Genetic Engineering and Biosynthesis Technology: Keys to Unlocking the Chains of Phage Therapy
Abstract
:1. Introduction
2. Phage Engineering and Biosynthesis Strategies
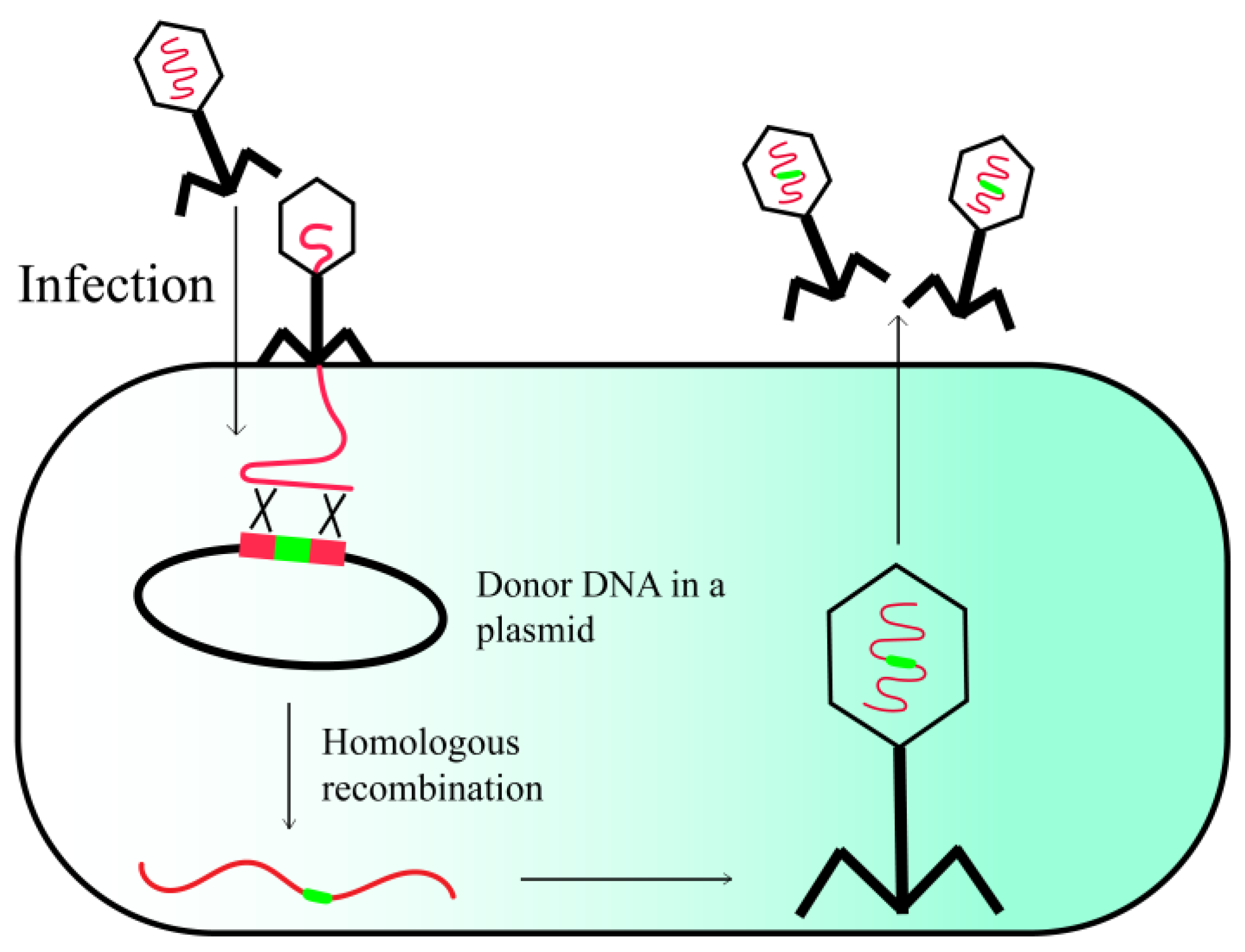
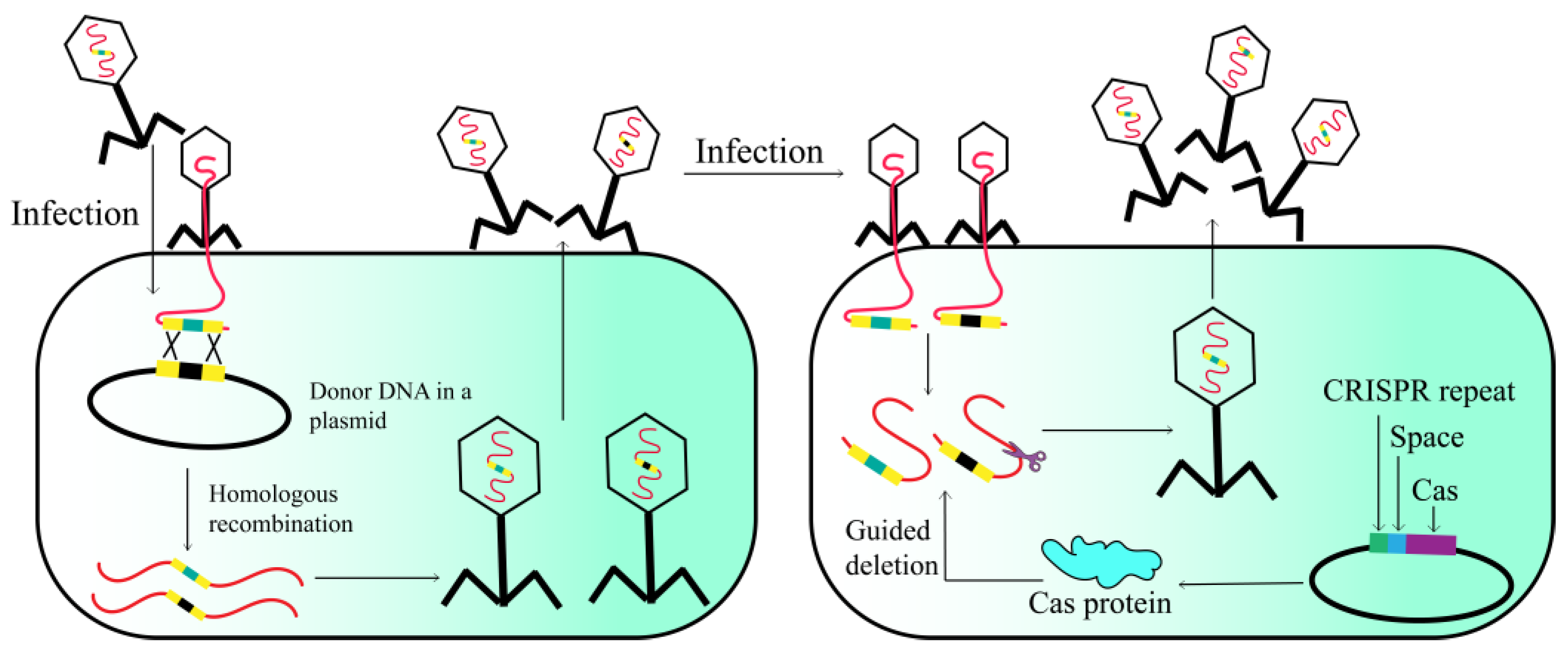
2.1. Host-Mediated Homologous Recombination
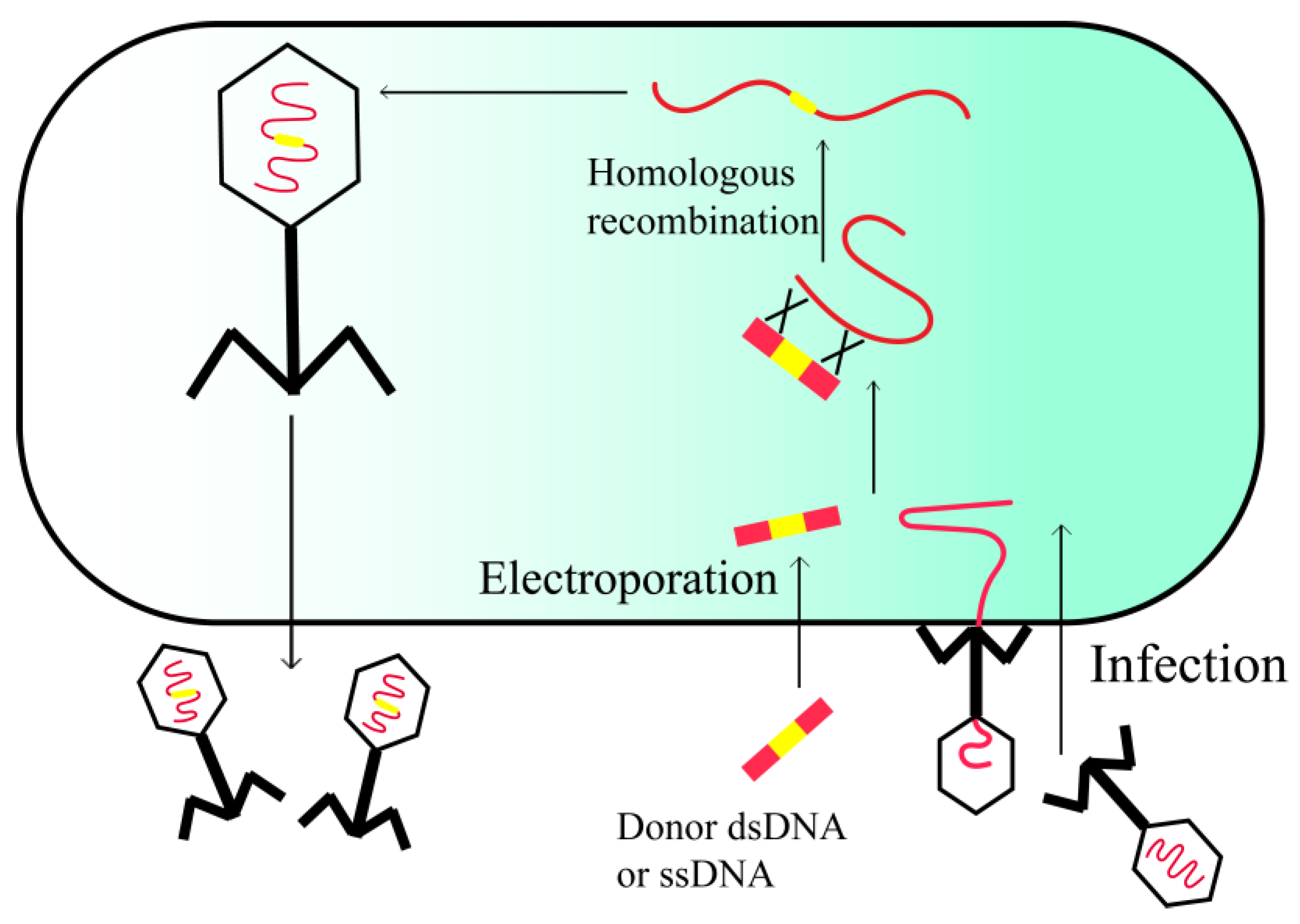
2.2. In Vivo Recombineering
2.3. BRED
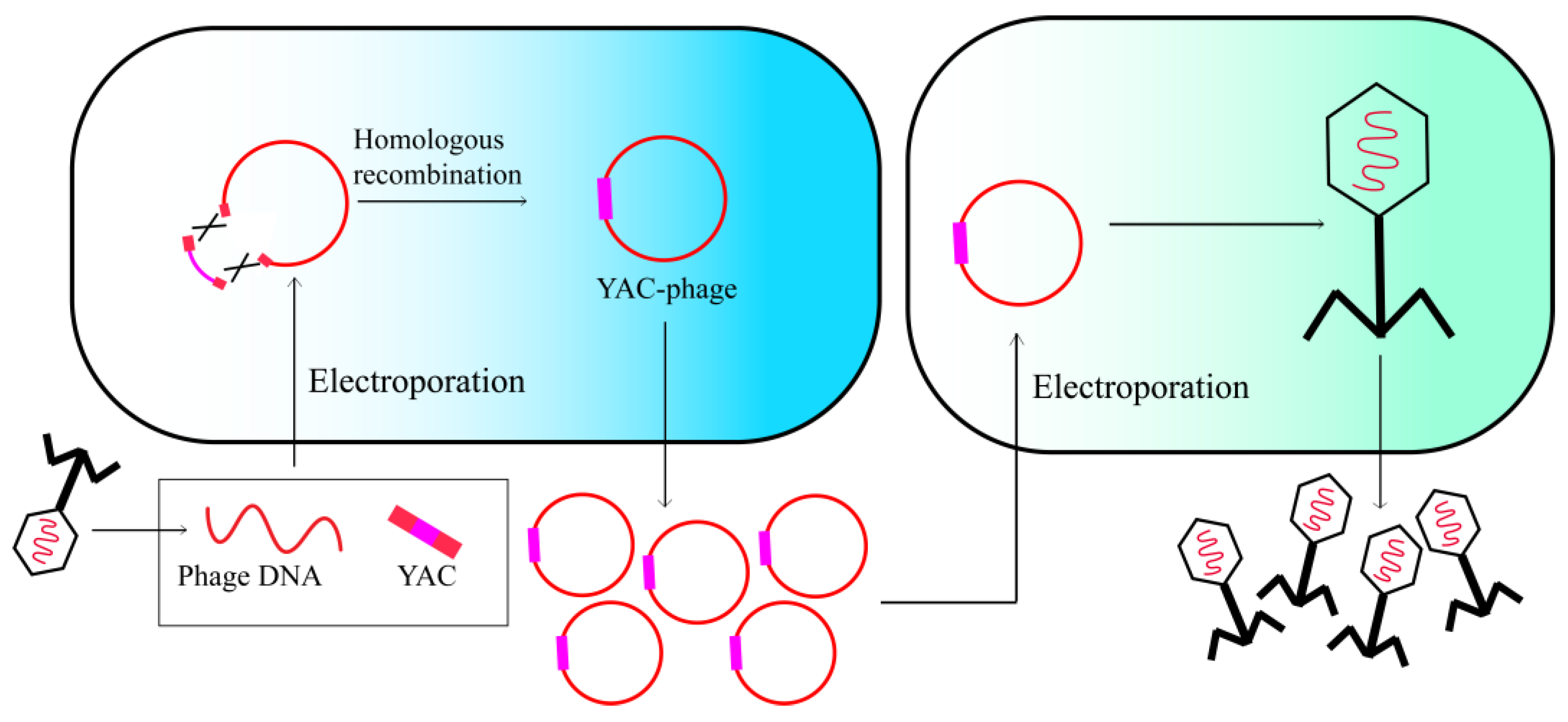
2.4. Yeast-Based Assembly of Phage Genomes
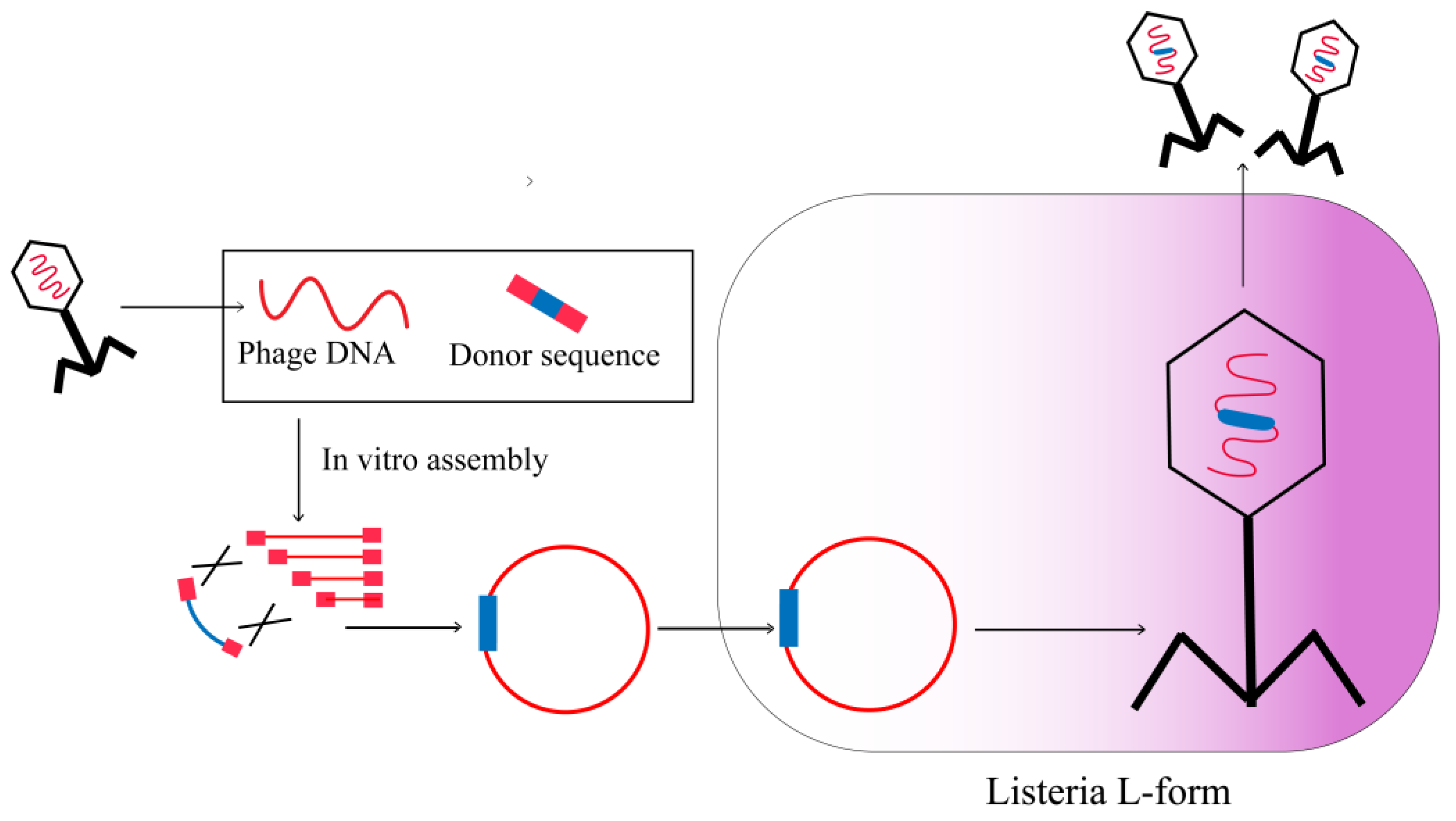
2.5. L-Form Bacteria
2.6. CRISPR-Cas
3. Application of Engineering Phage
3.1. Enhancing Bactericidal Activity
3.2. Restoring the Sensitivity of Drug-Resistant Bacteria to Antibiotics
3.3. Altering Phages’ Host Range
3.4. Increasing the Cleavage of Biofilm
3.5. Increasing the Half-Life of Phage In Vivo
3.6. Reducing Endotoxin Release
3.7. Engineering Phage as a Nanocarrier
3.8. Clinical Application of Engineering Bacteriophages
| Strain | Phage | Technology | Result | Ref. |
|---|---|---|---|---|
| E. coli | T3, T7 | Engineering phage genomes in Saccharomyces cerevisiae | Expanding phage host range | [28] |
| P. aeruginosa | P793 | Recombining with pGhost8 | Expanding phage host range | [73] |
| E. coli | T2 | Recombining with tail fiber gene | Expanding phage host range | [56] |
| P. aeruginosa | PaP1 | Recombining with ORF84 | Expanding phage host range | [74] |
| E. coli | T3 | Phage tail fiber mutagenesis | Expanding phage host range | [75] |
| E. coli | Fd | Recombining with tail fiber gene | Expanding phage host range | [58] |
| E. coli | T2, Fd | Recombining with tail fiber gene | Expanding phage host range | [76] |
| E. coli | PSA | Recombining with receptor binding proteins (RBPs) | Expanding phage host range | [77] |
| E. coli | T4 | Generating gp37 and gp38 variants | Expanding phage host range | [78] |
| E. coli | fd | Recombining with OrfU | Expanding phage host range | [59] |
| E. coli | T7 | Recombining with aiiA | Reducing biofilm formation | [63] |
| E. coli | T7 | Recombining with DspB | Reducing biofilm formation | [61] |
| E. coli | T7 | Recombining with peptide 1018 | Reducing biofilm formation | [79] |
| P. aeruginosa | Pf3 | Recombining with endonuclease BglII | Reducing endotoxin production | [66] |
| S. aureus | P954 | Recombining with chloramphenicol acetyl transferase (cat) gene | Reducing endotoxin production | [80] |
| E. coli | M13 | Recombining with antimicrobial peptides (AMPs) and protein toxins | Reducing endotoxin production | [81] |
| E. coli | λ | Integrating with Ndm-1 and Ctx-M-15 using CRISPR/Cas | Restoring antibiotic sensitivity | [51] |
| E. coli | M13 | Recombining with streptomycin sensitive genes | Restoring antibiotic sensitivity | [52] |
| L. monocytogenes | B025 | Removing lysogen module | Improving lytic ability | [34] |
| S. aureus | φMN1 | Integrating with CRISPR/Cas | Improving lytic ability | [48] |
| S. aureus | ØSaBov | Integrating with CRISPR/Cas | Improving lytic ability | [82] |
| E. amylovora | Y2 | Recombining with Depolymerase | Improving lytic ability | [83] |
| E. coli | M13 | CRISPR-cas9 target resistance genes and virulent genes | Improving lytic ability | [47] |
| E. coli | M13 | Recombining with peptide RGD and PmpD | Improving lytic ability | [84] |
| E. coli | T4 | HIV antigen was fused to outer capsid proteins | HIV vaccine | [85] |
| E. coli | T4 | Anthrax toxin proteins was fused to outer capsid proteins | Anthrax vaccine | [86] |
| E. coli | T4 | FMDV p1 protein was fused to outer capsid proteins | FMDV vaccine | [87] |
| E. coli | MS2 | Capsids radiolabeled with 64Cu | Targeted drug carriers | [88] |
| E. coli | λ | Recombining with integrin-binding peptide | Phage-mediated gene delivery and expression | [89] |
| S. typhimurium | P22 | Chemical modification by DTPA | Gd (III) carrier | [90] |
| E. coli | T7 | Recombining with gold-binding peptide | Gold nanorods carrier | [91] |
| E. coli | fd–tet | Self-assembled siRNA−nanophages | siRNA carrier | [92] |
| E. coli | M13 | Chemical modification to form Au-S bonds | Gold nanorods carrier | [68] |
| E. coli | M13 | Recombining with a biotin acceptor peptide (BAP) | Targeted drug carriers | [93] |
| E. coli | M13 | Recombining with a biotin acceptor peptide (BAP) | Targeted drug carriers | [94] |
| E. coli | fUSE5-ZZ | Recombining with IgG Fc-binding ZZ domain of protein A | Targeted drug carriers | [95] |
| E. coli | fUSE5-ZZ | Recombining with IgG Fc-binding ZZ domain of protein A | Antibacterial drug carriers | [96,97] |
| E. coli | f88 | Recombining with myelin oligodendrocyte glycoprotein (MOG) | Vector-mediated antigen delivery | [98] |
| E. coli | T4 | Recombining with GFP | Luciferase reporter phage | [99] |
| E. coli | T7 | Recombining with biotinylation peptide | Streptavidin-coated quantum dots reporter phage | [100] |
| B. anthracis | Wβ | Recombining with luxAB-2 | Bioluminescent reporter phage | [101,102] |
| E. coli | phiV10 | Recombining with luxCDABE operon | Luciferase reporter phage | [103] |
| L. monocytogenes | A511 | Recombining with nanoluciferase | Nanoluciferase (NLuc) reporter phage | [104] |
| E. coli | T7 | Recombining with nanoluciferase | Nanoluciferase (NLuc) reporter phage | [105] |
| E. coli | ΦV10 | Recombining with nanoluciferase | Nanoluciferase (NLuc) reporter phage | [106] |
| E. coli | K1E | Recombining with nanoluciferase | Nanoluciferase (NLuc) reporter phage | [107] |
| E. coli | T7 | Recombining with β-galactosidase | β-galactosidase reporter phage | [108] |
| M. smegmatis | TM4 | Recombining with GFP or ZsYellow | Fluorescent reporter phage | [109] |
| M. smegmatis | D29 | Recombining with Phsp60-egfp cassette using BRED | EGFP reporter phage | [24] |
| E. amylovora | Y2 | Homologous recombination with LuxAB | Luciferase reporter phage | [83] |
| E. coli | T7 | Homologous recombination with PhoE | Enhancing the half-life of phage | [65] |
| L. monocytogenes | A511 | Bacteriophages PEGylation | Enhancing the half-life of phage | [110] |
| S. typhi | Felix-O1 | Bacteriophages PEGylation | Enhancing the half-life of phage | [110] |
| E. faecalis | fEf11 | Recombining with defective prophage | Improving lytic ability and expanding phage host range | [111] |
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hansson, K.; Brenthel, A. Imagining a post-antibiotic era: A cultural analysis of crisis and antibiotic resistance. Med. Humanit. 2022, 48, 381–388. [Google Scholar] [CrossRef]
- Tarín-Pelló, A.; Suay-García, B.; Pérez-Gracia, M.T. Antibiotic resistant bacteria: Current situation and treatment options to accelerate the development of a new antimicrobial arsenal. Expert Rev. Anti Infect. Ther. 2022, 20, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Tagliabue, A.; Rappuoli, R. Changing Priorities in Vaccinology: Antibiotic Resistance Moving to the Top. Front. Immunol. 2018, 9, 1068. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bragg, R.; van der Westhuizen, W.; Lee, J.Y.; Coetsee, E.; Boucher, C. Bacteriophages as potential treatment option for antibiotic resistant bacteria. Adv. Exp. Med. Biol. 2014, 807, 97–110. [Google Scholar] [CrossRef]
- Royer, S.; Morais, A.P.; da Fonseca Batistão, D.W. Phage therapy as strategy to face post-antibiotic era: A guide to beginners and experts. Arch. Microbiol. 2021, 203, 1271–1279. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons Learned From the First 10 Consecutive Cases of Intravenous Bacteriophage Therapy to Treat Multidrug-Resistant Bacterial Infections at a Single Center in the United States. Open Forum Infect. Dis. 2020, 7, ofaa389. [Google Scholar] [CrossRef]
- Suh, G.A.; Lodise, T.P.; Van Tyne, D.; Tamma, P.D.; Sund, Z.; van Duin, D.; Rybak, M.J.; Patel, R.; Maresso, A.; Nussenblatt, V.; et al. Considerations for the Use of Phage Therapy in Clinical Practice. Antimicrob. Agents Chemother. 2022, 66, e0207121. [Google Scholar] [CrossRef]
- Gurney, J.; Brown, S.P.; Kaltz, O.; Hochberg, M.E. Steering Phages to Combat Bacterial Pathogens. Trends Microbiol. 2020, 28, 85–94. [Google Scholar] [CrossRef]
- Torres-Barceló, C.; Turner, P.E.; Buckling, A. Mitigation of evolved bacterial resistance to phage therapy. Curr. Opin. Virol. 2022, 53, 101201. [Google Scholar] [CrossRef]
- Stacey, H.J.; De Soir, S.; Jones, J.D. The Safety and Efficacy of Phage Therapy: A Systematic Review of Clinical and Safety Trials. Antibiotics 2022, 11, 1340. [Google Scholar] [CrossRef]
- Pires, D.P.; Meneses, L.; Brandão, A.C.; Azeredo, J. An overview of the current state of phage therapy for the treatment of biofilm-related infections. Curr. Opin. Virol. 2022, 53, 101209. [Google Scholar] [CrossRef] [PubMed]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [Green Version]
- Kilcher, S.; Loessner, M.J. Engineering Bacteriophages as Versatile Biologics. Trends Microbiol. 2019, 27, 355–367. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Hatfull, G.F.; Piuri, M. Recombineering: A powerful tool for modification of bacteriophage genomes. Bacteriophage 2012, 2, 5–14. [Google Scholar] [CrossRef]
- Del Val, E.; Nasser, W.; Abaibou, H.; Reverchon, S. RecA and DNA recombination: A review of molecular mechanisms. Biochem. Soc. Trans. 2019, 47, 1511–1531. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically Engineered Phages: A Review of Advances over the Last Decade. Microbiol. Mol. Biol. Rev. MMBR 2016, 80, 523–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Chamorro, L.; Boulanger, P.; Rossier, O. Strategies for Bacteriophage T5 Mutagenesis: Expanding the Toolbox for Phage Genome Engineering. Front. Microbiol. 2021, 12, 667332. [Google Scholar] [CrossRef]
- Sharan, S.K.; Thomason, L.C.; Kuznetsov, S.G.; Court, D.L. Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 2009, 4, 206–223. [Google Scholar] [CrossRef] [Green Version]
- Murphy, K.C. λ Recombination and Recombineering. EcoSal Plus 2016, 7, e0011-2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Łobocka, M.; Dąbrowska, K.; Górski, A. Engineered Bacteriophage Therapeutics: Rationale, Challenges and Future. BioDrugs 2021, 35, 255–280. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, L.J.; Piuri, M.; Swigonová, Z.; Balachandran, A.; Oldfield, L.M.; van Kessel, J.C.; Hatfull, G.F. BRED: A simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE 2008, 3, e3957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, J.L.; Piuri, M.; Broussard, G.; Marinelli, L.J.; Bastos, G.M.; Hirata, R.D.; Hatfull, G.F.; Hirata, M.H. Application of BRED technology to construct recombinant D29 reporter phage expressing EGFP. FEMS Microbiol. Lett. 2013, 344, 166–172. [Google Scholar] [CrossRef] [Green Version]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Fehér, T.; Karcagi, I.; Blattner, F.R.; Pósfai, G. Bacteriophage recombineering in the lytic state using the lambda red recombinases. Microb. Biotechnol. 2012, 5, 466–476. [Google Scholar] [CrossRef] [Green Version]
- Wetzel, K.S.; Guerrero-Bustamante, C.A.; Dedrick, R.M.; Ko, C.C.; Freeman, K.G.; Aull, H.G.; Divens, A.M.; Rock, J.M.; Zack, K.M.; Hatfull, G.F. CRISPY-BRED and CRISPY-BRIP: Efficient bacteriophage engineering. Sci. Rep. 2021, 11, 6796. [Google Scholar] [CrossRef]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Gibson, D.G. Oligonucleotide assembly in yeast to produce synthetic DNA fragments. Methods Mol. Biol. 2012, 852, 11–21. [Google Scholar] [CrossRef]
- Jaschke, P.R.; Lieberman, E.K.; Rodriguez, J.; Sierra, A.; Endy, D. A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast. Virology 2012, 434, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Assad-Garcia, N.; D’Souza, R.; Vashee, S.; Fouts, D.E.; Buzzeo, R.; Tripathi, A.; Oldfield, L.M. Cross-Genus “Boot-Up” of Synthetic Bacteriophage in Staphylococcus aureus by Using a New and Efficient DNA Transformation Method. Appl. Environ. Microbiol. 2022, 88, e0148621. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Deng, Z.; Tao, H.; Song, W.; Xing, B.; Liu, W.; Kong, L.; Yuan, S.; Ma, Y.; Wu, Y.; et al. Harnessing stepping-stone hosts to engineer, select, and reboot synthetic bacteriophages in one pot. Cell Rep. Methods 2022, 2, 100217. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Monteiro, R.; Mil-Homens, D.; Fialho, A.; Lu, T.K.; Azeredo, J. Designing P. aeruginosa synthetic phages with reduced genomes. Sci. Rep. 2021, 11, 2164. [Google Scholar] [CrossRef] [PubMed]
- Kilcher, S.; Studer, P.; Muessner, C.; Klumpp, J.; Loessner, M.J. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 567–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chikada, T.; Kanai, T.; Hayashi, M.; Kasai, T.; Oshima, T.; Shiomi, D. Direct Observation of Conversion From Walled Cells to Wall-Deficient L-Form and Vice Versa in Escherichia coli Indicates the Essentiality of the Outer Membrane for Proliferation of L-Form Cells. Front. Microbiol. 2021, 12, 645965. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007, 8, 172. [Google Scholar] [CrossRef] [Green Version]
- Martel, B.; Moineau, S. CRISPR-Cas: An efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 2014, 42, 9504–9513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, J.; Oromí-Bosch, A.; Mendoza, S.D.; Karambelkar, S.; Berry, J.D.; Bondy-Denomy, J. Bacteriophage genome engineering with CRISPR-Cas13a. Nat. Microbiol. 2022, 7, 1956–1966. [Google Scholar] [CrossRef]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005, 1, e60. [Google Scholar] [CrossRef]
- Shivram, H.; Cress, B.F.; Knott, G.J.; Doudna, J.A. Controlling and enhancing CRISPR systems. Nat. Chem. Biol. 2021, 17, 10–19. [Google Scholar] [CrossRef]
- Lemay, M.L.; Tremblay, D.M.; Moineau, S. Genome Engineering of Virulent Lactococcal Phages Using CRISPR-Cas9. ACS Synth. Biol. 2017, 6, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Kiro, R.; Shitrit, D.; Qimron, U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 2014, 11, 42–44. [Google Scholar] [CrossRef] [Green Version]
- Malone, L.M.; Birkholz, N.; Fineran, P.C. Conquering CRISPR: How phages overcome bacterial adaptive immunity. Curr. Opin. Biotechnol. 2021, 68, 30–36. [Google Scholar] [CrossRef]
- Liu, Y.; Dai, L.; Dong, J.; Chen, C.; Zhu, J.; Rao, V.B.; Tao, P. Covalent Modifications of the Bacteriophage Genome Confer a Degree of Resistance to Bacterial CRISPR Systems. J. Virol. 2020, 94, e01630-20. [Google Scholar] [CrossRef]
- Wu, X.; Zhu, J.; Tao, P.; Rao, V.B. Bacteriophage T4 Escapes CRISPR Attack by Minihomology Recombination and Repair. mBio 2021, 12, e0136121. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.; Kerven, J.; Chen, Y.; Sagona, A.P. Genetic Engineering of Bacteriophage K1F with Human Epidermal Growth Factor to Enhance Killing of Intracellular E. coli K1. ACS Synth. Biol. 2023, 12, 2094–2106. [Google Scholar] [CrossRef] [PubMed]
- Citorik, R.J.; Mimee, M.; Lu, T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014, 32, 1141–1145. [Google Scholar] [CrossRef] [Green Version]
- Bikard, D.; Euler, C.W.; Jiang, W.; Nussenzweig, P.M.; Goldberg, G.W.; Duportet, X.; Fischetti, V.A.; Marraffini, L.A. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014, 32, 1146–1150. [Google Scholar] [CrossRef] [Green Version]
- Azam, A.H.; Suzuki, M.; Penadés, J.R.; Cui, L.; Kiga, K.; Tan, X.E.; Ibarra-Chávez, R.; Watanabe, S.; Aiba, Y.; Sato’o, Y.; et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 2020, 11, 2934. [Google Scholar] [CrossRef]
- Meile, S.; Du, J.; Dunne, M.; Kilcher, S.; Loessner, M.J. Engineering therapeutic phages for enhanced antibacterial efficacy. Curr. Opin. Virol. 2022, 52, 182–191. [Google Scholar] [CrossRef]
- Yosef, I.; Manor, M.; Kiro, R.; Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 7267–7272. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Friedman, N.; Molshanski-Mor, S.; Qimron, U. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl. Environ. Microbiol. 2012, 78, 744–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and Evolutionary Determinants of Bacteriophage Host Range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Globus, R.; Molshanski-Mor, S.; Qimron, U. Extending the Host Range of Bacteriophage Particles for DNA Transduction. Mol. Cell 2017, 66, 721–728.e3. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Zhang, L.; Abdelgader, S.A.; Yu, L.; Xu, J.; Yao, H.; Lu, C.; Zhang, W. Alterations in gp37 Expand the Host Range of a T4-Like Phage. Appl. Environ. Microbiol. 2017, 83, e01576-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahichi, F.; Synnott, A.J.; Yamamichi, K.; Osada, T.; Tanji, Y. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol. Lett. 2009, 295, 211–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, S.; He, X.; Tan, Y.; Huang, G.; Zhang, L.; Lux, R.; Shi, W.; Hu, F. Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS ONE 2013, 8, e68562. [Google Scholar] [CrossRef] [Green Version]
- Marzari, R.; Sblattero, D.; Righi, M.; Bradbury, A. Extending filamentous phage host range by the grafting of a heterologous receptor binding domain. Gene 1997, 185, 27–33. [Google Scholar] [CrossRef]
- Heilpern, A.J.; Waldor, M.K. pIIICTX, a predicted CTXphi minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 2003, 185, 1037–1044. [Google Scholar] [CrossRef] [Green Version]
- Azeredo, J.; Sutherland, I.W. The use of phages for the removal of infectious biofilms. Curr. Pharm. Biotechnol. 2008, 9, 261–266. [Google Scholar] [CrossRef]
- Lu, T.K.; Collins, J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA 2007, 104, 11197–11202. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.H.; Wang, L.H.; Xu, J.L.; Zhang, H.B.; Zhang, X.F.; Zhang, L.H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Lamas-Samanamud, G.R. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl. Environ. Microbiol. 2014, 80, 5340–5348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, P.; Wang, L.; Sha, R.; Liu, L.; Qian, J.; Ishimwe, N.; Zhang, W.; Qian, J.; Zhang, Y.; Wen, L. A blood circulation-prolonging peptide anchored biomimetic phage-platelet hybrid nanoparticle system for prolonged blood circulation and optimized anti-bacterial performance. Theranostics 2021, 11, 2278–2296. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Costa, A.R.; Santos, J.F.; Siliakus, M.F.; van Lent, J.W.; Kengen, S.W.; Azeredo, J.; Kluskens, L.D. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci. Rep. 2016, 6, 39235. [Google Scholar] [CrossRef] [Green Version]
- Hagens, S.; Habel, A.; von Ahsen, U.; von Gabain, A.; Bläsi, U. Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob. Agents Chemother. 2004, 48, 3817–3822. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Kumari, A.; Kumari Negi, A.; Galav, V.; Thakur, S.; Agrawal, M.; Sharma, V. Nanotechnology Based Approaches in Phage Therapy: Overcoming the Pharmacological Barriers. Front. Pharmacol. 2021, 12, 699054. [Google Scholar] [CrossRef]
- Chen, I.A.; Peng, H.; Borg, R.E.; Dow, L.P.; Pruitt, B.L. Controlled phage therapy by photothermal ablation of specific bacterial species using gold nanorods targeted by chimeric phages. Proc. Natl. Acad. Sci. USA 2020, 117, 1951–1961. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Yu, G.; Wei, J.; Liao, X.; Zhang, Y.; Ren, Y.; Zhang, C.; Wang, Y.; Zhang, D.; Wang, J.; et al. A single thiolated-phage displayed nanobody-based biosensor for label-free detection of foodborne pathogen. J. Hazard. Mater. 2023, 443, 130157. [Google Scholar] [CrossRef]
- Peng, H.; Chen, I.A. Rapid Colorimetric Detection of Bacterial Species through the Capture of Gold Nanoparticles by Chimeric Phages. ACS Nano 2019, 13, 1244–1252. [Google Scholar] [CrossRef]
- Turner, K.H.; Everett, J.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014, 10, e1004518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nick, J.A.; Dedrick, R.M.; Gray, A.L.; Vladar, E.K.; Smith, B.E.; Freeman, K.G.; Malcolm, K.C.; Epperson, L.E.; Hasan, N.A.; Hendrix, J.; et al. Host and pathogen response to bacteriophage engineered against Mycobacterium abscessus lung infection. Cell 2022, 185, 1860–1874.e12. [Google Scholar] [CrossRef] [PubMed]
- Kot, W.; Hammer, K.; Neve, H.; Vogensen, F.K. Identification of the receptor-binding protein in lytic Leuconostoc pseudomesenteroides bacteriophages. Appl. Environ. Microbiol. 2013, 79, 3311–3314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacheco, L.G.; Slade, S.E.; Seyffert, N.; Santos, A.R.; Castro, T.L.; Silva, W.M.; Santos, A.V.; Santos, S.G.; Farias, L.M.; Carvalho, M.A.; et al. A combined approach for comparative exoproteome analysis of Corynebacterium pseudotuberculosis. BMC Microbiol. 2011, 11, 12. [Google Scholar] [CrossRef] [Green Version]
- Yehl, K.; Lemire, S.; Yang, A.C.; Ando, H.; Mimee, M.; Torres, M.T.; de la Fuente-Nunez, C.; Lu, T.K. Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 2019, 179, 459–469.e9. [Google Scholar] [CrossRef]
- Lin, T.Y.; Lo, Y.H.; Tseng, P.W.; Chang, S.F.; Lin, Y.T.; Chen, T.S. A T3 and T7 recombinant phage acquires efficient adsorption and a broader host range. PLoS ONE 2012, 7, e30954. [Google Scholar] [CrossRef] [Green Version]
- Dunne, M.; Rupf, B.; Tala, M.; Qabrati, X.; Ernst, P.; Shen, Y.; Sumrall, E.; Heeb, L.; Plückthun, A.; Loessner, M.J.; et al. Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Rep. 2019, 29, 1336–1350.e4. [Google Scholar] [CrossRef] [Green Version]
- Pouillot, F.; Blois, H.; Iris, F. Genetically engineered virulent phage banks in the detection and control of emergent pathogenic bacteria. Biosecurity Bioterrorism Biodefense Strategy Pract. Sci. 2010, 8, 155–169. [Google Scholar] [CrossRef] [Green Version]
- Lemon, D.J.; Kay, M.K.; Titus, J.K.; Ford, A.A.; Chen, W.; Hamlin, N.J.; Hwang, Y.Y. Construction of a genetically modified T7Select phage system to express the antimicrobial peptide 1018. J. Microbiol. 2019, 57, 532–538. [Google Scholar] [CrossRef]
- Paul, V.D.; Sundarrajan, S.; Rajagopalan, S.S.; Hariharan, S.; Kempashanaiah, N.; Padmanabhan, S.; Sriram, B.; Ramachandran, J. Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. BMC Microbiol. 2011, 11, 195. [Google Scholar] [CrossRef] [Green Version]
- Krom, R.J.; Bhargava, P.; Lobritz, M.A.; Collins, J.J. Engineered Phagemids for Nonlytic, Targeted Antibacterial Therapies. Nano Lett. 2015, 15, 4808–4813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.Y.; Moon, B.Y.; Park, J.W.; Thornton, J.A.; Park, Y.H.; Seo, K.S. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 2017, 7, 44929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Born, Y.; Fieseler, L.; Thöny, V.; Leimer, N.; Duffy, B.; Loessner, M.J. Engineering of Bacteriophages Y2::dpoL1-C and Y2::luxAB for Efficient Control and Rapid Detection of the Fire Blight Pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 2017, 83, e00341-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattarai, S.R.; Yoo, S.Y.; Lee, S.W.; Dean, D. Engineered phage-based therapeutic materials inhibit Chlamydia trachomatis intracellular infection. Biomaterials 2012, 33, 5166–5174. [Google Scholar] [CrossRef] [Green Version]
- Sathaliyawala, T.; Rao, M.; Maclean, D.M.; Birx, D.L.; Alving, C.R.; Rao, V.B. Assembly of human immunodeficiency virus (HIV) antigens on bacteriophage T4: A novel in vitro approach to construct multicomponent HIV vaccines. J. Virol. 2006, 80, 7688–7698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivachandra, S.B.; Li, Q.; Peachman, K.K.; Matyas, G.R.; Leppla, S.H.; Alving, C.R.; Rao, M.; Rao, V.B. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine 2007, 25, 1225–1235. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.J.; Tian, C.J.; Zhu, Q.S.; Zhao, M.Y.; Xin, A.G.; Nie, W.X.; Ling, S.R.; Zhu, M.W.; Wu, J.Y.; Lan, H.Y.; et al. Orally delivered foot-and-mouth disease virus capsid protomer vaccine displayed on T4 bacteriophage surface: 100% protection from potency challenge in mice. Vaccine 2008, 26, 1471–1481. [Google Scholar] [CrossRef]
- Aanei, I.L.; ElSohly, A.M.; Farkas, M.E.; Netirojjanakul, C.; Regan, M.; Taylor Murphy, S.; O’Neil, J.P.; Seo, Y.; Francis, M.B. Biodistribution of Antibody-MS2 Viral Capsid Conjugates in Breast Cancer Models. Mol. Pharm. 2016, 13, 3764–3772. [Google Scholar] [CrossRef] [Green Version]
- Lankes, H.A.; Zanghi, C.N.; Santos, K.; Capella, C.; Duke, C.M.; Dewhurst, S. In vivo gene delivery and expression by bacteriophage lambda vectors. J. Appl. Microbiol. 2007, 102, 1337–1349. [Google Scholar] [CrossRef] [Green Version]
- Min, J.; Jung, H.; Shin, H.H.; Cho, G.; Cho, H.; Kang, S. Implementation of P22 viral capsids as intravascular magnetic resonance T1 contrast conjugates via site-selective attachment of Gd(III)-chelating agents. Biomacromolecules 2013, 14, 2332–2339. [Google Scholar] [CrossRef]
- Zhou, X.; Cao, P.; Zhu, Y.; Lu, W.; Gu, N.; Mao, C. Phage-mediated counting by the naked eye of miRNA molecules at attomolar concentrations in a Petri dish. Nat. Mater. 2015, 14, 1058–1064. [Google Scholar] [CrossRef] [Green Version]
- Bedi, D.; Gillespie, J.W.; Petrenko, V.A., Jr.; Ebner, A.; Leitner, M.; Hinterdorfer, P.; Petrenko, V.A. Targeted delivery of siRNA into breast cancer cells via phage fusion proteins. Mol. Pharm. 2013, 10, 551–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DePorter, S.M.; McNaughton, B.R. Engineered M13 bacteriophage nanocarriers for intracellular delivery of exogenous proteins to human prostate cancer cells. Bioconjugate Chem. 2014, 25, 1620–1625. [Google Scholar] [CrossRef]
- Ghosh, D.; Kohli, A.G.; Moser, F.; Endy, D.; Belcher, A.M. Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS Synth. Biol. 2012, 1, 576–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar, H.; Yacoby, I.; Benhar, I. Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnol. 2008, 8, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yacoby, I.; Bar, H.; Benhar, I. Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob. Agents Chemother. 2007, 51, 2156–2163. [Google Scholar] [CrossRef] [Green Version]
- Yacoby, I.; Shamis, M.; Bar, H.; Shabat, D.; Benhar, I. Targeting antibacterial agents by using drug-carrying filamentous bacteriophages. Antimicrob. Agents Chemother. 2006, 50, 2087–2097. [Google Scholar] [CrossRef] [Green Version]
- Rakover, I.S.; Zabavnik, N.; Kopel, R.; Paz-Rozner, M.; Solomon, B. Antigen-specific therapy of EAE via intranasal delivery of filamentous phage displaying a myelin immunodominant epitope. J. Neuroimmunol. 2010, 225, 68–76. [Google Scholar] [CrossRef]
- Namura, M.; Hijikata, T.; Miyanaga, K.; Tanji, Y. Detection of Escherichia coli with fluorescent labeled phages that have a broad host range to E. coli in sewage water. Biotechnol. Prog. 2008, 24, 481–486. [Google Scholar] [CrossRef]
- Edgar, R.; McKinstry, M.; Hwang, J.; Oppenheim, A.B.; Fekete, R.A.; Giulian, G.; Merril, C.; Nagashima, K.; Adhya, S. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc. Natl. Acad. Sci. USA 2006, 103, 4841–4845. [Google Scholar] [CrossRef]
- Nguyen, C.; Makkar, R.; Sharp, N.J.; Page, M.A.; Molineux, I.J.; Schofield, D.A. Detection of Bacillus anthracis spores from environmental water using bioluminescent reporter phage. J. Appl. Microbiol. 2017, 123, 1184–1193. [Google Scholar] [CrossRef]
- Sharp, N.J.; Molineux, I.J.; Page, M.A.; Schofield, D.A. Rapid Detection of Viable Bacillus anthracis Spores in Environmental Samples by Using Engineered Reporter Phages. Appl. Environ. Microbiol. 2016, 82, 2380–2387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kim, M.; Kim, S.; Ryu, S. Sensitive detection of viable Escherichia coli O157:H7 from foods using a luciferase-reporter phage phiV10lux. Int. J. Food Microbiol. 2017, 254, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Meile, S.; Sarbach, A.; Du, J.; Schuppler, M.; Saez, C.; Loessner, M.J. Engineered Reporter Phages for Rapid Bioluminescence-Based Detection and Differentiation of Viable Listeria Cells. Appl. Environ. Microbiol. 2020, 86, e00442-20. [Google Scholar] [CrossRef] [PubMed]
- Hinkley, T.C.; Garing, S.; Singh, S.; Le Ny, A.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. Reporter bacteriophage T7(NLC) utilizes a novel NanoLuc::CBM fusion for the ultrasensitive detection of Escherichia coli in water. Analyst 2018, 143, 4074–4082. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Coronel-Aguilera, C.P.; Romero, P.L.; Perry, L.; Minocha, U.; Rosenfield, C.; Gehring, A.G.; Paoli, G.C.; Bhunia, A.K.; Applegate, B. The Use of a Novel NanoLuc-Based Reporter Phage for the Detection of Escherichia coli O157:H7. Sci. Rep. 2016, 6, 33235. [Google Scholar] [CrossRef] [Green Version]
- Dow, P.; Kotz, K.; Gruszka, S.; Holder, J.; Fiering, J. Acoustic separation in plastic microfluidics for rapid detection of bacteria in blood using engineered bacteriophage. Lab A Chip 2018, 18, 923–932. [Google Scholar] [CrossRef]
- Chen, J.; Picard, R.A.; Wang, D. Lyophilized Engineered Phages for Escherichia coli Detection in Food Matrices. ACS Sens. 2017, 2, 1573–1577. [Google Scholar] [CrossRef]
- Piuri, M.; Jacobs, W.R., Jr.; Hatfull, G.F. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis. PLoS ONE 2009, 4, e4870. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.P.; Cha, J.D.; Jang, E.H.; Klumpp, J.; Hagens, S.; Hardt, W.D.; Lee, K.Y.; Loessner, M.J. PEGylation of bacteriophages increases blood circulation time and reduces T-helper type 1 immune response. Microb. Biotechnol. 2008, 1, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Fouts, D.E.; DePew, J.; Stevens, R.H. Genetic modifications to temperate Enterococcus faecalis phage Ef11 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection. Microbiology 2013, 159, 1023–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalewska-Piątek, B.; Piątek, R. Bacteriophages as Potential Tools for Use in Antimicrobial Therapy and Vaccine Development. Pharmaceuticals 2021, 14, 331. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Costa, A.R.; Pinto, G.; Meneses, L.; Azeredo, J. Current challenges and future opportunities of phage therapy. FEMS Microbiol. Rev. 2020, 44, 684–700. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, S.; Wang, Y.; Jiang, K.; Guo, X.; Zhang, J.; Zhou, F.; Li, Q.; Jiang, Y.; Yang, C.; Teng, T. Genetic Engineering and Biosynthesis Technology: Keys to Unlocking the Chains of Phage Therapy. Viruses 2023, 15, 1736. https://doi.org/10.3390/v15081736
Lv S, Wang Y, Jiang K, Guo X, Zhang J, Zhou F, Li Q, Jiang Y, Yang C, Teng T. Genetic Engineering and Biosynthesis Technology: Keys to Unlocking the Chains of Phage Therapy. Viruses. 2023; 15(8):1736. https://doi.org/10.3390/v15081736
Chicago/Turabian StyleLv, Sixuan, Yuhan Wang, Kaixin Jiang, Xinge Guo, Jing Zhang, Fang Zhou, Qiming Li, Yuan Jiang, Changyong Yang, and Tieshan Teng. 2023. "Genetic Engineering and Biosynthesis Technology: Keys to Unlocking the Chains of Phage Therapy" Viruses 15, no. 8: 1736. https://doi.org/10.3390/v15081736
APA StyleLv, S., Wang, Y., Jiang, K., Guo, X., Zhang, J., Zhou, F., Li, Q., Jiang, Y., Yang, C., & Teng, T. (2023). Genetic Engineering and Biosynthesis Technology: Keys to Unlocking the Chains of Phage Therapy. Viruses, 15(8), 1736. https://doi.org/10.3390/v15081736








