Development of Zika Virus Mini-Replicon Based Single-Round Infectious Particles as Gene Delivery Vehicles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
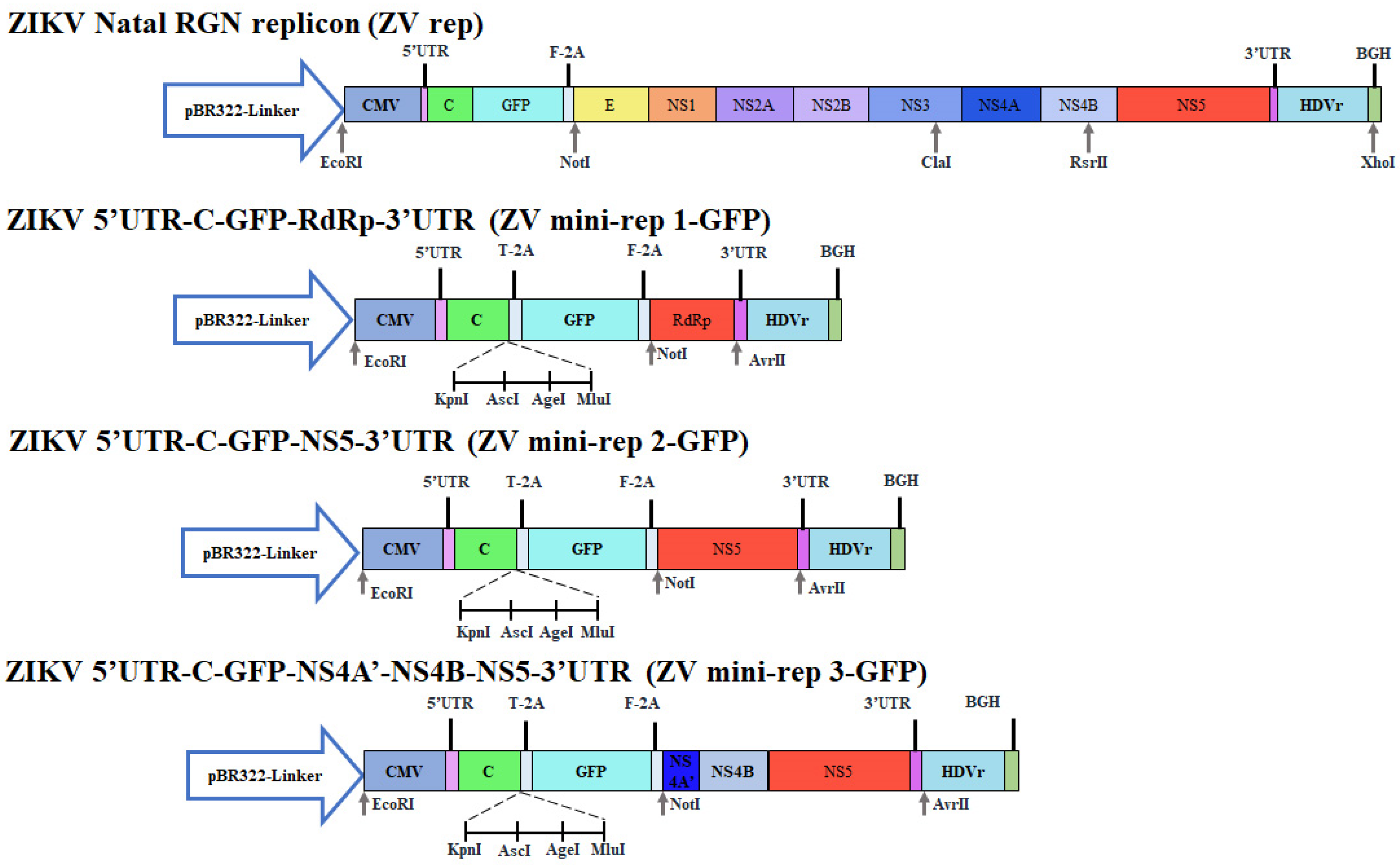
2.2. Construction of ZIKV Mini-Replicons Comprising the Green Fluorescent Protein Gene
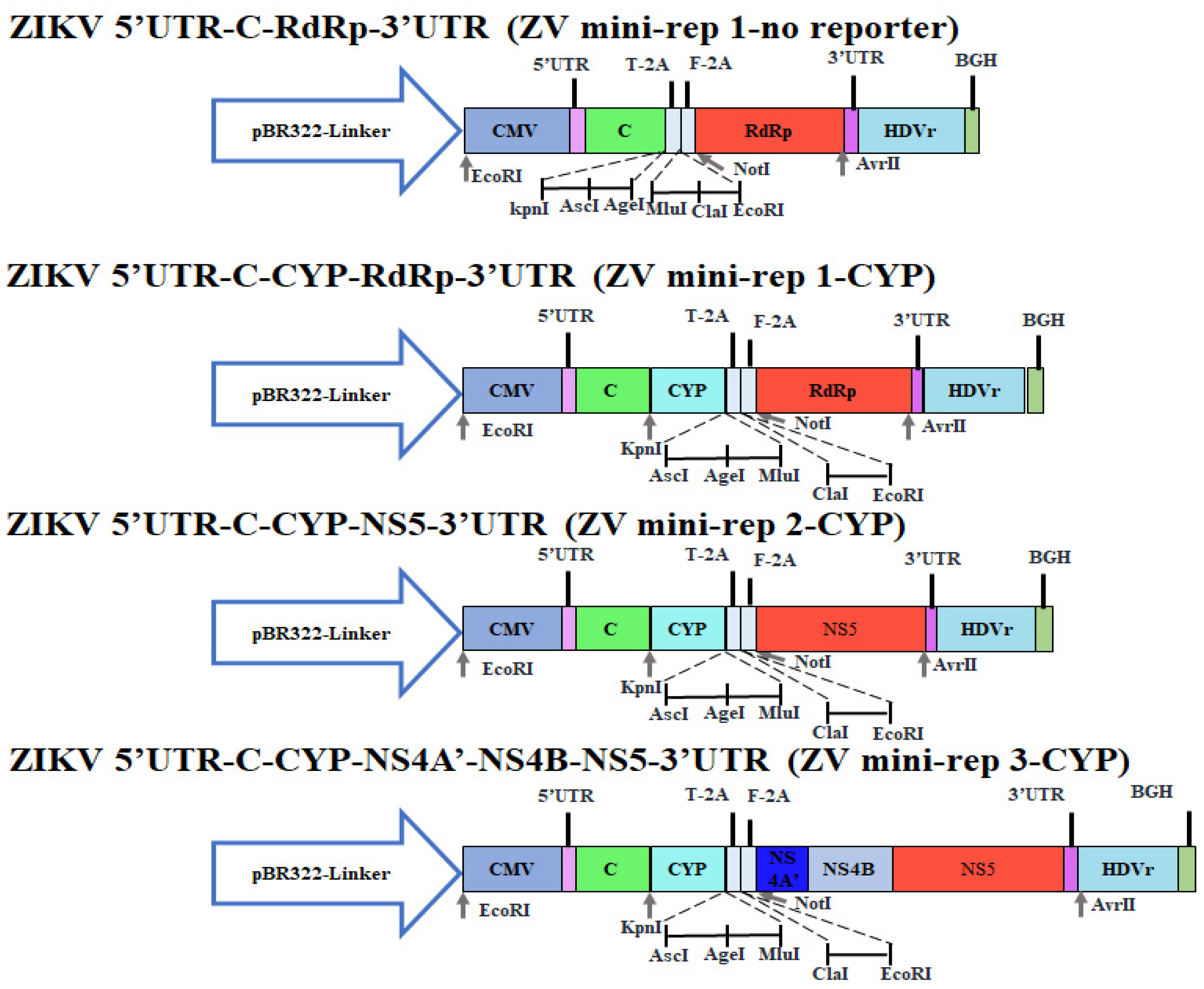
2.3. Construction of ZIKV Mini-Replicons Comprising the Cyan Fluorescent Protein-Linker- Yellow Fluorescent Protein (CFP/YFP, CYP) Fusion Gene or Human ACE2 Gene
2.4. Production of ZIKV Mini-Replicon-Driven RNA-Based SRIPs Expressing GFP, CYP, or hACE2
2.5. Transient Reporter Expression Kinetics and Cytopathic Ability by ZIKV Mini-rep 2-CYP and 3-CYP SRIPs
2.6. In Vivo Expression of hACE2 in the Mice with an Intravenous Injection of ZIKV prME/Mini-rep 3-hACE2 SRIPs
2.7. Statistical Analysis
3. Results
3.1. Construction of ZIKV Mini-Replicons Encoding the Gene of Interest
3.2. The Involvement of ZIKV NS4B and MTase in RdRp-Mediated Replication of Mini-Replicon-Driven RNAs
3.3. Engineering ZIKV Mini-Replicon Vectors
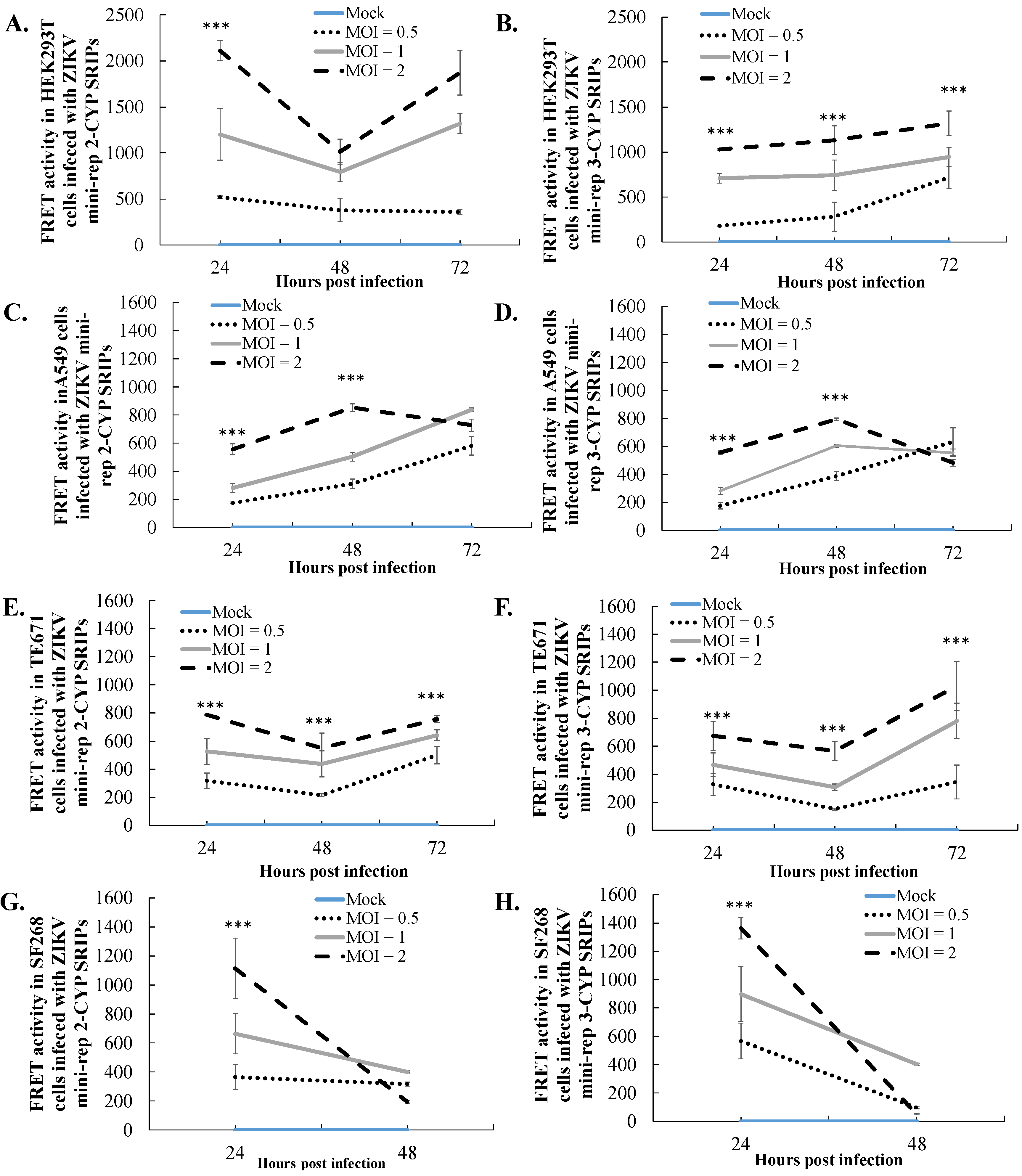
3.4. Comparative Analysis of Heterologous Gene Expression in Infected Cells with ZIKV SRIPs Carrying Mini-Replicon-Driven RNAs
3.5. In Vivo Expression of hACE2 Delivered by ZIKV Mini-rep 3 SRIPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baud, D.; Gubler, D.J.; Schaub, B.; Lanteri, M.C.; Musso, D. An update on Zika virus infection. Lancet 2017, 390, 2099–2109. [Google Scholar] [CrossRef] [PubMed]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Song, J.; Lu, X.; Deng, Y.Q.; Musyoki, A.M.; Cheng, H.; Zhang, Y.; Yuan, Y.; Song, H.; Haywood, J.; et al. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell Host Microbe 2016, 19, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yi, G.; Du, F.; Chuang, Y.C.; Vaughan, R.C.; Sankaran, B.; Kao, C.C.; Li, P. Structure and function of the Zika virus full-length NS5 protein. Nat. Commun. 2017, 8, 14762. [Google Scholar] [CrossRef]
- Ikeda, M.; Yi, M.; Li, K.; Lemon, S.M. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 2002, 76, 2997–3006. [Google Scholar] [CrossRef]
- Leardkamolkarn, V.; Sirigulpanit, W.; Chotiwan, N.; Kumkate, S.; Huang, C.Y. Development of Dengue type-2 virus replicons expressing GFP reporter gene in study of viral RNA replication. Virus Res. 2012, 163, 552–562. [Google Scholar] [CrossRef]
- Fayzulin, R.; Scholle, F.; Petrakova, O.; Frolov, I.; Mason, P.W. Evaluation of replicative capacity and genetic stability of West Nile virus replicons using highly efficient packaging cell lines. Virology 2006, 351, 196–209. [Google Scholar] [CrossRef]
- Pijlman, G.P.; Suhrbier, A.; Khromykh, A.A. Kunjin virus replicons: An RNA-based, non-cytopathic viral vector system for protein production, vaccine and gene therapy applications. Expert Opin. Biol. Ther. 2006, 6, 135–145. [Google Scholar] [CrossRef]
- Lu, C.Y.; Hour, M.J.; Wang, C.Y.; Huang, S.H.; Mu, W.X.; Chang, Y.C.; Lin, C.W. Single-Round Infectious Particle Antiviral Screening Assays for the Japanese Encephalitis Virus. Viruses 2017, 9, 76. [Google Scholar] [CrossRef]
- Lu, C.Y.; Lin, C.S.; Lai, H.C.; Yu, Y.W.; Liao, C.Y.; Su, W.C.; Ko, B.H.; Chang, Y.S.; Huang, S.H.; Lin, C.W. The Rescue and Characterization of Recombinant, Microcephaly-Associated Zika Viruses as Single-Round Infectious Particles. Viruses 2019, 11, 1005. [Google Scholar] [CrossRef]
- Lin, C.S.; Li, W.J.; Liao, C.Y.; Kan, J.Y.; Kung, S.H.; Huang, S.H.; Lai, H.C.; Lin, C.W. A Reverse Mutation E143K within the PrM Protein of Zika Virus Asian Lineage Natal RGN Strain Increases Infectivity and Cytopathicity. Viruses 2022, 14, 1572. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.C.; Tsai, M.H.; Hu, H.S.; Pu, S.Y.; Wu, R.H.; Wu, S.H.; Lin, H.M.; Song, J.S.; Chao, Y.S.; Yueh, A.; et al. Characterization of an efficient dengue virus replicon for development of assays of discovery of small molecules against dengue virus. Antivir. Res. 2013, 98, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, S.; Yang, P.; Wang, C.; Du, Y.; Yu, W.; Sun, Z. Replicon-based Japanese encephalitis virus vaccines elicit immune response in mice. J. Virol. Methods 2012, 179, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Zaks, T.Z.; Wang, R.F.; Irvine, K.R.; Kammula, U.S.; Marincola, F.M.; Leitner, W.W.; Restifo, N.P. Cancer therapy using a self-replicating RNA vaccine. Nat. Med. 1999, 5, 823–827. [Google Scholar] [CrossRef]
- Pushko, P.; Parker, M.; Ludwig, G.V.; Davis, N.L.; Johnston, R.E.; Smith, J.F. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: Expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 1997, 239, 389–401. [Google Scholar] [CrossRef]
- Varnavski, A.N.; Young, P.R.; Khromykh, A.A. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 2000, 74, 4394–4403. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, W.H.; Chen, H.F.; Huang, L.M.; Gao, J.Y.; Lin, C.W.; Wang, Y.C.; Yang, C.S.; Liu, Y.L.; Hou, M.H.; et al. Tafenoquine and its derivatives as inhibitors for the severe acute respiratory syndrome coronavirus 2. J. Biol. Chem. 2022, 298, 101658. [Google Scholar] [CrossRef]
- Lin, C.S.; Huang, S.H.; Yan, B.Y.; Lai, H.C.; Lin, C.W. Effective Antiviral Activity of the Tyrosine Kinase Inhibitor Sunitinib Malate against Zika Virus. Infect. Chemother. 2021, 53, 730–740. [Google Scholar] [CrossRef]
- Zhou, Y.; Ray, D.; Zhao, Y.; Dong, H.; Ren, S.; Li, Z.; Guo, Y.; Bernard, K.A.; Shi, P.Y.; Li, H.; et al. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 2007, 81, 3891–3903. [Google Scholar] [CrossRef]
- Lim, S.P.; Koh, J.H.; Seh, C.C.; Liew, C.W.; Davidson, A.D.; Chua, L.S.; Chandrasekaran, R.; Cornvik, T.C.; Shi, P.Y.; Lescar, J.; et al. A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J. Biol. Chem. 2013, 288, 31105–33114. [Google Scholar] [CrossRef]
- Potisopon, S.; Priet, S.; Collet, A.; Decroly, E.; Canard, B.; Selisko, B. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res. 2014, 42, 11642–11656. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Xie, X.; Lee, L.T.; Chandrasekaran, R.; Reynaud, A.; Yap, L.; Wang, Q.Y.; Dong, H.; Kang, C.; Yuan, Z.; et al. Dimerization of flavivirus NS4B protein. J. Virol. 2014, 88, 3379–3391. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Dalebout, T.J.; Lukashevich, I.S.; Bredenbeek, P.J.; Franco, D. Molecular and immunological characterization of a DNA-launched yellow fever virus 17D infectious clone. J. Gen. Virol. 2015, 96 Pt 4, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Mason, P.W.; Shustov, A.V.; Frolov, I. Production and characterizatio-n of vaccines based on flaviviruses defective in replication. Virology 2006, 351, 432–443. [Google Scholar] [CrossRef]
- Huang, Y.T.; Liao, J.T.; Yen, L.C.; Chang, Y.K.; Lin, Y.L.; Liao, C.L. Japanese encephalitis virus replicon-based vaccine expressing enterovirus-71 epitope confers dual protection from lethal challenges. J. Biomed. Sci. 2015, 22, 74. [Google Scholar] [CrossRef]
- Reynard, O.; Mokhonov, V.; Mokhonova, E.; Leung, J.; Page, A.; Mateo, M.; Pyankova, O.; Georges-Courbot, M.C.; Raoul, H.; Khromykh, A.A.; et al. Kunjin virus replicon-based vaccines expressing Ebola virus glycoprotein GP protect the guinea pig against lethal Ebola virus infection. J. Infect. Dis. 2011, 204 (Suppl. S3), S1060–S1065. [Google Scholar] [CrossRef]
- Giel-Moloney, M.; Vaine, M.; Zhang, L.; Parrington, M.; Gajewska, B.; Vogel, T.U.; Pougatcheva, S.O.; Duan, X.; Farrell, T.; Ustyugova, I. Application of replication-defective West Nile virus vector to non-flavivirus vaccine targets. Hum. Vaccin. Immunother. 2017, 13, 2982–2986. [Google Scholar] [CrossRef]
- Israelow, B.; Song, E.; Mao, T.; Lu, P.; Meir, A.; Liu, F.; Alfajaro, M.M.; Wei, J.; Dong, H.; Homer, R.J.; et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020, 217, e20201241. [Google Scholar] [CrossRef]
- Hassan, A.O.; Case, J.B.; Winkler, E.S.; Thackray, L.B.; Kafai, N.M.; Bailey, A.L.; McCune, B.T.; Fox, J.M.; Chen, R.E.; Alsoussi, W.B.; et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020, 182, 744–753.e4. [Google Scholar] [CrossRef]
- Rathnasinghe, R.; Strohmeier, S.; Amanat, F.; Gillespie, V.L.; Krammer, F.; García-Sastre, A.; Coughlan, L.; Schotsaert, M.; Uccellini, M.B. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020, 9, 2433–2445. [Google Scholar] [CrossRef]
- Rai, P.; Chuong, C.; LeRoith, T.; Smyth, J.W.; Panov, J.; Levi, M.; Kehn-Hall, K.; Duggal, N.K.; Lucarelli, J.W. Adenovirus transduction to express human ACE2 causes obesity-specific morbidity in mice, impeding studies on the effect of host nutritional status on SARS-CoV-2 pathogenesis. Virology 2021, 563, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.-S.; Kan, J.-Y.; Lai, H.-C.; Lin, C.-W. Development of Zika Virus Mini-Replicon Based Single-Round Infectious Particles as Gene Delivery Vehicles. Viruses 2023, 15, 1762. https://doi.org/10.3390/v15081762
Wu J-S, Kan J-Y, Lai H-C, Lin C-W. Development of Zika Virus Mini-Replicon Based Single-Round Infectious Particles as Gene Delivery Vehicles. Viruses. 2023; 15(8):1762. https://doi.org/10.3390/v15081762
Chicago/Turabian StyleWu, Joh-Sin, Ju-Ying Kan, Hsueh-Chou Lai, and Cheng-Wen Lin. 2023. "Development of Zika Virus Mini-Replicon Based Single-Round Infectious Particles as Gene Delivery Vehicles" Viruses 15, no. 8: 1762. https://doi.org/10.3390/v15081762




