Interaction between hTIM-1 and Envelope Protein Is Important for JEV Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Plasmids and Antibodies
2.3. Western Blot
2.4. Co-Immunoprecipitation (Co-IP) Assays
2.5. GST Pull-Down Assays
2.6. Plaque Assay
2.7. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.8. Immunofluorescence Assays (IFA) or Confocal Fluorescence Microscopy
2.9. Protein Neutralization Infection or Entry Assays
2.10. Antibodies Neutralization Assays
2.11. Statistical Analysis
3. Results
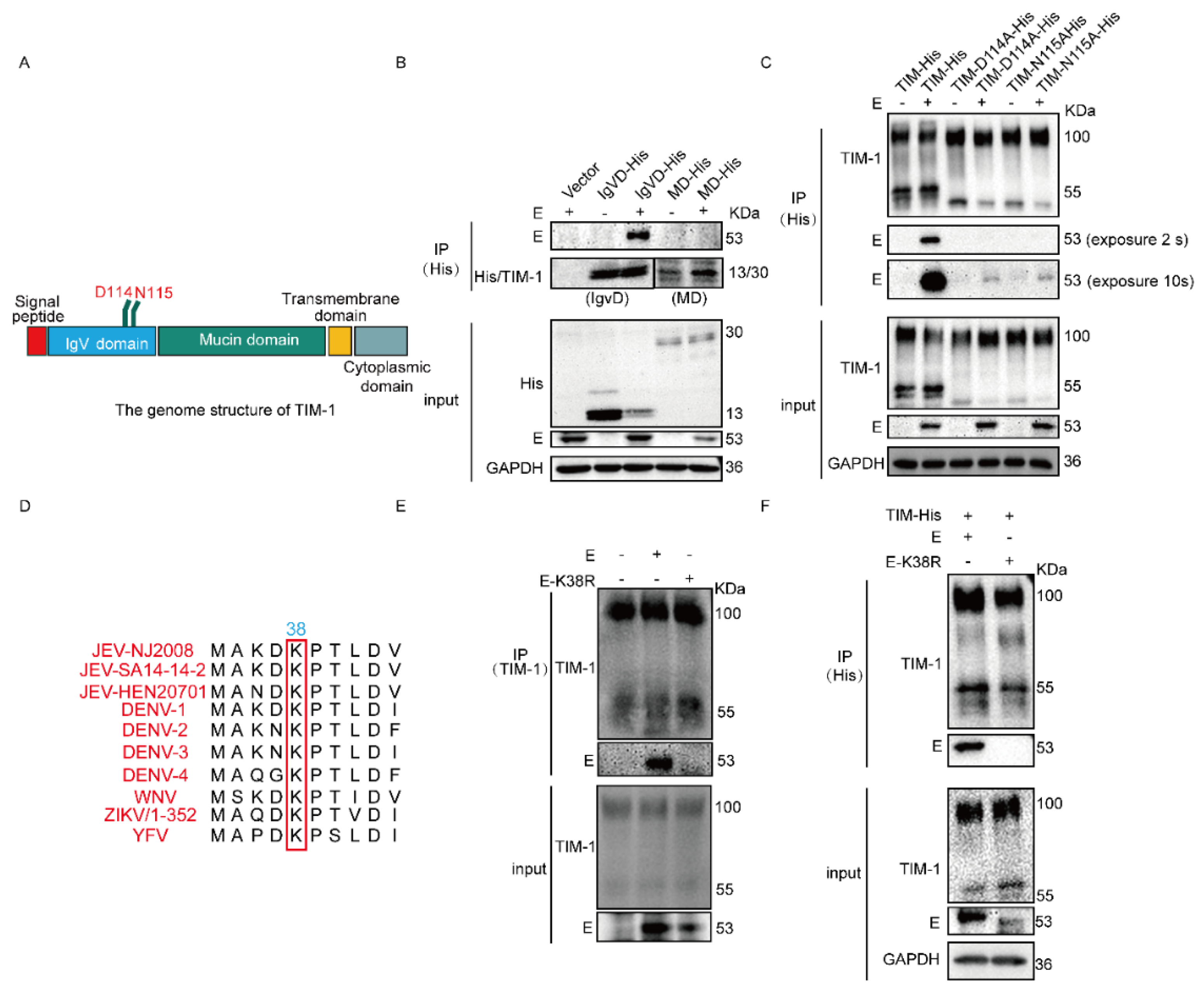
3.1. hTIM-1 Directly Interacts with JEV E Protein
3.2. N114 and D115 of hTIM-1 and K38 of JEV E Are Crucial for Their Interaction
3.3. JEV and hTIM-1 Are Internalized into Cells and Transported Together in Early and Late Endosomes
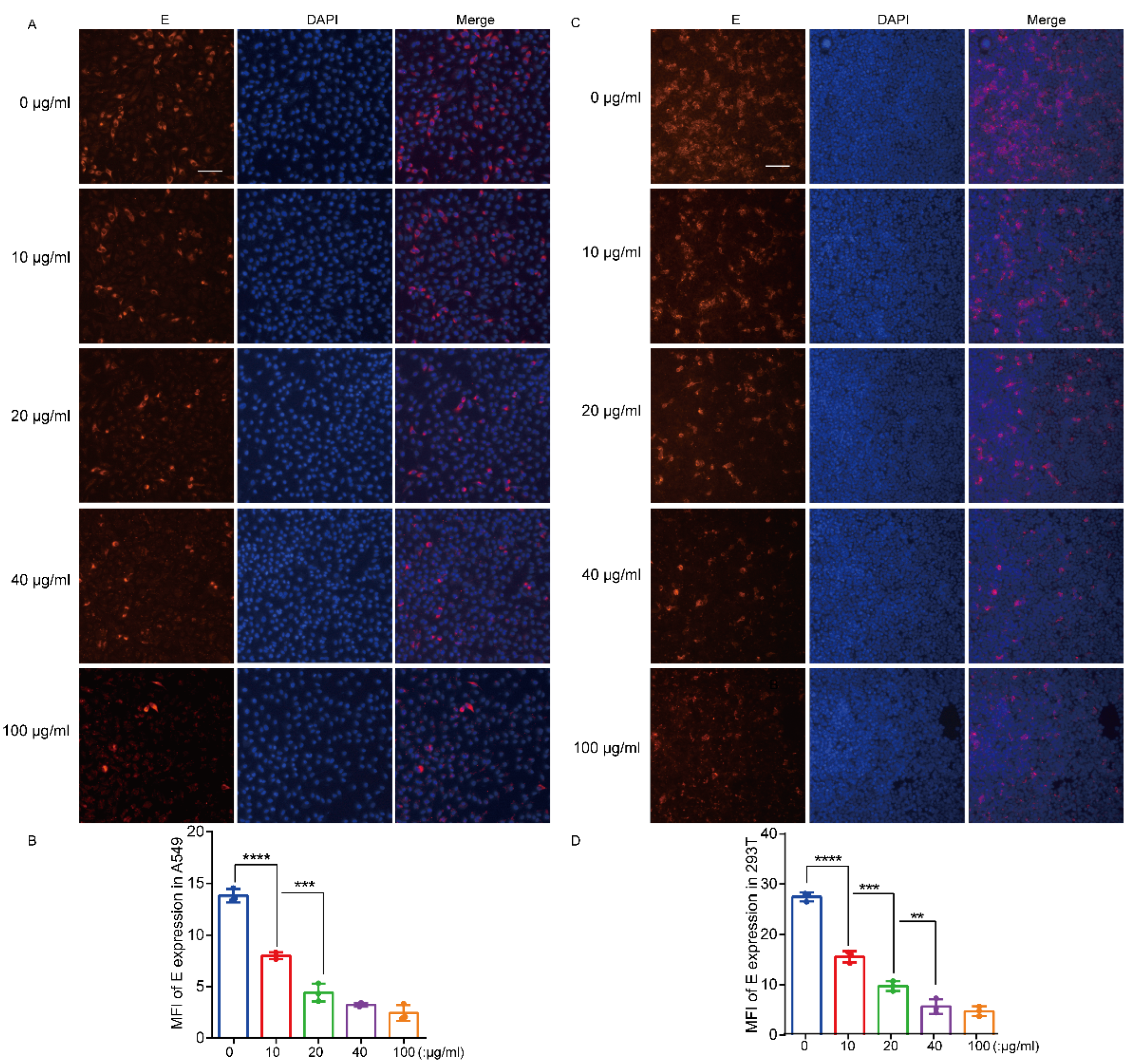
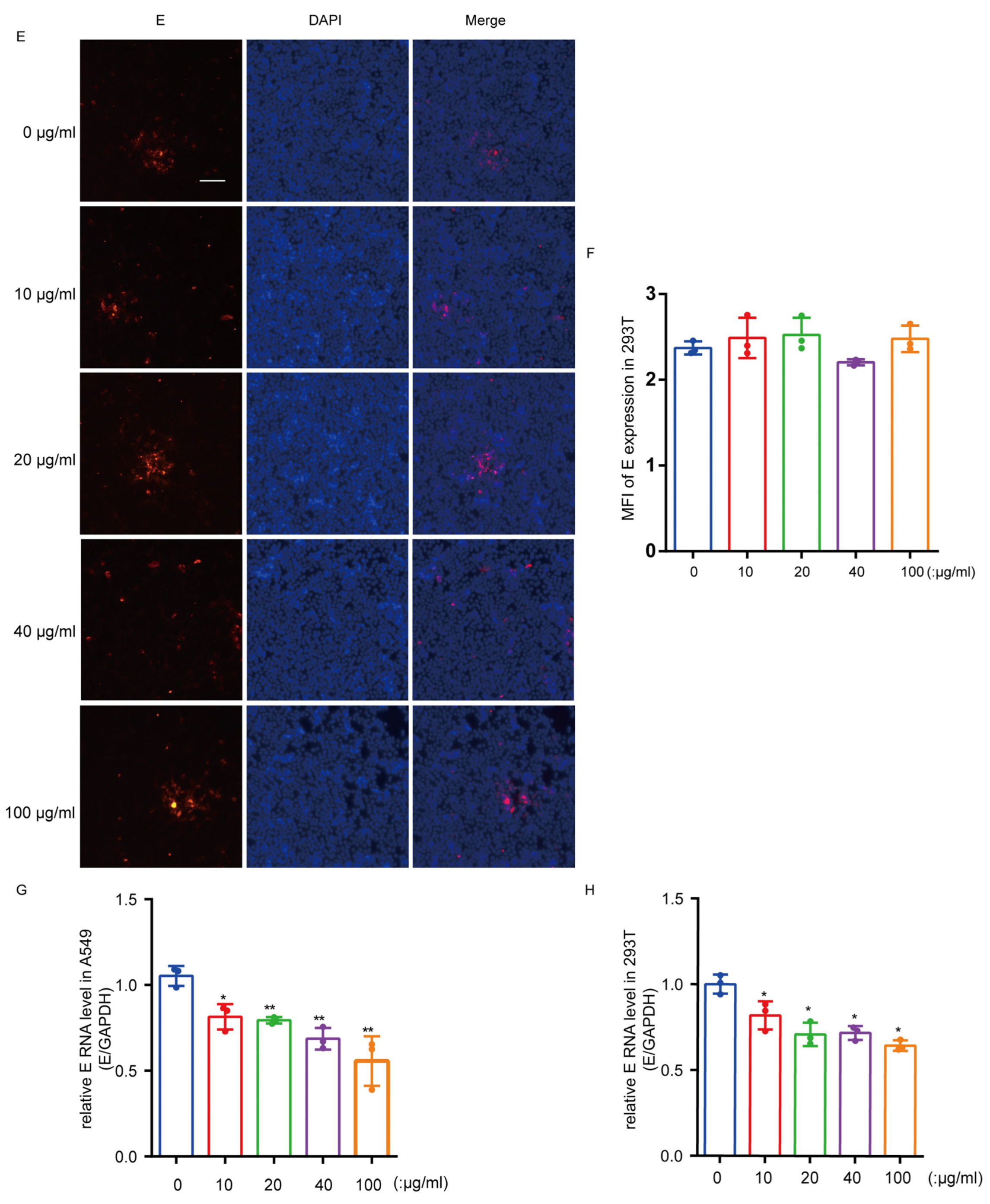
3.4. Recombinant hTIM-1 Ectodomain Protein Can Inhibit JEV Infection in Cells
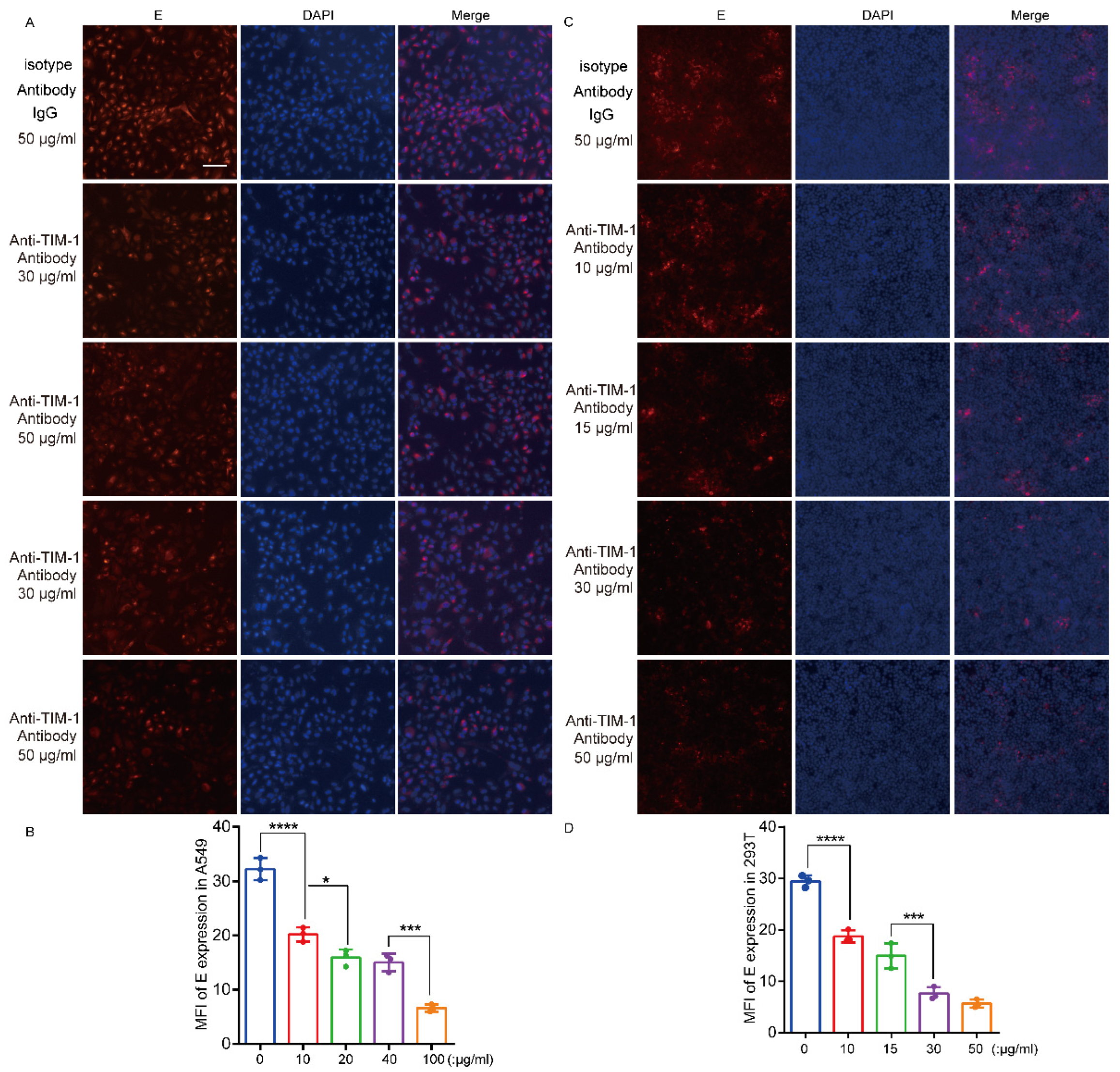
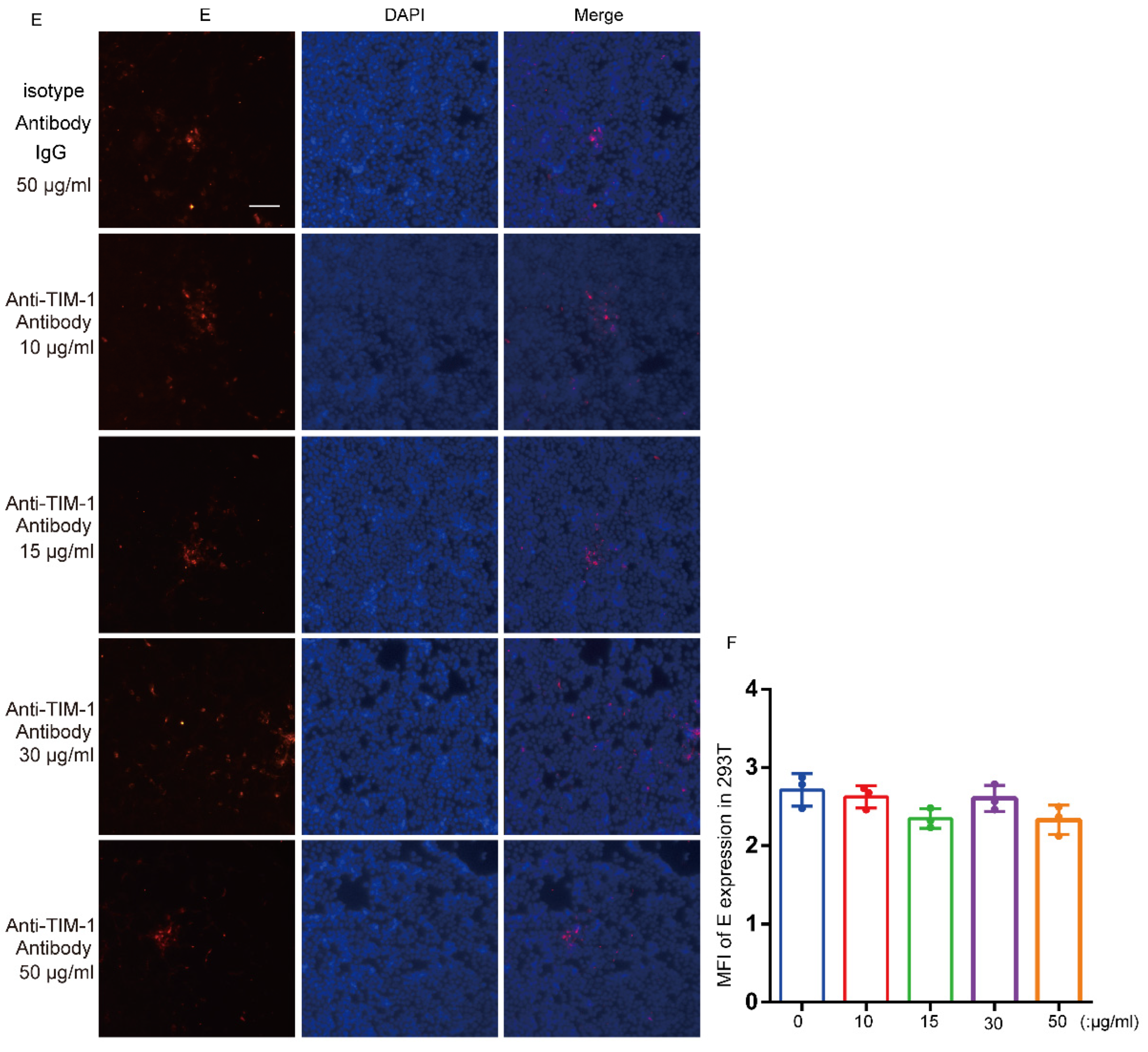
3.5. hTIM-1 Antibodies Block JEV Infection in Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campbell, G.L.; Hills, S.L.; Fischer, M.; Jacobson, J.A.; Hoke, C.H.; Hombach, J.M.; Marfin, A.A.; Solomon, T.; Tsai, T.F.; Tsu, V.D.; et al. Estimated global incidence of Japanese encephalitis: A systematic review. Bull. World Health Organ. 2011, 89, 766–774. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Japanese Encephalitis Vaccines: WHO position paper, February 2015–Recommendations. Vaccine 2016, 34, 302–303. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T. Control of Japanese encephalitis–Within our grasp? N. Engl. J. Med. 2006, 355, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Gould, E.A.; Solomon, T. Pathogenic flaviviruses. Lancet 2008, 371, 500–509. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Liao, K.C.; Prasanth, K.R.; Soto-Acosta, R.; Yeh, S.C.; Schott-Lerner, G.; Pompon, J.; Sessions, O.M.; Bradrick, S.S.; et al. Biochemistry and Molecular Biology of Flaviviruses. Chem. Rev. 2018, 118, 4448–4482. [Google Scholar] [CrossRef]
- Pearce, J.C.; Learoyd, T.P.; Langendorf, B.J.; Logan, J.G. Japanese encephalitis: The vectors, ecology and potential for expansion. J. Travel Med. 2018, 25, S16–S26. [Google Scholar] [CrossRef]
- Ricklin, M.E.; García-Nicolás, O.; Brechbühl, D.; Python, S.; Zumkehr, B.; Nougairede, A.; Charrel, R.N.; Posthaus, H.; Oevermann, A.; Summerfield, A. Vector-free transmission and persistence of Japanese encephalitis virus in pigs. Nat. Commun. 2016, 7, 10832. [Google Scholar] [CrossRef]
- Kim, J.K.; Kim, J.M.; Song, B.H.; Yun, S.I.; Yun, G.N.; Byun, S.J.; Lee, Y.M. Profiling of viral proteins expressed from the genomic RNA of Japanese encephalitis virus using a panel of 15 region-specific polyclonal rabbit antisera: Implications for viral gene expression. PLoS ONE 2015, 10, e0124318. [Google Scholar] [CrossRef]
- Srivastava, K.S.; Jeswani, V.; Pal, N.; Bohra, B.; Vishwakarma, V.; Bapat, A.A.; Patnaik, Y.P.; Khanna, N.; Shukla, R. Japanese Encephalitis Virus: An Update on the Potential Antivirals and Vaccines. Vaccines 2023, 11, 742. [Google Scholar] [CrossRef]
- Stocks, C.E.; Lobigs, M. Signal peptidase cleavage at the flavivirus C-prM junction: Dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J. Virol. 1998, 72, 2141–2149. [Google Scholar] [CrossRef]
- Zhu, Y.; He, Z.; Qi, Z. Virus-host Interactions in Early Japanese Encephalitis Virus Infection. Virus Res. 2023, 331, 199120. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Laxminarayana, S.V.; Chandra, N.; Ravi, V.; Desai, A. Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology 2009, 385, 47–57. [Google Scholar] [CrossRef]
- Zhu, Y.Z.; Cao, M.M.; Wang, W.B.; Wang, W.; Ren, H.; Zhao, P.; Qi, Z.T. Association of heat-shock protein 70 with lipid rafts is required for Japanese encephalitis virus infection in Huh7 cells. J. Gen. Virol. 2012, 93, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Nain, M.; Mukherjee, S.; Karmakar, S.P.; Paton, A.W.; Paton, J.C.; Abdin, M.Z.; Basu, A.; Kalia, M.; Vrati, S. GRP78 Is an Important Host Factor for Japanese Encephalitis Virus Entry and Replication in Mammalian Cells. J. Virol. 2017, 91, e02274-16. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ravi, V.; Desai, A. Japanese encephalitis virus interacts with vimentin to facilitate its entry into porcine kidney cell line. Virus Res. 2011, 160, 404–408. [Google Scholar] [CrossRef]
- Wang, P.; Hu, K.; Luo, S.; Zhang, M.; Deng, X.; Li, C.; Jin, W.; Hu, B.; He, S.; Li, M.; et al. DC-SIGN as an attachment factor mediates Japanese encephalitis virus infection of human dendritic cells via interaction with a single high-mannose residue of viral E glycoprotein. Virology 2016, 488, 108–119. [Google Scholar] [CrossRef]
- Fan, W.; Qian, P.; Wang, D.; Zhi, X.; Wei, Y.; Chen, H.; Li, X. Integrin αvβ3 promotes infection by Japanese encephalitis virus. Res. Vet. Sci. 2017, 111, 67–74. [Google Scholar] [CrossRef]
- Kuchroo, V.K.; Dardalhon, V.; Xiao, S.; Anderson, A.C. New roles for TIM family members in immune regulation. Nat. Rev. Immunol. 2008, 8, 577–580. [Google Scholar] [CrossRef]
- Kondratowicz, A.S.; Lennemann, N.J.; Sinn, P.L.; Davey, R.A.; Hunt, C.L.; Moller-Tank, S.; Meyerholz, D.K.; Rennert, P.; Mullins, R.F.; Brindley, M.; et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA 2011, 108, 8426–8431. [Google Scholar] [CrossRef]
- Santiago, C.; Ballesteros, A.; Martínez-Muñoz, L.; Mellado, M.; Kaplan, G.G.; Freeman, G.J.; Casasnovas, J.M. Structures of T cell immunoglobulin mucin protein 4 show a metal-Ion-dependent ligand binding site where phosphatidylserine binds. Immunity 2007, 27, 941–951. [Google Scholar] [CrossRef]
- Santiago, C.; Ballesteros, A.; Tami, C.; Martínez-Muñoz, L.; Kaplan, G.G.; Casasnovas, J.M. Structures of T Cell immunoglobulin mucin receptors 1 and 2 reveal mechanisms for regulation of immune responses by the TIM receptor family. Immunity 2007, 26, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Moller-Tank, S.; Maury, W. Phosphatidylserine receptors: Enhancers of enveloped virus entry and infection. Virology 2014, 468, 565–580. [Google Scholar] [CrossRef]
- Moller-Tank, S.; Albritton, L.M.; Rennert, P.D.; Maury, W. Characterizing functional domains for TIM-mediated enveloped virus entry. J. Virol. 2014, 88, 6702–6713. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, G.; Totsuka, A.; Thompson, P.; Akatsuka, T.; Moritsugu, Y.; Feinstone, S.M. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. Embo. J. 1996, 15, 4282–4296. [Google Scholar] [CrossRef]
- Feigelstock, D.; Thompson, P.; Mattoo, P.; Zhang, Y.; Kaplan, G.G. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J. Virol. 1998, 72, 6621–6628. [Google Scholar] [CrossRef] [PubMed]
- Meertens, L.; Carnec, X.; Lecoin, M.P.; Ramdasi, R.; Guivel-Benhassine, F.; Lew, E.; Lemke, G.; Schwartz, O.; Amara, A. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 2012, 12, 544–557. [Google Scholar] [CrossRef]
- Jemielity, S.; Wang, J.J.; Chan, Y.K.; Ahmed, A.A.; Li, W.; Monahan, S.; Bu, X.; Farzan, M.; Freeman, G.J.; Umetsu, D.T.; et al. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 2013, 9, e1003232. [Google Scholar] [CrossRef]
- Moller-Tank, S.; Kondratowicz, A.S.; Davey, R.A.; Rennert, P.D.; Maury, W. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J. Virol. 2013, 87, 8327–8341. [Google Scholar] [CrossRef]
- Morizono, K.; Chen, I.S. Role of phosphatidylserine receptors in enveloped virus infection. J. Virol. 2014, 88, 4275–4290. [Google Scholar] [CrossRef]
- Yuan, S.; Cao, L.; Ling, H.; Dang, M.; Sun, Y.; Zhang, X.; Chen, Y.; Zhang, L.; Su, D.; Wang, X.; et al. TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope. Protein Cell 2015, 6, 814–824. [Google Scholar] [CrossRef]
- Dejarnac, O.; Hafirassou, M.L.; Chazal, M.; Versapuech, M.; Gaillard, J.; Perera-Lecoin, M.; Umana-Diaz, C.; Bonnet-Madin, L.; Carnec, X.; Tinevez, J.Y.; et al. TIM-1 Ubiquitination Mediates Dengue Virus Entry. Cell Rep. 2018, 23, 1779–1793. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.I.; Xia, H.; Aguilera-Aguirre, L.; Hage, A.; van Tol, S.; Shan, C.; Xie, X.; Sturdevant, G.L.; Robertson, S.J.; McNally, K.L.; et al. Envelope protein ubiquitination drives entry and pathogenesis of Zika virus. Nature 2020, 585, 414–419. [Google Scholar] [CrossRef]
- Wang, X.; Li, S.H.; Zhu, L.; Nian, Q.G.; Yuan, S.; Gao, Q.; Hu, Z.; Ye, Q.; Li, X.F.; Xie, D.Y.; et al. Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability. Nat. Commun. 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jia, R.; Shen, H.; Wang, M.; Yin, Z.; Cheng, A. Structures and Functions of the Envelope Glycoprotein in Flavivirus Infections. Viruses 2017, 9, 338. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Lei, Y.; Yang, P.; Gao, Q.; Wang, N.; Cao, L.; Yuan, S.; Huang, X.; Deng, Y.; Ma, W.; et al. Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies. Nat. Microbiol. 2018, 3, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Arocho, A.; Chen, B.; Ladanyi, M.; Pan, Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. Am. J. Surg. Pathol. Part B 2006, 15, 56–61. [Google Scholar] [CrossRef]
- Zhang, X.; Liang, C.; Wang, H.; Guo, Z.; Rong, H.; Pan, J.; Li, W.; Pei, R.; Chen, X.; Zhang, Z.; et al. T-Cell Immunoglobulin and Mucin Domain 1 (TIM-1) Is a Functional Entry Factor for Tick-Borne Encephalitis Virus. mBio 2022, 13, e0286021. [Google Scholar] [CrossRef] [PubMed]
- Kirui, J.; Abidine, Y.; Lenman, A.; Islam, K.; Gwon, Y.D.; Lasswitz, L.; Evander, M.; Bally, M.; Gerold, G. The Phosphatidylserine Receptor TIM-1 Enhances Authentic Chikungunya Virus Cell Entry. Cells 2021, 10, 1828. [Google Scholar] [CrossRef]
- Evans, J.P.; Liu, S.L. Multifaceted Roles of TIM-Family Proteins in Virus-Host Interactions. Trends Microbiol. 2020, 28, 224–235. [Google Scholar] [CrossRef]
- Bohan, D.; Van Ert, H.; Ruggio, N.; Rogers, K.J.; Badreddine, M.; Aguilar Briseño, J.A.; Elliff, J.M.; Rojas Chavez, R.A.; Gao, B.; Stokowy, T.; et al. Phosphatidylserine receptors enhance SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009743. [Google Scholar] [CrossRef]
- Niu, J.; Jiang, Y.; Xu, H.; Zhao, C.; Zhou, G.; Chen, P.; Cao, R. TIM-1 Promotes Japanese Encephalitis Virus Entry and Infection. Viruses 2018, 10, 630. [Google Scholar] [CrossRef] [PubMed]
- Brunton, B.; Rogers, K.; Phillips, E.K.; Brouillette, R.B.; Bouls, R.; Butler, N.S.; Maury, W. TIM-1 serves as a receptor for Ebola virus in vivo, enhancing viremia and pathogenesis. PLoS Negl. Trop. Dis. 2019, 13, e0006983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, X.; Hu, L.; Zhang, Y.; Zheng, H.; Wu, H.; Wang, J.; Luo, L.; Xiao, H.; Qiao, C.; et al. TIM-1 Augments Cellular Entry of Ebola Virus Species and Mutants, Which Is Blocked by Recombinant TIM-1 Protein. Microbiol. Spectr. 2022, 10, e0221221. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Primer Sequence |
|---|---|
| hTIM-1-F | 5′-AACTGTCTCTACCTTTGTTCCTCC-3′ |
| hTIM-1-R | 5′-GTTCTCTCCTTATTGCTCCCTG-3′ |
| JEV(NJ2008/SA14-14-2)-E-F | 5′-GGCAAACGACAAACCAACATT-3′ |
| JEV(NJ2008/SA14-14-2)-E-R | 5′-ATCAGCTCGCTTCTCGTTGTG-3′ |
| JEV(HEN0701)-E-F | 5′-AGAGATTCCTACTCTCGACATGC-3′ |
| JEV(HEN0701)-E-R | 5′-GCCCACAGTATCCTGCAACC-3′ |
| GAPDH-F | 5′-GAGTCAACGGATTTGGTCGT-3′ |
| GAPDH-R | 5′-GACAAGCTTCCCGTTCTCAG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Z.; Pan, J.; Xie, S.; Yang, X.; Cao, R. Interaction between hTIM-1 and Envelope Protein Is Important for JEV Infection. Viruses 2023, 15, 1589. https://doi.org/10.3390/v15071589
Liang Z, Pan J, Xie S, Yang X, Cao R. Interaction between hTIM-1 and Envelope Protein Is Important for JEV Infection. Viruses. 2023; 15(7):1589. https://doi.org/10.3390/v15071589
Chicago/Turabian StyleLiang, Zhenjie, Junhui Pan, Shengda Xie, Xingmiao Yang, and Ruibing Cao. 2023. "Interaction between hTIM-1 and Envelope Protein Is Important for JEV Infection" Viruses 15, no. 7: 1589. https://doi.org/10.3390/v15071589
APA StyleLiang, Z., Pan, J., Xie, S., Yang, X., & Cao, R. (2023). Interaction between hTIM-1 and Envelope Protein Is Important for JEV Infection. Viruses, 15(7), 1589. https://doi.org/10.3390/v15071589





