Rhinovirus Genotypes Circulating in Bulgaria, 2018–2021
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population and Specimen Collection
2.2. Clinical Data and Statistics
2.3. Extraction of Nucleic Acids and Detection of RV
2.4. RT-PCR and Sanger Sequencing
2.5. Genotyping and Sequence Analysis
3. Results
3.1. RV Detections and Clinical Characteristics
3.1.1. RV Detections
3.1.2. Clinical Data
- ARI (n = 5);
- Acute respiratory distress (ARD) (n = 1);
- ARI/URTI/Laryngitis (n = 1);
- ARI/LRTI/Bronchiolitis (n = 13);
- ARI/LRTI/Croup (n = 2);
- ILI (n = 4);
- ILI/ARI-possible COVID-19 (n = 18);
- Other (n = 3).
3.1.3. Diagnosis and Virus Type
3.1.4. Virus Types before and after the COVID-19 Pandemic
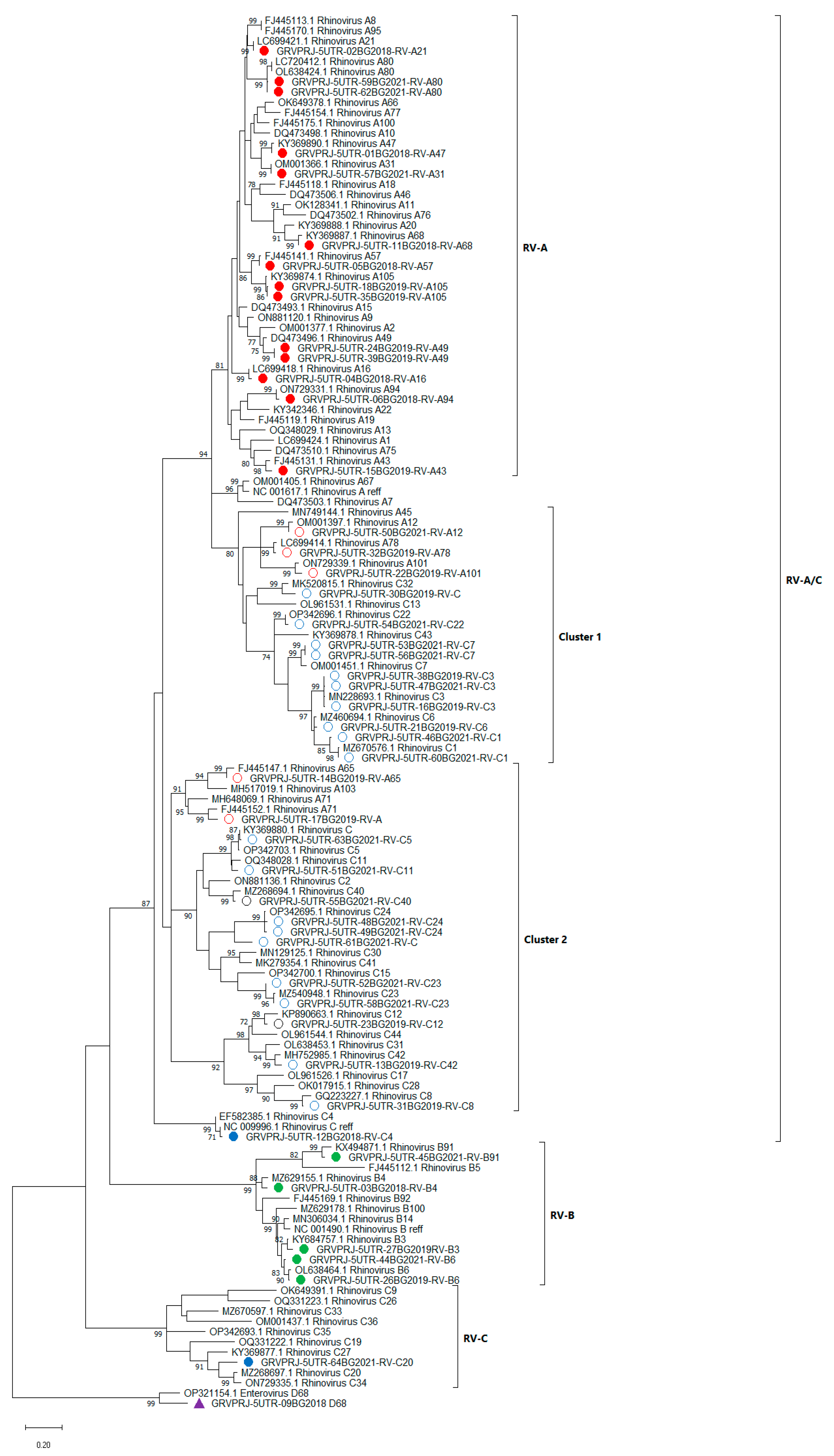
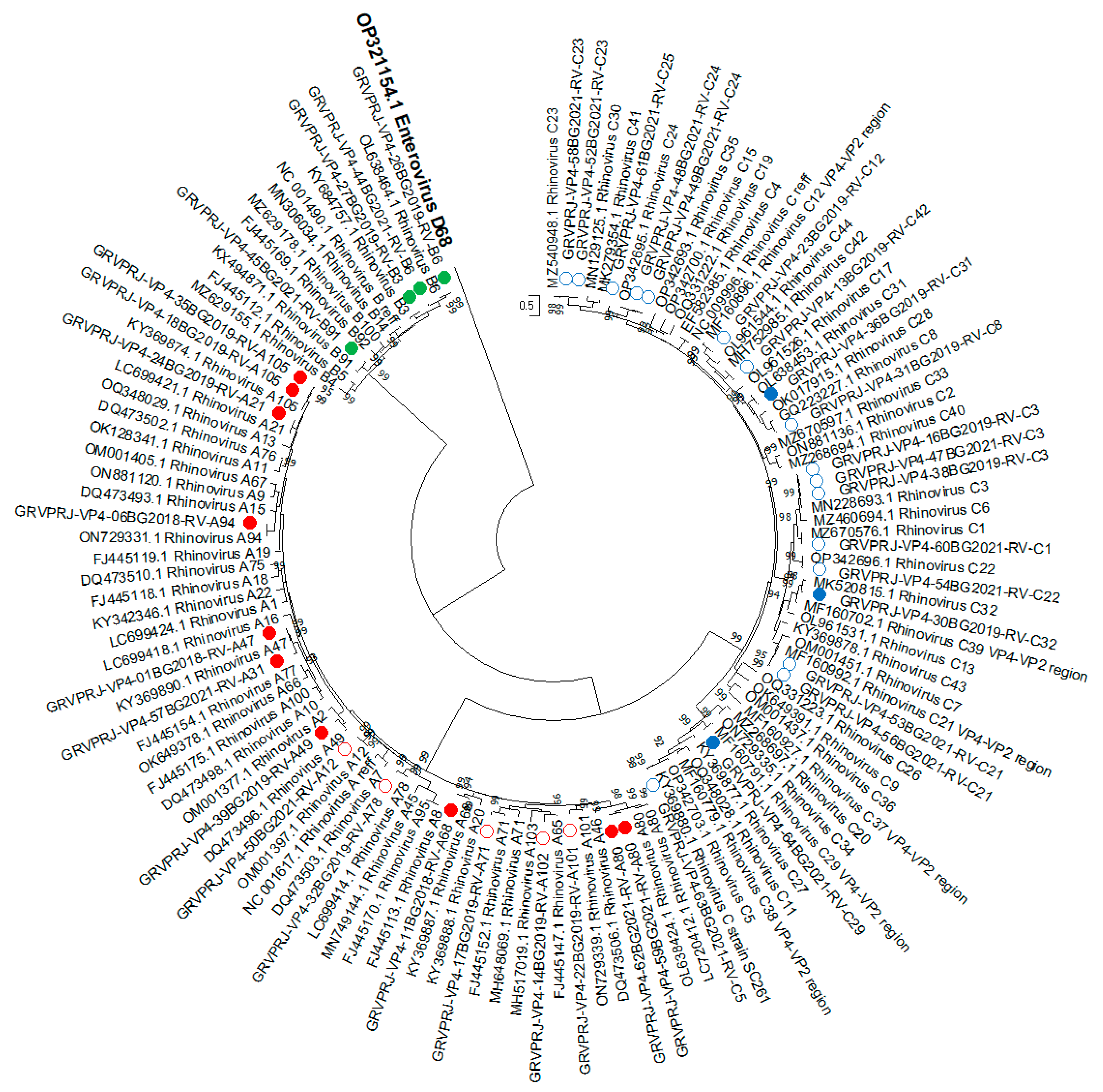
3.1.5. Phylogenetic Analysis and Genotype Assignments of RV Strains
3.1.6. Recombination Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Makela, M.J.; Puhakka, T.; Ruuskanen, O.; Leinonen, M.; Saikku, P.; Kimpimaki, M.; Blomqvist, S.; Hyypia, T.; Arstila, P. Viruses and Bacteria in the Etiology of the Common Cold. J. Clin. Microbiol. 1998, 36, 539–542. [Google Scholar] [CrossRef]
- Fendrick, A.M.; Monto, A.S.; Nightengale, B.; Sarnes, M. The Economic Burden of Non–Influenza-Related Viral Respiratory Tract Infection in the United States. Arch. Intern. Med. 2003, 163, 487–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayden, F.G. Rhinovirus and the Lower Respiratory Tract. Rev. Med. Virol. 2004, 14, 17–31. [Google Scholar] [CrossRef]
- McIntyre, C.L.; Knowles, N.J.; Simmonds, P. Proposals for the Classification of Human Rhinovirus Species A, B and C into Genotypically Assigned Types. J. Gen. Virol. 2013, 94, 1791–1806. [Google Scholar] [CrossRef]
- Simmonds, P.; McIntyre, C.; Savolainen-Kopra, C.; Tapparel, C.; Mackay, I.M.; Hovi, T. Proposals for the Classification of Human Rhinovirus Species C into Genotypically Assigned Types. J. Gen. Virol. 2010, 91, 2409–2419. [Google Scholar] [CrossRef]
- Kiang, D.; Kalra, I.; Yagi, S.; Louie, J.K.; Boushey, H.; Boothby, J.; Schnurr, D.P. Assay for 5′ Noncoding Region Analysis of All Human Rhinovirus Prototype Strains. J. Clin. Microbiol. 2008, 46, 3736–3745. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Holloway, B.; Dare, R.K.; Kuypers, J.; Yagi, S.; Williams, J.V.; Hall, C.B.; Erdman, D.D. Real-Time Reverse Transcription-PCR Assay for Comprehensive Detection of Human Rhinoviruses. J. Clin. Microbiol. 2008, 46, 533–539. [Google Scholar] [CrossRef] [Green Version]
- Bochkov, Y.A.; Grindle, K.; Vang, F.; Evans, M.D.; Gern, J.E. Improved Molecular Typing Assay for Rhinovirus Species A, B, and C. J. Clin. Microbiol. 2014, 52, 2461–2471. [Google Scholar] [CrossRef] [Green Version]
- Wisdom, A.; Leitch, E.C.M.; Gaunt, E.; Harvala, H.; Simmonds, P. Screening Respiratory Samples for Detection of Human Rhinoviruses (HRVs) and Enteroviruses: Comprehensive VP4-VP2 Typing Reveals High Incidence and Genetic Diversity of HRV Species C. J. Clin. Microbiol. 2009, 47, 3958–3967. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.-M.; Kiesner, C.; Pappas, T.; Lee, I.; Grindle, K.; Jartti, T.; Jakiela, B.; Lemanske, R.F.; Shult, P.A.; Gern, J.E. A Diverse Group of Previously Unrecognized Human Rhinoviruses Are Common Causes of Respiratory Illnesses in Infants. PLoS ONE 2007, 2, e966. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Kroneman, A.; Vennema, H.; Deforche, K.; Avoort, H.v.d.; Peñaranda, S.; Oberste, M.S.; Vinjé, J.; Koopmans, M. An Automated Genotyping Tool for Enteroviruses and Noroviruses. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2011, 51, 121–125. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evol. Int. J. Org. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: London, UK, 2000; ISBN 978-0-19-513585-5. [Google Scholar]
- Miller, E.K.; Khuri-Bulos, N.; Williams, J.V.; Shehabi, A.A.; Faouri, S.; Al Jundi, I.; Chen, Q.; Heil, L.; Mohamed, Y.; Morin, L.-L.; et al. Human Rhinovirus C Associated with Wheezing in Hospitalised Children in the Middle East. J. Clin. Virol. 2009, 46, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Bizzintino, J.; Lee, W.-M.; Laing, I.A.; Vang, F.; Pappas, T.; Zhang, G.; Martin, A.C.; Khoo, S.-K.; Cox, D.W.; Geelhoed, G.C.; et al. Association between Human Rhinovirus C and Severity of Acute Asthma in Children. Eur. Respir. J. 2011, 37, 1037–1042. [Google Scholar] [CrossRef]
- Lamson, D.; Renwick, N.; Kapoor, V.; Liu, Z.; Palacios, G.; Ju, J.; Dean, A.; St George, K.; Briese, T.; Lipkin, W.I. MassTag Polymerase-Chain-Reaction Detection of Respiratory Pathogens, including a New Rhinovirus Genotype, That Caused Influenza-like Illness in New York State during 2004–2005. J. Infect. Dis. 2006, 194, 1398–1402. [Google Scholar] [CrossRef] [Green Version]
- Linder, J.E.; Kraft, D.C.; Mohamed, Y.; Lu, Z.; Heil, L.; Tollefson, S.; Saville, B.R.; Wright, P.F.; Williams, J.V.; Miller, E.K. Human Rhinovirus C: Age, Season, and Lower Respiratory Illness over the Past 3 Decades. J. Allergy Clin. Immunol. 2013, 131, e1–e6. [Google Scholar] [CrossRef]
- Ratnamohan, V.M.; Zeng, F.; Donovan, L.; MacIntyre, C.R.; Kok, J.; Dwyer, D.E. Phylogenetic Analysis of Human Rhinoviruses Collected over Four Successive Years in Sydney, Australia. Influenza Other Respir. Viruses 2016, 10, 493–503. [Google Scholar] [CrossRef] [Green Version]
- McIntyre, C.L.; McWilliam Leitch, E.C.; Savolainen-Kopra, C.; Hovi, T.; Simmonds, P. Analysis of Genetic Diversity and Sites of Recombination in Human Rhinovirus Species C. J. Virol. 2010, 84, 10297–10310. [Google Scholar] [CrossRef] [Green Version]
- Harvala, H.; Broberg, E.; Benschop, K.; Berginc, N.; Ladhani, S.; Susi, P.; Christiansen, C.; McKenna, J.; Allen, D.; Makiello, P.; et al. Recommendations for Enterovirus Diagnostics and Characterisation within and beyond Europe. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2018, 101, 11–17. [Google Scholar] [CrossRef]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Bamford, D.H.; Breitbart, M.; Davison, A.J.; Ghabrial, S.A.; Gorbalenya, A.E.; Knowles, N.J.; Krell, P.; et al. Ratification Vote on Taxonomic Proposals to the International Committee on Taxonomy of Viruses (2015). Arch. Virol. 2015, 160, 1837–1850. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; He, J.; Liu, Y.; Xu, L.; Guan, W.; Hu, Y. Molecular genotyping of human rhinovirus by using PCR and Sanger sequencing. Methods Mol. Biol. 2015, 1221, 39–47. [Google Scholar] [CrossRef]
- Palmenberg, A.C.; Spiro, D.; Kuzmickas, R.; Wang, S.; Djikeng, A.; Rathe, J.A.; Fraser-Liggett, C.M.; Liggett, S.B. Sequencing and Analyses of All Known Human Rhinovirus Genomes Reveal Structure and Evolution. Science 2009, 324, 55–59. [Google Scholar] [CrossRef] [Green Version]

| Diagnosis | RV-A n = 19 | p 1 | RV-B n = 5 | p 1 | RV-C n = 24 | p 1 |
|---|---|---|---|---|---|---|
| ARI | 2 (10.5%) | n.s. | 1 (20%) | n.s. | 2 (8.3%) | n.s. |
| ARD | 0 | - | 1 (20%) | n.s. | 0 | |
| ARI/URI/Laryngitis | 0 | - | 1 (20%) | n.s. | 0 | |
| ARI/LRTI/Bronchiolitis * | 6 (31.6%) | n.s. | 0 | - | 6 (25%) | n.s. |
| ARI/LRTI/Croup | 0 | - | 0 | - | 2 (8.3%) | n.s. |
| ILI | 4 (21.1%) | 0.0199 | 0 | - | 0 | - |
| ILI/ARI-possible COVID-19 | 4 (21.1%) | 0.0349 | 2 (40%) | n.s. | 14 (58.3%) | 0.0392 |
| Other | 3 (15.8%) | n.s. | 0 | - | 0 | - |
| Detection Period | RV-A n = 19 | RV-B n = 5 | RV-C n = 24 |
|---|---|---|---|
| Before COVID-19 | 15 | 3 | 9 |
| After COVID-19 | 4 | 2 | 15 |
| Fisher’s exact test value | p = 0.0168 | n.s. | p = 0.0189 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgieva, I.; Stoyanova, A.; Angelova, S.; Korsun, N.; Stoitsova, S.; Nikolaeva-Glomb, L. Rhinovirus Genotypes Circulating in Bulgaria, 2018–2021. Viruses 2023, 15, 1608. https://doi.org/10.3390/v15071608
Georgieva I, Stoyanova A, Angelova S, Korsun N, Stoitsova S, Nikolaeva-Glomb L. Rhinovirus Genotypes Circulating in Bulgaria, 2018–2021. Viruses. 2023; 15(7):1608. https://doi.org/10.3390/v15071608
Chicago/Turabian StyleGeorgieva, Irina, Asya Stoyanova, Svetla Angelova, Neli Korsun, Savina Stoitsova, and Lubomira Nikolaeva-Glomb. 2023. "Rhinovirus Genotypes Circulating in Bulgaria, 2018–2021" Viruses 15, no. 7: 1608. https://doi.org/10.3390/v15071608
APA StyleGeorgieva, I., Stoyanova, A., Angelova, S., Korsun, N., Stoitsova, S., & Nikolaeva-Glomb, L. (2023). Rhinovirus Genotypes Circulating in Bulgaria, 2018–2021. Viruses, 15(7), 1608. https://doi.org/10.3390/v15071608







