Clinical Validity of a Machine Learning Decision Support System for Early Detection of Hepatitis B Virus: A Binational External Validation Study
Abstract
:1. Introduction
2. Methods
2.1. Cohort Selection/External Validation Dataset
2.2. Outcome Definition and Assessment
2.3. HepB LiveTest Model
2.4. Statistics and Case-Mix Effect
3. Results
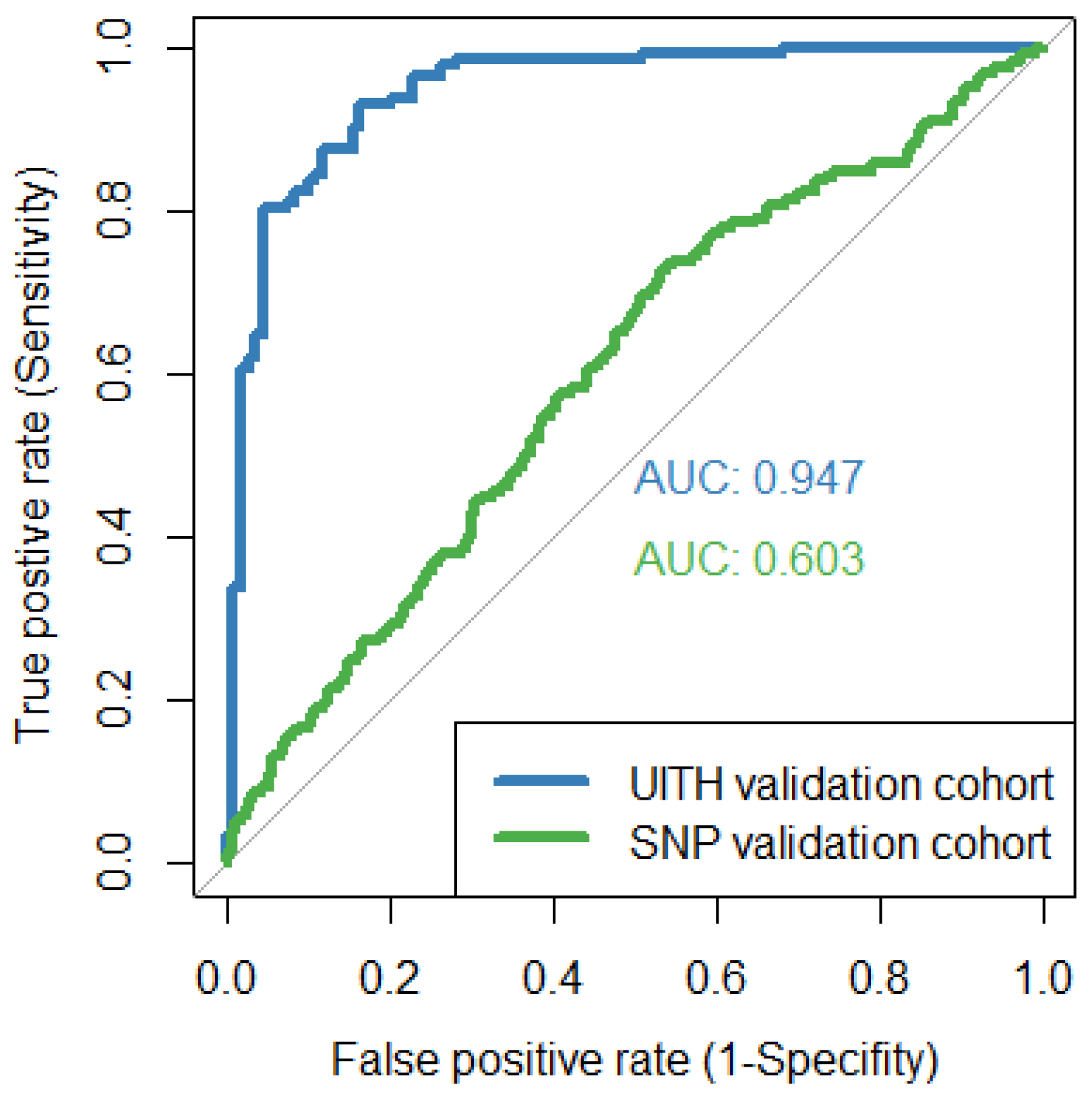
3.1. Performance of HepB LiveTest on External Patient Cohorts
3.2. Inspection of Dataset Shift on Case-Mix Effect
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Hepatitis B Key Facts. 2022. Available online: https://www.who.int/newsroom/factsheets/detail/hepatitis-b (accessed on 29 September 2022).
- Spearman, C.W.; Afihene, M.; Ally, R.; Apica, B.; Awuku, Y.; Cunha, L.; Dusheiko, G.; Gogela, N.; Kassianides, C.; Kew, M.; et al. Hepatitis B in sub-Saharan Africa: Strategies to achieve the 2030 elimination targets. Lancet Gastroenterol. Hepatol. 2017, 2, 900–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ajuwon, B.I.; Yujuico, I.; Roper, K.; Richardson, A.; Sheel, M.; Lidbury, B.A. Hepatitis B virus infection in Nigeria: A systematic review and meta-analysis of data published between 2010 and 2019. BMC Infect. Dis. 2021, 21, 1120. [Google Scholar] [CrossRef] [PubMed]
- Allard, N.L.; MacLachlan, J.H.; Tran, L.; Yussf, N.; Cowie, B.C. Time for universal hepatitis B screening for Australian adults. Med. J. Aust. 2021, 215, 103–105.e1. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Health Sector Strategy on Viral Hepatitis 2016–2021. Towards Ending Viral Hepatitis. Available online: https://apps.who.int/iris/handle/10665/246177 (accessed on 15 December 2022).
- Shang, G.; Richardson, A.; Gahan, M.E.; Easteal, S.; Ohms, S.; Lidbury, B.A. Predicting the presence of hepatitis B virus surface antigen in Chinese patients by pathology data mining. J. Med. Virol. 2013, 85, 1334–1339. [Google Scholar] [CrossRef]
- Richardson, A.M.; Lidbury, B.A. Enhancement of hepatitis virus immunoassay outcome predictions in imbalanced routine pathology data by data balancing and feature selection before the application of support vector machines. BMC Med. Inform. Decis. Mak. 2017, 17, 121. [Google Scholar] [CrossRef] [Green Version]
- Ramrakhiani, N.S.; Chen, V.L.; Le, M.; Yeo, Y.H.; Barnett, S.D.; Waljee, A.K.; Zhu, J.; Nguyen, M.H. Optimizing hepatitis B virus screening in the United States using a simple demographics-based model. Hepatology 2022, 75, 430–437. [Google Scholar] [CrossRef]
- Ajuwon, B.I.; Richardson, A.; Roper, K.; Sheel, M.; Audu, R.; Salako, B.L.; Bojuwoye, M.O.; Katibi, I.A.; Lidbury, B.A. The development of a machine learning algorithm for early detection of viral hepatitis B infection in Nigerian patients. Sci. Rep. 2023, 13, 3244. [Google Scholar] [CrossRef]
- Moons, K.G.; Altman, D.G.; Reitsma, J.B.; Ioannidis, J.P.; Macaskill, P.; Steyerberg, E.W.; Vickers, A.J.; Ransohoff, D.F.; Collins, G.S. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 2015, 162, W1–W73. [Google Scholar] [CrossRef] [Green Version]
- Steyerberg, E.W.; Vergouwe, Y. Towards better clinical prediction models: Seven steps for development and an ABCD for validation. Eur. Heart J. 2014, 35, 1925–1931. [Google Scholar] [CrossRef] [Green Version]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. BMC Med. 2015, 13, 1. [Google Scholar] [CrossRef] [Green Version]
- Steyerberg, E.W.; Moons, K.G.; Van der Windt, D.A.; Hayden, J.A.; Perel, P.; Schroter, S.; Riley, R.D.; Hemingway, H.; Altman, D.G.; PROGRESS Group. Prognosis Research Strategy (PROGRESS) 3: Prognostic model research. PLoS Med. 2013, 10, e1001381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleeker, S.E.; Moll, H.A.; Steyerberg, E.W.; Donders, A.R.; Derksen-Lubsen, G.; Grobbee, D.E.; Moons, K.G. External validation is necessary in prediction research: A clinical example. J. Clin. Epidemiol. 2003, 56, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E.W.; Harrell, F.E., Jr. Prediction models need appropriate internal, internal-external, and external validation. J. Clin. Epidemiol. 2016, 69, 245–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Calster, B.; Wynants, L.; Timmerman, D.; Steyerberg, E.W.; Collins, G.S. Predictive analytics in health care: How can we know it works? J. Am. Med. Inform. Assoc. 2019, 26, 1651–1654. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, J.C.; Altman, D.G.J.B. Commentary: Prognostic models: Clinically useful or quickly forgotten? BMJ 1995, 311, 1539–1541. [Google Scholar] [CrossRef]
- Luijken, K.; Wynants, L.; Van Smeden, M.; Van Calster, B.; Steyerberg, E.W.; Groenwold, R.H.H.; Collaborators. Changing predictor measurement procedures affected the performance of prediction models in clinical examples. J. Clin. Epidemiol. 2020, 119, 7–18. [Google Scholar] [CrossRef] [Green Version]
- Luijken, K.; Groenwold, R.H.H.; Van Calster, B.; Steyerberg, E.W.; Van Smeden, M. Impact of predictor measurement heterogeneity across settings on the performance of prediction models: A measurement error perspective. Stat. Med. 2019, 38, 3444–3459. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention. Guidelines for Viral Hepatitis Surveillance and Case Management. 2015. Available online: https://www.cdc.gov/hepatitis/statistics/surveillanceguidelines.htm (accessed on 15 January 2023).
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Cook, N.R. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation 2007, 115, 928–935. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Xu, W. Fast Implementation of DeLong’s Algorithm for Comparing the Areas Under Correlated Receiver Operating Characteristic Curves. IEEE Signal Process. Lett. 2014, 21, 1389–1393. [Google Scholar] [CrossRef]
- R Core Team: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018.
- Collins, G.S.; Ogundimu, E.O.; Altman, D.G. Sample size considerations for the external validation of a multivariable prognostic model: A resampling study. Stat. Med. 2016, 35, 214–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debray, T.P.; Vergouwe, Y.; Koffijberg, H.; Nieboer, D.; Steyerberg, E.W.; Moons, K.G. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J. Clin. Epidemiol. 2015, 68, 279–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennells, L.; Kaptoge, S.; White, I.R.; Thompson, S.G.; Wood, A.M. Assessing risk prediction models using individual participant data from multiple studies. Am. J. Epidemiol. 2014, 179, 621–632. [Google Scholar] [CrossRef] [Green Version]
- Wells, P.S.; Anderson, D.R.; Rodger, M.; Ginsberg, J.S.; Kearon, C.; Gent, M.; Turpie, A.G.; Bormanis, J.; Weitz, J.; Chamberlain, M.; et al. Derivation of a simple clinical model to categorize patients probability of pulmonary embolism: Increasing the models utility with the SimpliRED D-dimer. Thromb. Haemost. 2000, 83, 416–420. [Google Scholar] [PubMed]
- Wynants, L.; Timmerman, D.; Bourne, T.; Van Huffel, S.; Van Calster, B. Screening for data clustering in multicenter studies: The residual intraclass correlation. BMC Med. Res. Methodol. 2013, 13, 128. [Google Scholar] [CrossRef]
- Vergouwe, Y.; Moons, K.G.; Steyerberg, E.W. External validity of risk models: Use of benchmark values to disentangle a case-mix effect from incorrect coefficients. Am. J. Epidemiol. 2010, 172, 971–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ransohoff, D.F.; Feinstein, A.R. Problems of spectrum and bias in evaluating the efficacy of diagnostic tests. N. Engl. J. Med. 1978, 299, 926–930. [Google Scholar] [CrossRef]
- Debray, T.P.; Riley, R.D.; Rovers, M.M.; Reitsma, J.B.; Moons, K.G. Individual participant data (IPD) meta-analyses of diagnostic and prognostic modeling studies: Guidance on their use. PLoS Med. 2015, 12, e1001886. [Google Scholar] [CrossRef] [Green Version]
- Van Calster, B.; Steyerberg, E.W.; Wynants, L.; van Smeden, M. There is no such thing as a validated prediction model. BMC Med. 2023, 21, 70. [Google Scholar] [CrossRef]
- Siontis, G.C.; Tzoulaki, I.; Castaldi, P.J.; Ioannidis, J.P. External validation of new risk prediction models is infrequent and reveals worse prognostic discrimination. J. Clin. Epidemiol. 2015, 68, 25–34. [Google Scholar] [CrossRef]
- Berndt, E.R.; Gibbons, R.S.; Kolotilin, A.; Taub, A.L. The heterogeneity of concentrated prescribing behavior: Theory and evidence from antipsychotics. J. Health Econ. 2015, 40, 26–39. [Google Scholar] [CrossRef]
- Agniel, D.; Kohane, I.S.; Weber, G.M. Biases in electronic health record data due to processes within the healthcare system: Retrospective observational study. BMJ 2018, 361, k1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferraro, S.; Borille, S.; Carnevale, A.; Frusciante, E.; Bassani, N.; Panteghini, M. Verification of the harmonization of human epididymis protein 4 assays. Clin. Chem. Lab. Med. 2016, 54, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- White, E. Measurement error in biomarkers: Sources, assessment, and impact on studies. IARC Sci. Publ. 2011, 163, 143–161. [Google Scholar]
- Lim, E.M.; Cembrowski, G.; Cembrowski, M.; Clarke, G. Race-specific WBC and neutrophil count reference intervals. Int. J. Lab. Hematol. 2010, 32, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Franzini, C. Relevance of analytical and biological variations to quality and interpretation of test results: Examples of application to haematology. Ann. Ist. Super. Sanita 1995, 31, 9–13. [Google Scholar]
- Miller, W.G. Harmonization: Its Time Has Come. Clin. Chem. 2017, 63, 1184–1186. [Google Scholar] [CrossRef]
- Velkov, S.; Ott, J.J.; Protzer, U.; Michler, T. The global hepatitis B virus genotype distribution approximated from available genotyping data. Genes 2018, 9, 495. [Google Scholar] [CrossRef] [Green Version]
- Coskun, A.; Braga, F.; Carobene, A.; Tejedor Ganduxe, X.; Aarsand, A.K.; Fernández-Calle, P.; Díaz-Garzón Marco, J.; Bartlett, W.; Jonker, N.; Aslan, B.; et al. Systematic review and meta-analysis of within-subject and between-subject biological variation estimates of 20 haematological parameters. Clin. Chem. Lab. Med. 2019, 58, 25–32. [Google Scholar] [CrossRef] [Green Version]
| Patient Characteristics | HepB LiveTest Derivation Cohort (n = 916) | UITH-Nigerian Validation Cohort (n = 258) | SNP-Australian Validation Cohort (n = 9102) | p-Value |
|---|---|---|---|---|
| Demographics | ||||
| Age, years | 38.8 ± 12.5 | 40.8 ± 13.5 | 45.5 ± 18.3 | <0.001 a |
| Sex, male, (n, %) | 540 (58.9%) | 154 (59.6%) | 4811 (52.8%) | <0.001 b |
| Pathology markers | ||||
| ALT, U/L | 101.0 ± 225.2 | 182.5 ± 344.1 | 57.9 ± 199.3 | <0.001 a |
| AST, U/L | 79.4 ± 173.7 | 128.4 ± 251.2 | — | |
| ALKP, U/L | 84.5 ± 40.1 | 85.7 ± 45.4 | 92.4 ± 83.1 | 0.008 a |
| Crea, µmol/L | 84.3 ± 48.5 | 81.8 ± 28.7 | 86.9 ± 56.2 | 0.148 a |
| TBil, µmol/L | 16.3 ± 35.2 | 18.8 ± 41.5 | 14.7 ± 28.3 | 0.029 a |
| GGT, U/L | 27.8 ± 17.5 | 29.2 ± 19.7 | 84.1 ± 213.3 | <0.001 a |
| ALB, g/L | 37.2 ± 8.1 | 40.0 ± 6.5 | 43.2 ± 5.4 | <0.001 a |
| Hb, g/L | 139.5 ± 19.0 | 137.8 ± 19.2 | 140.3 ± 18.0 | 0.046 a |
| Hct, L/L | 0.41 ± 0.05 | 0.4 ± 0.05 | 0.41 ± 0.05 | 0.006 a |
| WBC, 109/L | 6.4 ± 3.0 | 6.9 ± 3.2 | 7.9 ± 6.5 | <0.001 a |
| PLT, 109/L | 252.6 ± 92.0 | 251.6 ± 102.9 | 261.9 ± 89.9 | 0.003 a |
| MCHC, g/L | 340.7 ± 8.1 | 340.4 ± 8.2 | 342.2 ± 7.2 | <0.001 a |
| MCH, pg/RBC | 30.3 ± 2.6 | 30.3 ± 2.6 | 30.6 ± 2.2 | <0.001 a |
| MCV, fL | 88.9 ± 7.0 | 88.9 ± 7.0 | 89.4 ± 5.9 | 0.028 a |
| RBC, 1012/L | 4.6 ± 0.6 | 4.5 ± 0.6 | 4.6 ± 0.6 | 0.030 a |
| RDW, % | 14.1 ± 2.0 | 14.3 ± 2.0 | 13.8 ± 1.6 | <0.001 a |
| Neut, % | 4.96 ± 4.7 | 4.91 ± 2.7 | 4.9 ± 2.9 | 1.000 a |
| Lymph, % | 2.1 ± 1.0 | 2.1 ± 0.9 | 2.0 ± 1.6 | 0.112 a |
| Presence of HBsAg, n (%) | 636 (69.4%) | 148 (57.3%) | 173 (1.9%) | <0.001 b |
| HepB LiveTest Performance | Sensitivity (%) | Specificity (%) | ACC (95 CI%) |
|---|---|---|---|
| UITH-Nigerian validation cohort | 91.2 | 83.6 | 87.9 (83.3–91.6) |
| SNP-Australian validation cohort | 66.4 | 50.9 | 51.2 (50.2–52.2) |
| Clinical Attribute | Change in Mean Value % | |
|---|---|---|
| NIMR-Derivation Cohort and UITH-Nigerian Validation Cohort | NIMR-Derivation Cohort and SNP-Australian Validation Cohort | |
| Age, years | 5.2 | 17.2 |
| ALT, U/L | 80.7 | −42.7 |
| AST, U/L | 61.7 | — |
| ALKP, U/L | 1.4 | 9.3 |
| Crea, µmol/L | −3.0 | 3.08 |
| TBil, µmol/L | 15.3 | −9.8 |
| GGT, U/L | 5.0 | 202.5 |
| ALB, g/L | 7.5 | 16.1 |
| Hb, g/L | −1.2 | 0.6 |
| Hct, L/L | −2.4 | 0.0 |
| WBC, 109/L | 7.8 | 23.4 |
| PLT, 109/L | −0.4 | 3.7 |
| MCHC, g/L | −0.1 | 0.4 |
| MCH, pg/RBC | 0.0 | 1.0 |
| MCV, fL | 0.0 | 0.6 |
| RBC, 1012/L | −2.2 | 0.0 |
| RDW, % | 1.4 | −2.1 |
| Neut, % | −1.0 | −1.2 |
| Lymph, % | 0.0 | −4.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajuwon, B.I.; Richardson, A.; Roper, K.; Lidbury, B.A. Clinical Validity of a Machine Learning Decision Support System for Early Detection of Hepatitis B Virus: A Binational External Validation Study. Viruses 2023, 15, 1735. https://doi.org/10.3390/v15081735
Ajuwon BI, Richardson A, Roper K, Lidbury BA. Clinical Validity of a Machine Learning Decision Support System for Early Detection of Hepatitis B Virus: A Binational External Validation Study. Viruses. 2023; 15(8):1735. https://doi.org/10.3390/v15081735
Chicago/Turabian StyleAjuwon, Busayo I., Alice Richardson, Katrina Roper, and Brett A. Lidbury. 2023. "Clinical Validity of a Machine Learning Decision Support System for Early Detection of Hepatitis B Virus: A Binational External Validation Study" Viruses 15, no. 8: 1735. https://doi.org/10.3390/v15081735






