived from the Intracellular Membrane and Contains Trans-Golgi Network Protein 2. Arch. Virol. 2014, 159, 979–991. [Google Scholar] [CrossRef]
- Ma, H.; Song, X.; Harrison, T.J.; Li, R.; Huang, G.; Zhang, H.; Kong, W.; Wang, Y. Immunogenicity and Efficacy of a Bacterially Expressed HEV ORF3 Peptide, Assessed by Experimental Infection of Primates. Arch. Virol. 2009, 154, 1641–1648. [Google Scholar] [CrossRef]
- Syed, S.F.; Sun, Y.; Du, T.; Chen, Y.; Liu, B.; Wang, X.; Li, H.; Nan, Y.; Zhou, E.-M.; Zhao, Q. Evaluation of Recombinant Chinese Avian Hepatitis E Virus (CaHEV) ORF2 and ORF3 Proteins for Protection of Chickens against CaHEV Infection. Vaccine 2017, 35, 3482–3489. [Google Scholar] [CrossRef]
- Zahmanova, G.G.; Mazalovska, M.; Takova, K.H.; Toneva, V.T.; Minkov, I.N.; Mardanova, E.S.; Ravin, N.V.; Lomonossoff, G.P. Rapid High-Yield Transient Expression of Swine Hepatitis E ORF2 Capsid Proteins in Nicotiana Benthamiana Plants and Production of Chimeric Hepatitis E Virus-Like Particles Bearing the M2e Influenza Epitope. Plants 2020, 9, 29. [Google Scholar] [CrossRef] [Green Version]
- Takova, K.; Koynarski, T.; Minkov, G.; Toneva, V.; Mardanova, E.; Ravin, N.; Lukov, G.L.; Zahmanova, G. Development and Optimization of an Enzyme Immunoassay to Detect Serum Antibodies against the Hepatitis E Virus in Pigs, Using Plant-Derived ORF2 Recombinant Protein. Vaccines 2021, 9, 991. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Takova, K.H.; Toneva, V.T.; Zahmanova, G.G.; Tsybalova, L.M.; Ravin, N.V. A Plant-Based Transient Expression System for the Rapid Production of Highly Immunogenic Hepatitis E Virus-like Particles. Biotechnol. Lett. 2020, 42, 2441–2446. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.-C.; Zhang, J.; Zhang, X.-F.; Zhou, C.; Wang, Z.-Z.; Huang, S.-J.; Wang, H.; Yang, C.-L.; Jiang, H.-M.; Cai, J.-P.; et al. Efficacy and Safety of a Recombinant Hepatitis E Vaccine in Healthy Adults: A Large-Scale, Randomised, Double-Blind Placebo-Controlled, Phase 3 Trial. Lancet Lond. Engl. 2010, 376, 895–902. [Google Scholar] [CrossRef]
- Cao, Y.-F.; Tao, H.; Hu, Y.-M.; Shi, C.-B.; Wu, X.; Liang, Q.; Chi, C.-P.; Li, L.; Liang, Z.-L.; Meng, J.-H.; et al. A Phase 1 Randomized Open-Label Clinical Study to Evaluate the Safety and Tolerability of a Novel Recombinant Hepatitis E Vaccine. Vaccine 2017, 35, 5073–5080. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Meng, J. Expression, purification and immunogenicity of a novel hepatitis E virus-like particle. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi Chin. J. Cell. Mol. Immunol. 2006, 22, 339–342. [Google Scholar]
- Wu, X.; Chen, P.; Lin, H.; Hao, X.; Liang, Z. Hepatitis E Virus: Current Epidemiology and Vaccine. Hum. Vaccines Immunother. 2016, 12, 2603–2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norwegian Institute of Public Health. An Effectiveness Trial (Phase IV) to Evaluate Protection of Pregnant Women by Hepatitis E Virus (HEV) Vaccine in Bangladesh and Risk Factors for Severe HEV Infection. Available online: https://clinicaltrials.gov/ct2/show/NCT02759991 (accessed on 14 July 2023).
- Purdy, M.A.; McCaustland, K.A.; Krawczynski, K.; Spelbring, J.; Reyes, G.R.; Bradley, D.W. Preliminary Evidence That a TrpE-HEV Fusion Protein Protects Cynomolgus Macaques against Challenge with Wild-Type Hepatitis e Virus (HEV). J. Med. Virol. 1993, 41, 90–94. [Google Scholar] [CrossRef]
- Im, S.W.K.; Zhang, J.Z.; Zhuang, H.; Che, X.Y.; Zhu, W.F.; Xu, G.M.; Li, K.; Xia, N.S.; Ng, M.H. A Bacterially Expressed Peptide Prevents Experimental Infection of Primates by the Hepatitis E Virus. Vaccine 2001, 19, 3726–3732. [Google Scholar] [CrossRef]
- Ge, S.; Zhang, J.; Huang, G.; Pang, S.; Zhou, K.; Xia, N. The immuno-protect study of a hepatitis E virus ORF2 peptide expressed in E. coli. Wei Sheng Wu Xue Bao 2003, 43, 35–42. [Google Scholar]
- Li, S.-W.; Zhang, J.; He, Z.-Q.; Gu, Y.; Liu, R.-S.; Lin, J.; Chen, Y.-X.; Ng, M.-H.; Xia, N.-S. Mutational Analysis of Essential Interactions Involved in the Assembly of Hepatitis E Virus Capsid*. J. Biol. Chem. 2005, 280, 3400–3406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.W.; Zhang, J.; Li, Y.M.; Ou, S.H.; Huang, G.Y.; He, Z.Q.; Ge, S.X.; Xian, Y.L.; Pang, S.Q.; Ng, M.H.; et al. A Bacterially Expressed Particulate Hepatitis E Vaccine: Antigenicity, Immunogenicity and Protectivity on Primates. Vaccine 2005, 23, 2893–2901. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-N.; Cao, Y.-F.; Chen, Z.-Y.; Li, Y.-T.; Liu, Y.-L. Reproductive Toxicity Test of Recombinant Hepatitis E Vaccine. Chin. J. Biol. 2014, 27, 1503–1507. [Google Scholar]
- Zheng, M.; Jiang, J.; Zhang, X.; Wang, N.; Wang, K.; Li, Q.; Li, T.; Lin, Q.; Wang, Y.; Yu, H.; et al. Characterization of Capsid Protein (P495) of Hepatitis E Virus Expressed in Escherichia Coli and Assembling into Particles in Vitro. Vaccine 2018, 36, 2104–2111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Emerson, S.U.; Nguyen, H.; Engle, R.E.; Govindarajan, S.; Gerin, J.L.; Purcell, R.H. Immunogenicity and Protective Efficacy of a Vaccine Prepared from 53 KDa Truncated Hepatitis E Virus Capsid Protein Expressed in Insect Cells. Vaccine 2001, 20, 853–857. [Google Scholar] [CrossRef]
- Robinson, R.A.; Burgess, W.H.; Emerson, S.U.; Leibowitz, R.S.; Sosnovtseva, S.A.; Tsarev, S.; Purcell, R.H. Structural Characterization of Recombinant Hepatitis E Virus ORF2 Proteins in Baculovirus-Infected Insect Cells. Protein Expr. Purif. 1998, 12, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Purcell, R.H.; Nguyen, H.; Shapiro, M.; Engle, R.E.; Govindarajan, S.; Blackwelder, W.C.; Wong, D.C.; Prieels, J.-P.; Emerson, S.U. Pre-Clinical Immunogenicity and Efficacy Trial of a Recombinant Hepatitis E Vaccine. Vaccine 2003, 21, 2607–2615. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-C.; Takeda, N.; Miyamura, T.; Matsuura, Y.; Wang, J.C.Y.; Engvall, H.; Hammar, L.; Xing, L.; Cheng, R.H. Essential Elements of the Capsid Protein for Self-Assembly into Empty Virus-like Particles of Hepatitis E Virus. J. Virol. 2005, 79, 12999–13006. [Google Scholar] [CrossRef] [Green Version]
- Li, T.-C.; Suzaki, Y.; Ami, Y.; Dhole, T.N.; Miyamura, T.; Takeda, N. Protection of Cynomolgus Monkeys against HEV Infection by Oral Administration of Recombinant Hepatitis E Virus-like Particles. Vaccine 2004, 22, 370–377. [Google Scholar] [CrossRef]
- Wu, T.; Li, S.-W.; Zhang, J.; Ng, M.-H.; Xia, N.-S.; Zhao, Q. Hepatitis E Vaccine Development. Hum. Vaccines Immunother. 2012, 8, 823–827. [Google Scholar] [CrossRef]
- Wen, J.; Behloul, N.; Dai, X.; Dong, C.; Liang, J.; Zhang, M.; Shi, C.; Meng, J. Immunogenicity Difference between Two Hepatitis E Vaccines Derived from Genotype 1 and 4. Antiviral Res. 2016, 128, 36–42. [Google Scholar] [CrossRef]
- Li, S.-W.; Zhao, Q.; Wu, T.; Chen, S.; Zhang, J.; Xia, N.-S. The Development of a Recombinant Hepatitis E Vaccine HEV 239. Hum. Vaccines Immunother. 2015, 11, 908–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Wu, X.; Ou, S.; Lin, C.; Cheng, T.; Li, S.; Ng, M.H.; Zhang, J.; Xia, N. Difference of T Cell and B Cell Activation in Two Homologous Proteins with Similar Antigenicity but Great Distinct Immunogenicity. Mol. Immunol. 2007, 44, 3261–3266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; McAtee, P.; Yarbough, P.O.; Tam, A.W.; Fuerst, T. Expression, Characterization, and Immunoreactivities of a Soluble Hepatitis E Virus Putative Capsid Protein Species Expressed in Insect Cells. Clin. Diagn. Lab. Immunol. 1997, 4, 423–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAtee, C.P.; Zhang, Y.; Yarbough, P.O.; Bird, T.; Fuerst, T.R. Purification of a Soluble Hepatitis E Open Reading Frame 2-Derived Protein with Unique Antigenic Properties. Protein Expr. Purif. 1996, 8, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Schofield, D.J.; Glamann, J.; Emerson, S.U.; Purcell, R.H. Identification by Phage Display and Characterization of Two Neutralizing Chimpanzee Monoclonal Antibodies to the Hepatitis E Virus Capsid Protein. J. Virol. 2000, 74, 5548–5555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrestha, M.P.; Scott, R.M.; Joshi, D.M.; Mammen, M.P.; Thapa, G.B.; Thapa, N.; Myint, K.S.A.; Fourneau, M.; Kuschner, R.A.; Shrestha, S.K.; et al. Safety and Efficacy of a Recombinant Hepatitis E Vaccine. N. Engl. J. Med. 2007, 356, 895–903. [Google Scholar] [CrossRef]
- Andrews, J.U.S. Military Sponsored Vaccine Trials and La Resistance in Nepal. Am. J. Bioeth. AJOB 2005, 5, W1–W3. [Google Scholar] [CrossRef]
- Basu, S.; Lurie, P. Hepatitis E Vaccine. N. Engl. J. Med. 2007, 356, 2421–2422. [Google Scholar] [CrossRef]
- Gupta, J.; Kaul, S.; Srivastava, A.; Kaushik, N.; Ghosh, S.; Sharma, C.; Batra, G.; Banerjee, M.; Shalimar; Nayak, B.; et al. Expression, Purification and Characterization of the Hepatitis E Virus Like-Particles in the Pichia Pastoris. Front. Microbiol. 2020, 11, 107817. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, S.R.; Bobrowicz, P.; Bobrowicz, B.; Davidson, R.C.; Li, H.; Mitchell, T.; Nett, J.H.; Rausch, S.; Stadheim, T.A.; Wischnewski, H.; et al. Production of Complex Human Glycoproteins in Yeast. Science 2003, 301, 1244–1246. [Google Scholar] [CrossRef]
- Hermeling, S.; Crommelin, D.J.A.; Schellekens, H.; Jiskoot, W. Structure-Immunogenicity Relationships of Therapeutic Proteins. Pharm. Res. 2004, 21, 897–903. [Google Scholar] [CrossRef]
- Simanavicius, M.; Tamosiunas, P.L.; Petraityte-Burneikiene, R.; Johne, R.; Ulrich, R.G.; Zvirbliene, A.; Kucinskaite-Kodze, I. Generation in Yeast and Antigenic Characterization of Hepatitis E Virus Capsid Protein Virus-like Particles. Appl. Microbiol. Biotechnol. 2018, 102, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-X.; Gu, M.-R.; Zhang, P.; Jin, Z.-J.; Meng, F.-H.; Chen, E.-J.; Yang, Z.; Liu, Y.; Wang, Y.-C. Expression of ORF2 protein of HEV genotype IV in Hansenula polymorpha. Sheng Wu Gong Cheng Xue Bao Chin. J. Biotechnol. 2007, 23, 73–78. [Google Scholar]
- Su, C.; Li, L.; Jin, Z.; Han, X.; Zhao, P.; Wang, L.; Jiang, C.; Wang, Y.; Wang, W.; Xu, D.; et al. Fermentation, purification and immunogenicity evaluation of hepatitis E virus-like particles expressed in Hansenula polymorpha. Sheng Wu Gong Cheng Xue Bao Chin. J. Biotechnol. 2017, 33, 653–663. [Google Scholar] [CrossRef]
- Ward, B.J.; Makarkov, A.; Séguin, A.; Pillet, S.; Trépanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, Immunogenicity, and Safety of a Plant-Derived, Quadrivalent, Virus-like Particle Influenza Vaccine in Adults (18–64 Years) and Older Adults (≥65 Years): Two Multicentre, Randomised Phase 3 Trials. Lancet 2020, 396, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.-Y.; Couture, M.; D’Aoust, M.-A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 Randomized Trial of a Plant-Derived Virus-like Particle Vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef]
- Schillberg, S.; Finnern, R. Plant Molecular Farming for the Production of Valuable Proteins—Critical Evaluation of Achievements and Future Challenges. J. Plant Physiol. 2021, 258–259, 153359. [Google Scholar] [CrossRef] [PubMed]
- Zahmanova, G.; Takova, K.; Valkova, R.; Toneva, V.; Minkov, I.; Andonov, A.; Lukov, G.L. Plant-Derived Recombinant Vaccines against Zoonotic Viruses. Life 2022, 12, 156. [Google Scholar] [CrossRef]
- Zahmanova, G.; Aljabali, A.A.; Takova, K.; Toneva, V.; Tambuwala, M.M.; Andonov, A.P.; Lukov, G.L.; Minkov, I. The Plant Viruses and Molecular Farming: How Beneficial They Might Be for Human and Animal Health? Int. J. Mol. Sci. 2023, 24, 1533. [Google Scholar] [CrossRef]
- Jung, J.-W.; Zahmanova, G.; Minkov, I.; Lomonossoff, G.P. Plant-Based Expression and Characterization of SARS-CoV-2 Virus-like Particles Presenting a Native Spike Protein. Plant Biotechnol. J. 2022, 20, 1363–1372. [Google Scholar] [CrossRef]
- Ma, Y.; Lin, S.-Q.; Gao, Y.; Li, M.; Luo, W.-X.; Zhang, J.; Xia, N.-S. Expression of ORF2 Partial Gene of Hepatitis E Virus in Tomatoes and Immunoactivity of Expression Products. World J. Gastroenterol. 2003, 9, 2211–2215. [Google Scholar] [CrossRef]
- Zhou, Y.-X.; Lee, M.Y.-T.; Ng, J.M.-H.; Chye, M.-L.; Yip, W.-K.; Zee, S.-Y.; Lam, E. A Truncated Hepatitis E Virus ORF2 Protein Expressed in Tobacco Plastids Is Immunogenic in Mice. World J. Gastroenterol. WJG 2006, 12, 306–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maloney, B.J.; Takeda, N.; Suzaki, Y.; Ami, Y.; Li, T.C.; Miyamura, T.; Arntzen, C.J.; Mason, H.S. Challenges in Creating a Vaccine to Prevent Hepatitis E. Vaccine 2005, 23, 1870–1874. [Google Scholar] [CrossRef]
- Mazalovska, M.; Varadinov, N.; Koynarski, T.; Minkov, I.; Teoharov, P.; Lomonossoff, G.P.; Zahmanova, G. Detection of Serum Antibodies to Hepatitis E Virus Based on HEV Genotype 3 ORF2 Capsid Protein Expressed in Nicotiana Benthamiana. Ann. Lab. Med. 2017, 37, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behloul, N.; Baha, S.; Liu, Z.; Wei, W.; Zhu, Y.; Rao, Y.; Shi, R.; Meng, J. Design and Development of a Chimeric Vaccine Candidate against Zoonotic Hepatitis E and Foot-and-Mouth Disease. Microb. Cell Factories 2020, 19, 137. [Google Scholar] [CrossRef] [PubMed]
- Zahmanova, G.; Mazalovska, M.; Takova, K.; Toneva, V.; Minkov, I.; Peyret, H.; Lomonossoff, G. Efficient Production of Chimeric Hepatitis B Virus-Like Particles Bearing an Epitope of Hepatitis E Virus Capsid by Transient Expression in Nicotiana Benthamiana. Life 2021, 11, 64. [Google Scholar] [CrossRef]
- Baha, S.; Zhang, M.; Behloul, N.; Liu, Z.; Wei, W.; Meng, J. Efficient Production and Characterization of Immunogenic HEV-PCV2 Chimeric Virus-like Particles. Vet. Microbiol. 2022, 268, 109410. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Kotlyarov, R.Y.; Stuchinskaya, M.D.; Nikolaeva, L.I.; Zahmanova, G.; Ravin, N.V. High-Yield Production of Chimeric Hepatitis E Virus-Like Particles Bearing the M2e Influenza Epitope and Receptor Binding Domain of SARS-CoV-2 in Plants Using Viral Vectors. Int. J. Mol. Sci. 2022, 23, 15684. [Google Scholar] [CrossRef]
- Xiang, K.; Kusov, Y.; Ying, G.; Yan, W.; Shan, Y.; Jinyuan, W.; Na, Y.; Yan, Z.; Hongjun, L.; Maosheng, S. A Recombinant HAV Expressing a Neutralization Epitope of HEV Induces Immune Response against HAV and HEV in Mice. Viruses 2017, 9, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Su, Q.; Yi, Y.; Jia, Z.; Wang, H.; Lu, X.; Qiu, F.; Bi, S. Enhanced Mucosal Immune Responses Induced by a Combined Candidate Mucosal Vaccine Based on Hepatitis A Virus and Hepatitis E Virus Structural Proteins Linked to Tuftsin. PLoS ONE 2015, 10, e0123400. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Cassi, X.; Martínez-Puchol, S.; Silva-Sales, M.; Cornejo, T.; Bartolome, R.; Bofill-Mas, S.; Girones, R. Unveiling Viruses Associated with Gastroenteritis Using a Metagenomics Approach. Viruses 2020, 12, 1432. [Google Scholar] [CrossRef] [PubMed]
- Mardanova, E.S.; Kotlyarov, R.Y.; Ravin, N.V. Rapid Transient Expression of Receptor-Binding Domain of SARS-CoV-2 and the Conserved M2e Peptide of Influenza A Virus Linked to Flagellin in Nicotiana Benthamiana Plants Using Self-Replicating Viral Vector. Plants 2022, 11, 3425. [Google Scholar] [CrossRef]
- Xia, M.; Wei, C.; Wang, L.; Cao, D.; Meng, X.-J.; Jiang, X.; Tan, M. A Trivalent Vaccine Candidate against Hepatitis E Virus, Norovirus, and Astrovirus. Vaccine 2016, 34, 905–913. [Google Scholar] [CrossRef] [Green Version]
- Trabelsi, K.; Kamen, A.; Kallel, H. Development of a Vectored Vaccine against Hepatitis E Virus. Vaccine 2014, 32, 2808–2811. [Google Scholar] [CrossRef]
- Kamili, S.; Spelbring, J.; Carson, D.; Krawczynski, K. Protective Efficacy of Hepatitis E Virus DNA Vaccine Administered by Gene Gun in the Cynomolgus Macaque Model of Infection. J. Infect. Dis. 2004, 189, 258–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamili, S.; Spelbring, J.; Krawczynski, K. DNA Vaccination against Hepatitis E Virus Infection in Cynomolgus Macaques. J. Gastroenterol. Hepatol. 2002, 17, S365–S369. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, T.M.; Lole, K.S.; Tripathy, A.S.; Arankalle, V.A. Immunogenicity of Candidate Hepatitis E Virus DNA Vaccine Expressing Complete and Truncated ORF2 in Mice. Vaccine 2007, 25, 4350–4360. [Google Scholar] [CrossRef]
- Arankalle, V.A.; Lole, K.S.; Deshmukh, T.M.; Srivastava, S.; Shaligram, U.S. Challenge Studies in Rhesus Monkeys Immunized with Candidate Hepatitis E Vaccines: DNA, DNA-Prime-Protein-Boost and DNA-Protein Encapsulated in Liposomes. Vaccine 2009, 27, 1032–1039. [Google Scholar] [CrossRef]
- Sanford, B.J.; Opriessnig, T.; Kenney, S.P.; Dryman, B.A.; Córdoba, L.; Meng, X.-J. Assessment of the Cross-Protective Capability of Recombinant Capsid Proteins Derived from Pig, Rat, and Avian Hepatitis E Viruses (HEV) against Challenge with a Genotype 3 HEV in Pigs. Vaccine 2012, 30, 6249–6255. [Google Scholar] [CrossRef] [Green Version]
- Sanford, B.J.; Dryman, B.A.; Huang, Y.-W.; Feagins, A.R.; Leroith, T.; Meng, X.-J. Prior Infection of Pigs with a Genotype 3 Swine Hepatitis E Virus (HEV) Protects against Subsequent Challenges with Homologous and Heterologous Genotypes 3 and 4 Human HEV. Virus Res. 2011, 159, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Sridhar, S.; Yip, C.C.-Y.; Wu, S.; Chew, N.F.-S.; Leung, K.-H.; Chan, J.F.-W.; Zhao, P.S.; Chan, W.-M.; Poon, R.W.-S.; Tsoi, H.-W.; et al. Transmission of Rat Hepatitis E Virus Infection to Humans in Hong Kong: A Clinical and Epidemiological Analysis. Hepatol. Baltim. Md 2021, 73, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Situ, J.; Cai, J.-P.; Yip, C.C.-Y.; Wu, S.; Zhang, A.J.-X.; Wen, L.; Chew, N.F.-S.; Chan, W.-M.; Poon, R.W.-S.; et al. Multimodal Investigation of Rat Hepatitis E Virus Antigenicity: Implications for Infection, Diagnostics, and Vaccine Efficacy. J. Hepatol. 2021, 74, 1315–1324. [Google Scholar] [CrossRef] [PubMed]


| Africa | Asia | Europe | North America | South America | Oceania | ||
|---|---|---|---|---|---|---|---|
| IgG | % positive | 21.76% | 15.80% | 9.31% | 8.05% | 7.28% | 5.99% |
| n individuals | 22,377 | 681,373 | 132,419 | 71,989 | 14,586 | 1563 | |
| IgM | % positive | 3.09% | 1.86% | 0.79% | 0.22% | 2.43% | - |
| n individuals | 5001 | 141,565 | 146,322 | 12,197 | 2680 | - | |
| HEV RNA | % positive | 0.00% | 0.93% | 0.08% | 0.00% | 0.18% | 0.00% |
| n individuals | 278 | 727,744 | 2,441,774 | 34,761 | 1054 | 74,131 |
| Name | HEV Genotype/ ORF2 Length | Protein Size/VLPs Formation | Expression System | Immunological Studies | References |
|---|---|---|---|---|---|
| trpE-C2 | HEV-1 Burmese strain 221–660 aa | fusion protein | E. coli | Monkeys are fully protected after challenged with a HEV-1 Burmese strain | [223] |
| E2 | HEV-1 394–606 aa | 23 kDa homodimer | E. coli | Rhesus monkeys are fully protected against infection with a HEV-1 virus. | [224,225] |
| p239 | HEV-1 368–606 aa | 30 kDa VLPs | E. coli | Passed clinical trial phases II and III, showed 100% efficacy | [226,227] |
| p179 | HEV-4 439–617 aa | 20kDa VLPs | E. coli | passed phase I clinical trial | [219,228] |
| p495 | HEV-1 112–606 aa | 53 kDa VLPs | E. coli and Tn5 insect cells | Mouse immunization results showed that p495 VLPs extracted from E. coli and insect cells had comparable immunogenicity, as well as p239 | [229] |
| Full-length ORF2 | HEV-1 Sar-55 strain (Pakistan) aa 1–660 | 72 kDa no VLPs 63 kDa no VLPs 56 kDa form VLPs 53 kDa no VLPs | Insect cells | Rhesus macaques immunized with the 53 kDa protein are not fully protected 56 kD Sar-55-ORF2 with adjuvant-induced protection against HEV in monkeys challenged with HEV-1, 2, or 3 strain 56 kDa Sar-55 ORF2 was evaluated in human clinical trials phase I and II. | [230,231,232] |
| ORF2 fragment | HEV-1 Burmese strain aa 112–608 | 50 kDa in size forms VLPs | Insect cells | Oral administration of five 10 mg doses of VLPs can confer protection from hepatitis for immunized macaques upon challenge with 10,000 50% monkey infectious dose (MID50) of homogenous genotype 1 strain | [36,233,234] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahmanova, G.; Takova, K.; Tonova, V.; Koynarski, T.; Lukov, L.L.; Minkov, I.; Pishmisheva, M.; Kotsev, S.; Tsachev, I.; Baymakova, M.; et al. The Re-Emergence of Hepatitis E Virus in Europe and Vaccine Development. Viruses 2023, 15, 1558. https://doi.org/10.3390/v15071558
Zahmanova G, Takova K, Tonova V, Koynarski T, Lukov LL, Minkov I, Pishmisheva M, Kotsev S, Tsachev I, Baymakova M, et al. The Re-Emergence of Hepatitis E Virus in Europe and Vaccine Development. Viruses. 2023; 15(7):1558. https://doi.org/10.3390/v15071558
Chicago/Turabian StyleZahmanova, Gergana, Katerina Takova, Valeria Tonova, Tsvetoslav Koynarski, Laura L. Lukov, Ivan Minkov, Maria Pishmisheva, Stanislav Kotsev, Ilia Tsachev, Magdalena Baymakova, and et al. 2023. "The Re-Emergence of Hepatitis E Virus in Europe and Vaccine Development" Viruses 15, no. 7: 1558. https://doi.org/10.3390/v15071558
APA StyleZahmanova, G., Takova, K., Tonova, V., Koynarski, T., Lukov, L. L., Minkov, I., Pishmisheva, M., Kotsev, S., Tsachev, I., Baymakova, M., & Andonov, A. P. (2023). The Re-Emergence of Hepatitis E Virus in Europe and Vaccine Development. Viruses, 15(7), 1558. https://doi.org/10.3390/v15071558










