Epidemiology Update of Hepatitis E Virus (HEV) in Uruguay: Subtyping, Environmental Surveillance and Zoonotic Transmission
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA Extraction and Reverse-Transcription Nested PCR (RT-nPCR) from Domestic Pigs Samples
2.2. Wastewater and Surface Water Concentration, RNA Extraction and HEV Detection
2.3. Full-Length Genome Sequencing by Next Generation Sequencing (NGS), Sequence Analysis and Phylogenetic Reconstruction
3. Results
3.1. HEV Detection in Domestic Pig Stool Samples via RT-nPCR
3.2. Whole-Genome Analysis of a Swine HEV Strain
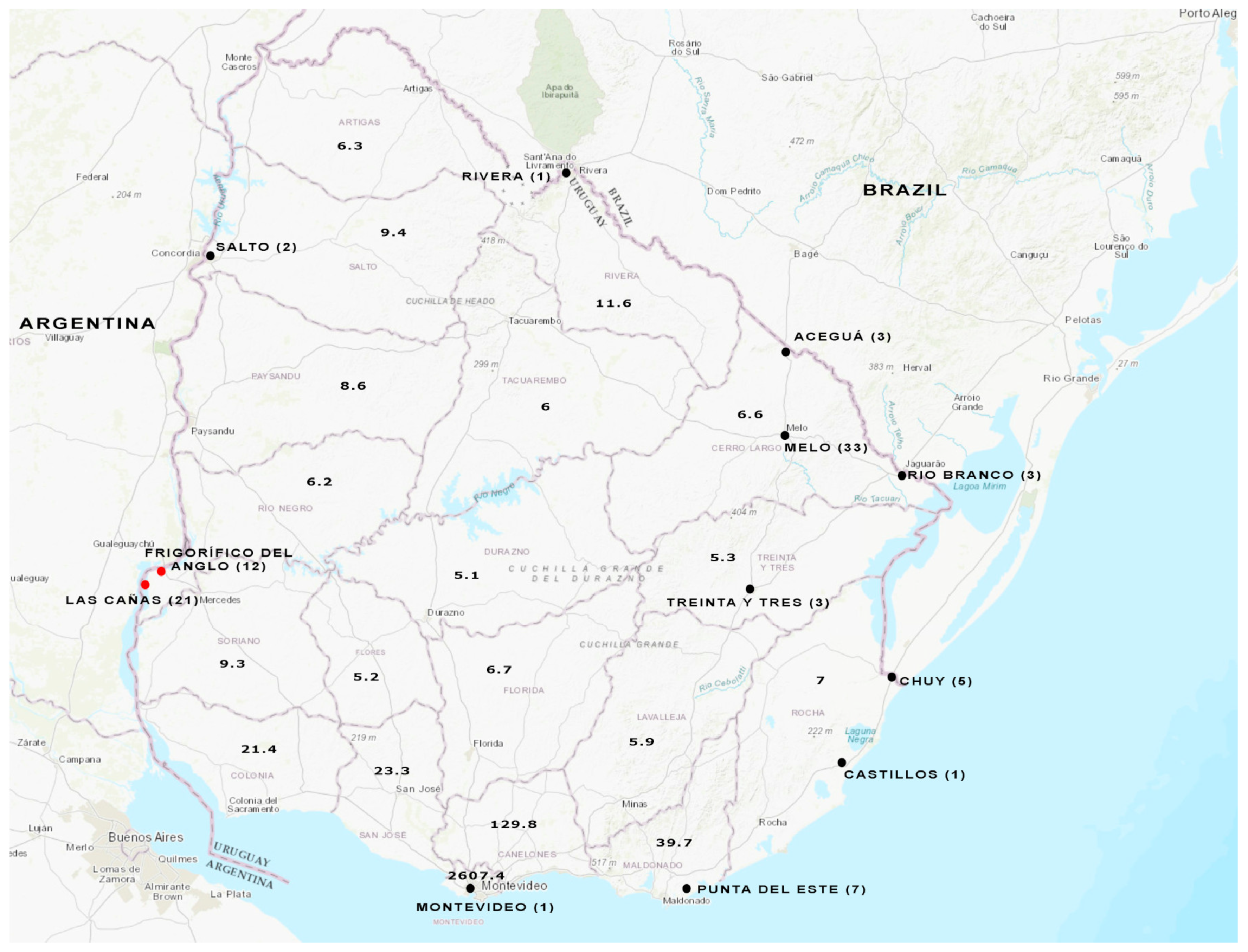
3.3. Analysis of Wastewater and Surface Water Samples
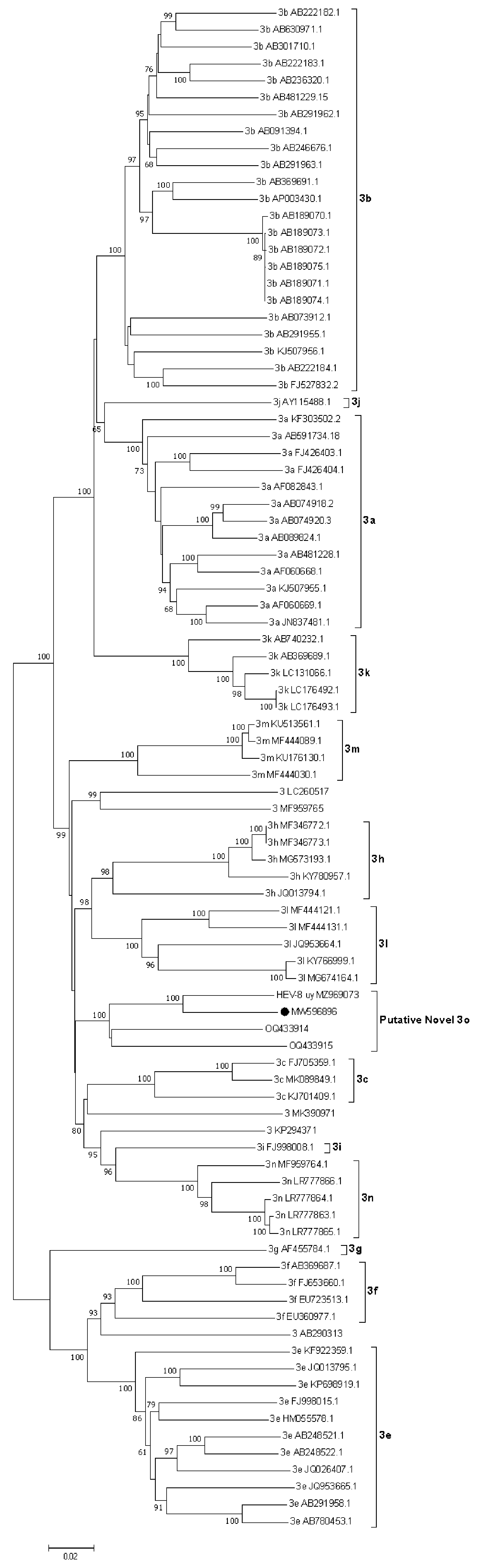
3.4. HEV Subtyping
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Hepatitis Report. 2017. Available online: https://www.who.int/publications/i/item/9789241565455 (accessed on 10 January 2022).
- Ji, H.; Chen, S.; He, Q.; Wang, W.; Gong, S.; Qian, Z.; Zhang, Y.; Wei, D.; Yu, W.; Huang, F. The different replication between nonenveloped and quasi-enveloped hepatitis E virus. J. Med. Virol. 2021, 93, 6267–6277. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Subhadra, S.; Singh, B.; Panda, B.K. Hepatitis E virus: The current scenario. Int. J. Infect. Dis. 2013, 17, e228–e233. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Zhang, Y.-J. Molecular Biology and Infection of Hepatitis E Virus. Front. Microbiol. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Purdy, M.A.; Drexler, J.F.; Meng, X.J.; Norder, H.; Okamoto, H.; Van der Poel, W.H.M.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 1778. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Izopet, J.; Nicot, F.; Simmonds, P.; Jameel, S.; Meng, X.-J.; Norder, H.; Okamoto, H.; van Der Poel, W.H.M.; Reuter, G.; et al. Update: Proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A). J. Gen. Virol. 2020, 101, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.Y.; Han, H. Hepatitis E viral infection in solid organ transplant patients. Curr. Opin. Organ Transplant. 2017, 22, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Abravanel, F.; Lhomme, S.; El Costa, H.; Schvartz, B.; Peron, J.-M.; Kamar, N.; Izopet, J. Rabbit Hepatitis E Virus Infections in Humans, France. Emerg. Infect. Dis. 2017, 23, 1191–1193. [Google Scholar] [CrossRef]
- Oliveira-Filho, E.F.; König, M.; Thiel, H.-J. Genetic variability of HEV isolates: Inconsistencies of current classification. Vet. Microbiol. 2013, 165, 148–154. [Google Scholar] [CrossRef]
- Smith, D.B.; Simmonds, P.; Izopet, J.; Oliveira-Filho, E.F.; Ulrich, R.G.; Johne, R.; Koenig, M.; Jameel, S.; Harrison, T.J.; Meng, X.-J.; et al. Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 2016, 97, 537–542. [Google Scholar] [CrossRef]
- Nicot, F.; Dimeglio, C.; Migueres, M.; Jeanne, N.; Latour, J.; Abravanel, F.; Ranger, N.; Harter, A.; Dubois, M.; Lameiras, S.; et al. Classification of the Zoonotic Hepatitis E Virus Genotype 3 Into Distinct Subgenotypes. Front. Microbiol. 2021, 11, 634430. [Google Scholar] [CrossRef]
- Takuissu, G.R.; Kenmoe, S.; Ndip, L.; Ebogo-Belobo, J.T.; Kengne-Ndé, C.; Mbaga, D.S.; Bowo-Ngandji, A.; Oyono, M.G.; Kenfack-Momo, R.; Tchatchouang, S.; et al. Hepatitis E Virus in Water Environments: A Systematic Review and Meta-analysis. Food Environ. Virol. 2022, 14, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, H.; Chassaing, M.; Berger, S.; Gantzer, C.; Bertrand, I.; Schvoerer, E. Transmission of hepatitis E virus by water: An issue still pending in industrialized countries. Water Res. 2019, 151, 144–157. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Waterborne Outbreaks of Hepatitis E: Recognition, Investigation and Control; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Mirazo, S.; Ramos, N.; Russi, J.C.; Arbiza, J. Genetic heterogeneity and subtyping of human Hepatitis E virus isolates from Uruguay. Virus Res. 2013, 173, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Mirazo, S.; Gardinali, N.R.; Cecilia, D.A.; Verger, L.; Ottonelli, F.; Ramos, N.; Castro, G.; Pinto, M.A.; Ré, V.; Pisano, B.; et al. Serological and virological survey of hepatitis E virus (HEV) in animal reservoirs from Uruguay reveals elevated prevalences and a very close phylogenetic relationship between swine and human strains. Vet. Microbiol. 2018, 213, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Ferreiro, I.; Herrera, M.L.; González, I.; Cancela, F.; Leizagoyen, C.; Loureiro, M.; Arellano, H.; Echaides, C.; Bon, B.; Castro, G.; et al. Hepatitis E Virus (HEV) infection in captive white-collared peccaries (Pecari tajacu) from Uruguay. Transbound. Emerg. Dis. 2020, 68, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- Cancela, F.; Panzera, Y.; Mainardi, V.; Gerona, S.; Ramos, N.; Pérez, R.; Arbiza, J.; Mirazo, S. Complete Genome Sequence of Hepatitis E Virus Genotype 3 Obtained from a Chronically Infected Individual in Uruguay. Microbiol. Resour. Announc. 2021, 10, e0036721. [Google Scholar] [CrossRef] [PubMed]
- MSP. Dirección General de la Salud División Epidemiología Departamento de Vigilancia en Salud. Montevideo, Uruguay. Report Provided upon Request for Information Made by the Author (received on 10 December 2022). Available online: https://www.gub.uy/ministerio-salud-publica/tramites-y-servicios/formularios/formularios-division-epidemiologia-departamento-vigilancia-salud (accessed on 18 September 2023).
- Bangueses, F.; Abin-Carriquiry, J.A.; Cancela, F.; Curbelo, J.; Mirazo, S. Serological and molecular prevalence of hepatitis E virus among blood donors from Uruguay. J. Med. Virol. 2020, 93, 4010–4014. [Google Scholar] [CrossRef]
- Songtanin, B.; Molehin, A.J.; Brittan, K.; Manatsathit, W.; Nugent, K. Hepatitis E Virus Infections: Epidemiology, Genetic Diversity, and Clinical Considerations. Viruses 2023, 15, 1389. [Google Scholar] [CrossRef]
- Velavan, T.P.; Pallerla, S.R.; Johne, R.; Todt, D.; Steinmann, E.; Schemmerer, M.; Wenzel, J.J.; Hofmann, J.; Shih, J.W.K.; Wedemeyer, H.; et al. Hepatitis E: An update on One Health and clinical medicine. Liver Int. 2021, 41, 1462–1473. [Google Scholar] [CrossRef]
- Kumar, M.; Jiang, G.; Thakur, A.K.; Chatterjee, S.; Bhattacharya, T.; Mohapatra, S.; Chaminda, T.; Tyagi, V.K.; Vithanage, M.; Bhattacharya, P.; et al. Lead time of early warning by wastewater surveillance for COVID-19: Geographical variations and impacting factors. Chem. Eng. J. 2022, 441, 135936. [Google Scholar] [CrossRef]
- Lo Castro, I.; Espul, C.; de Paula, V.S.; Altabert, N.R.; Gonzalez, J.E.; Lago, B.V.; Villar, L.M. High prevalence of hepatitis A and E viruses in environmental and clinical samples from West Argentina. Braz. J. Infect. Dis. 2023, 27, 102738. [Google Scholar] [CrossRef] [PubMed]
- Baez, P.A.; Lopez, M.C.; Duque-Jaramillo, A.; Pelaez, D.; Molina, F.; Navas, M.C. First evidence of the Hepatitis E virus in environmental waters in Colombia. PLoS ONE 2017, 12, e0177525. [Google Scholar] [CrossRef] [PubMed]
- Pisano, M.B.; Lugo, B.C.; Poma, R.; Cristobal, H.A.; Raskovsky, V.; Martinez Wassaf, M.G.; Rajal, V.B.; Re, V.E. Environmental hepatitis E virus detection supported by serological evidence in the northwest of Argentina. Trans. R. Soc. Trop. Med. Hyg. 2018, 112, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Heldt, F.H.; Staggmeier, R.; Gularte, J.S.; Demoliner, M.; Henzel, A.; Spilki, F.R. Hepatitis E Virus in Surface Water, Sediments, and Pork Products Marketed in Southern Brazil. Food Environ. Virol. 2016, 8, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Pisano, M.B.; Mirazo, S.; Re, V.E. Hepatitis E Virus Infection: Is It Really a Problem in Latin America? Clin. Liver Dis. 2020, 16, 108. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Simpson, S.L.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Blackall, L.L.; Bofill-Mas, S.; Bosh, A.; Brandao, J.; Choi, P.M.; et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022, 805, 149877. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cataluña, A.; Cuevas-Ferrando, E.; Randazzo, W.; Falcó, I.; Allende, A.; Sánchez, G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021, 758, 143870. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12, 10-1128. [Google Scholar] [CrossRef]
- Cancela, F.; Ramos, N.; Smyth, D.S.; Etchebehere, C.; Berois, M.; Rodríguez, J.; Rufo, C.; Alemán, A.; Borzacconi, L.; López, J.; et al. Wastewater surveillance of SARS-CoV-2 genomic populations on a country-wide scale through targeted sequencing. PLoS ONE 2023, 18, e0284483. [Google Scholar] [CrossRef]
- Bortagaray, V.; Gamazo, P.; Castro, S.; Grilli, M.; Colina, R.; Victoria, M. Comparison of the risk of infection of human rotavirus and astrovirus according to fishing and swimming activities at Las Cañas beach, Uruguay. J. Appl. Microbiol. 2022, 133, 3523–3533. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, D.R.L.; Durães-Carvalho, R.; Gardinali, N.R.; Machado, L.C.; de Paula, V.S.; da Luz Wallau, G.; de Oliveira, J.M.; Pena, L.J.; Pinto, M.A.; Gil, L.H.V.G.; et al. Uncovering neglected subtypes and zoonotic transmission of Hepatitis E virus (HEV) in Brazil. Virol. J. 2023, 20, 83. [Google Scholar] [CrossRef] [PubMed]
- Sayed, I.M. Dual Infection of Hepatitis A Virus and Hepatitis E Virus- What Is Known? Viruses 2023, 15, 298. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Nasheri, N. Animal reservoirs for hepatitis E virus within the Paslahepevirus genus. Vet. Microbiol. 2023, 278, 109618. [Google Scholar] [CrossRef] [PubMed]
- Mirazo, S.; Ramos, N.; Mainardi, V.; Gerona, S.; Arbiza, J. Transmission, diagnosis, and management of hepatitis E: An update. Hepatic Med. 2014, 6, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Cancela, F.; Cravino, A.; Icasuriaga, R.; González, P.; Bentancor, F.; Leizagoyen, C.; Echaides, C.; Ferreiro, I.; Cabrera, A.; Arbiza, J.; et al. Co-circulation of Hepatitis E Virus (HEV) Genotype 3 and Moose-HEV-Like Strains in Free-Ranging-Spotted Deer (Axis axis) in Uruguay. Food Environ. Virol. 2023, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Uruguay Wastewater Treatment. Available online: http://www.ose.com.uy/saneamiento/tratamientos (accessed on 18 June 2023).
- Mirazo, S.; Mir, D.; Bello, G.; Ramos, N.; Musto, H.; Arbiza, J. New insights into the hepatitis E virus genotype 3 phylodynamics and evolutionary history. Infect. Genet. Evol. 2016, 43, 267–273. [Google Scholar] [CrossRef]
- Dell’Amico, M.C.; Cavallo, A.; Gonzales, J.L.; Bonelli, S.I.; Valda, Y.; Pieri, A.; Segund, H.; Ibañez, R.; Mantella, A.; Bartalesi, F.; et al. Hepatitis E virus genotype 3 in humans and Swine, Bolivia. Emerg. Infect. Dis. 2011, 17, 1488–1490. [Google Scholar] [CrossRef]
- Pisano, M.B.; Culasso, A.C.A.; Altabert, N.; Wassaf, M.G.M.; Nates, S.V.; González, J.; Contigiani, M.S.; Campos, R.; Ré, V.E. Phylogeography and evolutionary history of hepatitis E virus genotype 3 in Argentina. Trans. R. Soc. Trop. Med. Hyg. 2022, 116, 34–42. [Google Scholar] [CrossRef]
- Lu, L.; Li, C.; Hagedorn, C.H. Phylogenetic analysis of global hepatitis E virus sequences: Genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 2006, 16, 5–36. [Google Scholar] [CrossRef]
- Wu, J.-C.; Chen, C.-M.; Chiang, T.-Y.; Tsai, W.-H.; Jeng, W.-J.; Sheen, I.-J.; Lin, C.-C.; Meng, X.-J. Spread of hepatitis E virus among different-aged pigs: Two-year survey in Taiwan. J. Med. Virol. 2002, 66, 488–492. [Google Scholar] [CrossRef]
- Mirazo, S.; Ramos, N.; Russi, J.C.; Gagliano, G.; Arbiza, J. Detection and molecular characterization of sporadic cases of acute human hepatitis E virus infection in Uruguay. Arch. Virol. 2011, 156, 1451–1454. [Google Scholar] [CrossRef]
- Salvador, D.; Neto, C.; Benoliel, M.J.; Caeiro, M.F. Assessment of the Presence of Hepatitis E virus in Surface Water and Drinking Water in Portugal. Microorganisms 2020, 8, 761. [Google Scholar] [CrossRef]

| Accession Number | Subtype | Complete Genome a | Partial ORF2 a |
|---|---|---|---|
| AF082843 | 3a | 0.147–0.150 | 0.131–0.141 |
| AP003430 | 3b | 0.144–0.145 | 0.133–0.146 |
| FJ705359 | 3c | 0.136–0.140 | 0.134–0.147 |
| AF296165-7 | 3d b | - | 0.132–0.149 |
| AB248521 | 3e | 0.171–0.177 | 0.137–0.163 |
| AB369687 | 3f | 0.174–0.176 | 0.143–0.158 |
| AF455784 | 3g | 0.169–0.174 | 0.160–0.170 |
| JQ013794 | 3h | 0.132–0.137 | 0.114–0.128 |
| FJ998008 | 3i | 0.132 | 0.120–0.136 |
| AY115488 | 3j | 0.151–0.155 | 0.133–0.143 |
| AB369689 | 3k | 0.146–0.148 | 0.130–0.136 |
| JQ953664 | 3l | 0.136–0.139 | 0.124–0.143 |
| KU513561 | 3m | 0.134–0.135 | 0.120–0.143 |
| MF959764 | 3n | 0.133–0.137 | 0.120–0.146 |
| PRsw1 OQ433914 * | Putative 3o | 0.118–0.119 | 0.110–0.126 |
| RJ-sw1 OQ433915 * | Putative 3o | 0.121–0.125 | 0.115–0.122 |
| KP294371 | Unassigned | 0.136 | 0.126–0.136 |
| LC260517 | Unassigned | 0.142–0.146 | 0.117–0.148 |
| MF959765 | Unassigned | 0.134–0.141 | 0.143–0.153 |
| MK390971 | Unassigned | 0.142–0.145 | 0.116–0.136 |
| AB290313 | Unassigned | 0.175–0.180 | 0.150–0.168 |
| Strains | Subtype | Complete Genome | Partial ORF2 * |
|---|---|---|---|
| Uruguayan isolates | Putative 3o | 0.075 | 0.000–0.096 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cancela, F.; Icasuriaga, R.; Cuevas, S.; Hergatacorzian, V.; Olivera, M.; Panzera, Y.; Pérez, R.; López, J.; Borzacconi, L.; González, E.; et al. Epidemiology Update of Hepatitis E Virus (HEV) in Uruguay: Subtyping, Environmental Surveillance and Zoonotic Transmission. Viruses 2023, 15, 2006. https://doi.org/10.3390/v15102006
Cancela F, Icasuriaga R, Cuevas S, Hergatacorzian V, Olivera M, Panzera Y, Pérez R, López J, Borzacconi L, González E, et al. Epidemiology Update of Hepatitis E Virus (HEV) in Uruguay: Subtyping, Environmental Surveillance and Zoonotic Transmission. Viruses. 2023; 15(10):2006. https://doi.org/10.3390/v15102006
Chicago/Turabian StyleCancela, Florencia, Romina Icasuriaga, Santiago Cuevas, Valentina Hergatacorzian, Mauricio Olivera, Yanina Panzera, Ruben Pérez, Julieta López, Liliana Borzacconi, Elizabeth González, and et al. 2023. "Epidemiology Update of Hepatitis E Virus (HEV) in Uruguay: Subtyping, Environmental Surveillance and Zoonotic Transmission" Viruses 15, no. 10: 2006. https://doi.org/10.3390/v15102006
APA StyleCancela, F., Icasuriaga, R., Cuevas, S., Hergatacorzian, V., Olivera, M., Panzera, Y., Pérez, R., López, J., Borzacconi, L., González, E., Montaldo, N., Gaitán, M., López-Verges, S., Bortagaray, V., Victoria, M., Colina, R., Arbiza, J., Berois, M., & Mirazo, S. (2023). Epidemiology Update of Hepatitis E Virus (HEV) in Uruguay: Subtyping, Environmental Surveillance and Zoonotic Transmission. Viruses, 15(10), 2006. https://doi.org/10.3390/v15102006










