Viruses in Laboratory Drosophila and Their Impact on Host Gene Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Virus Detection and Quantification of Host Expression
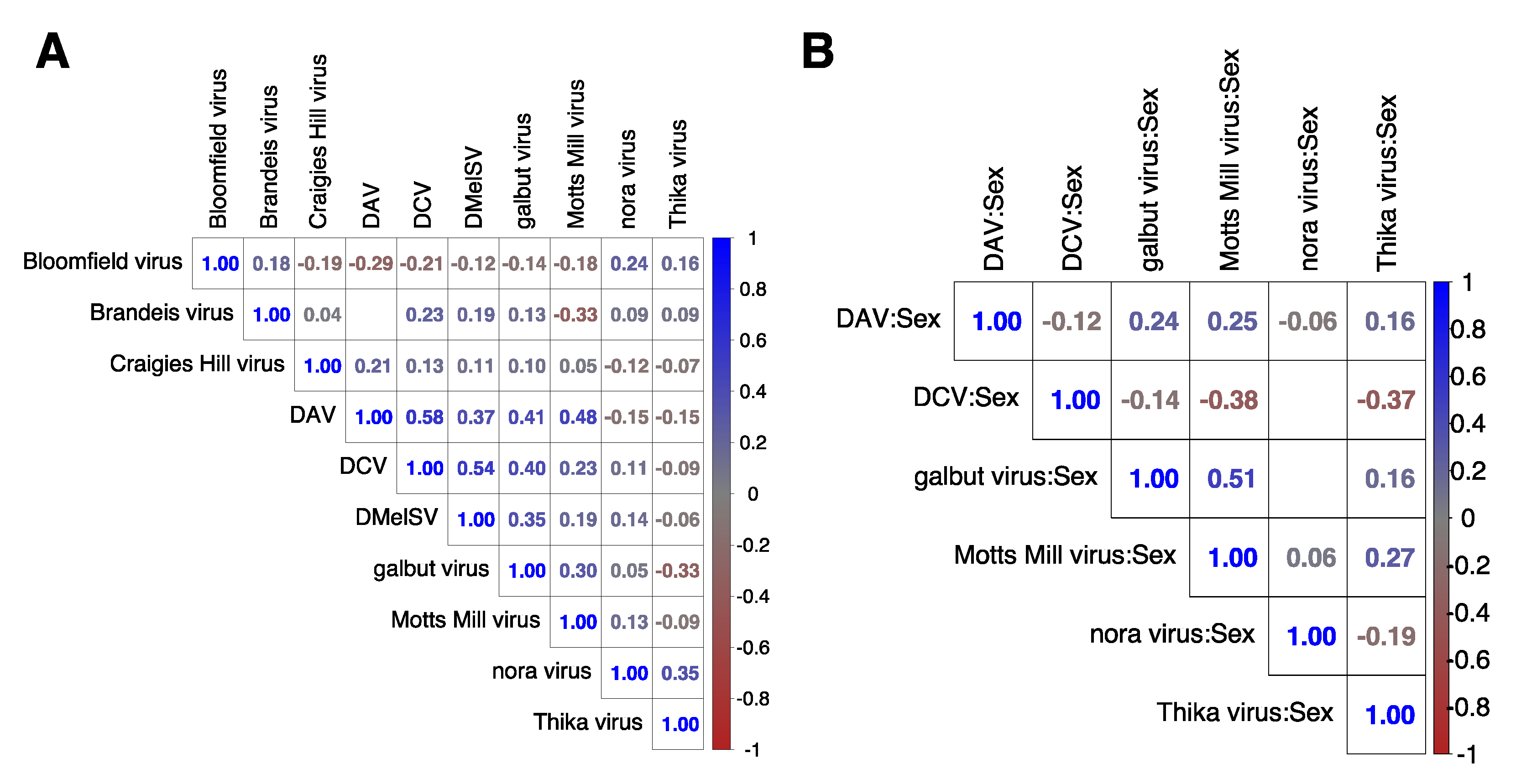
2.3. Association between Coinfecting Viruses in Each Dataset
2.4. Virus Prevalence and Diversity
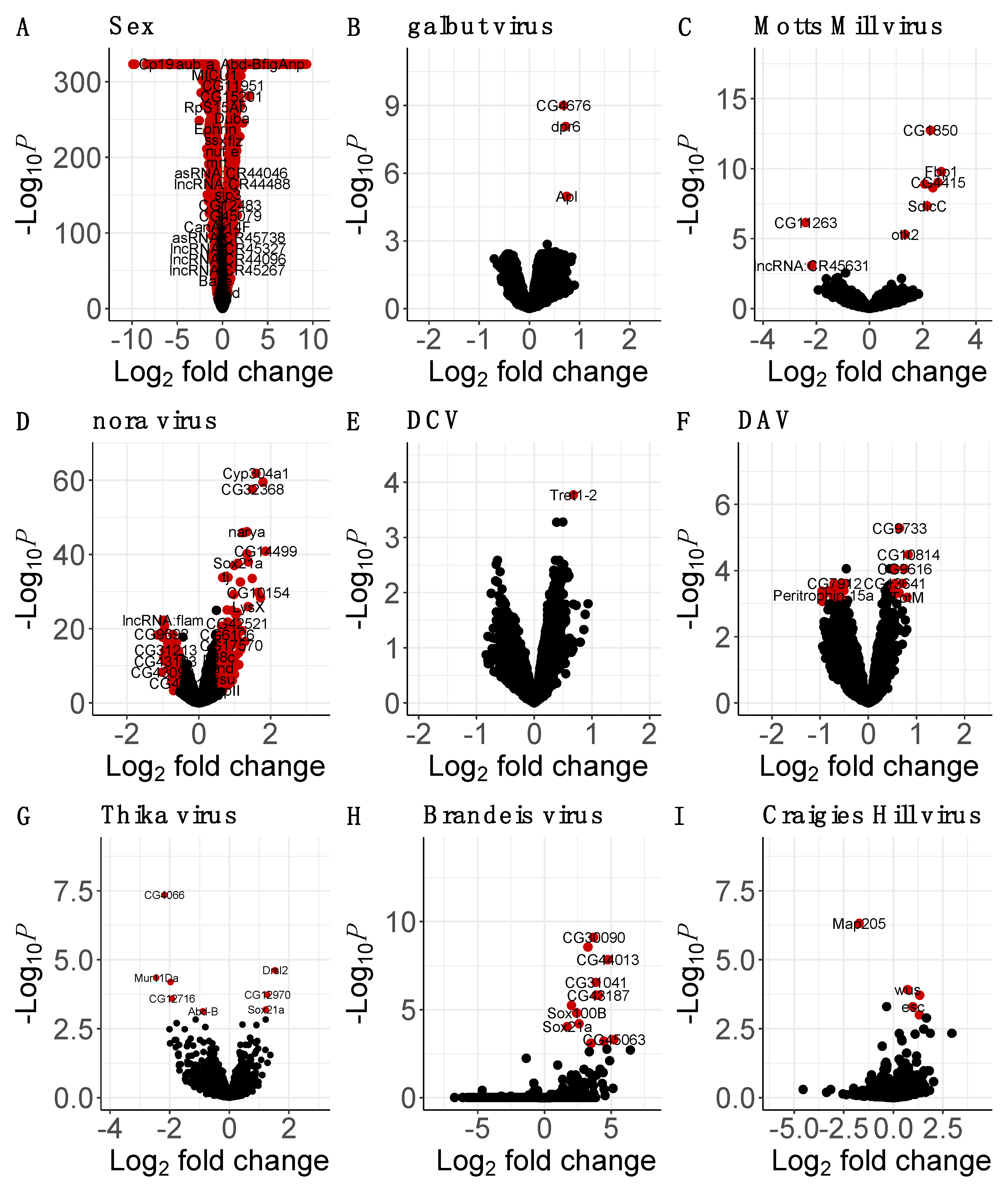
2.5. Differential Host Gene Expression
2.6. Correlation Analysis
3. Results
3.1. Ten RNA Viruses Were Detectable in Laboratory RNAseq Datasets
3.2. Laboratory Viruses Are Closely Related to Each Other
3.3. Many Laboratory Virus Infections Did Not Induce Detectable Changes in Gene Expression
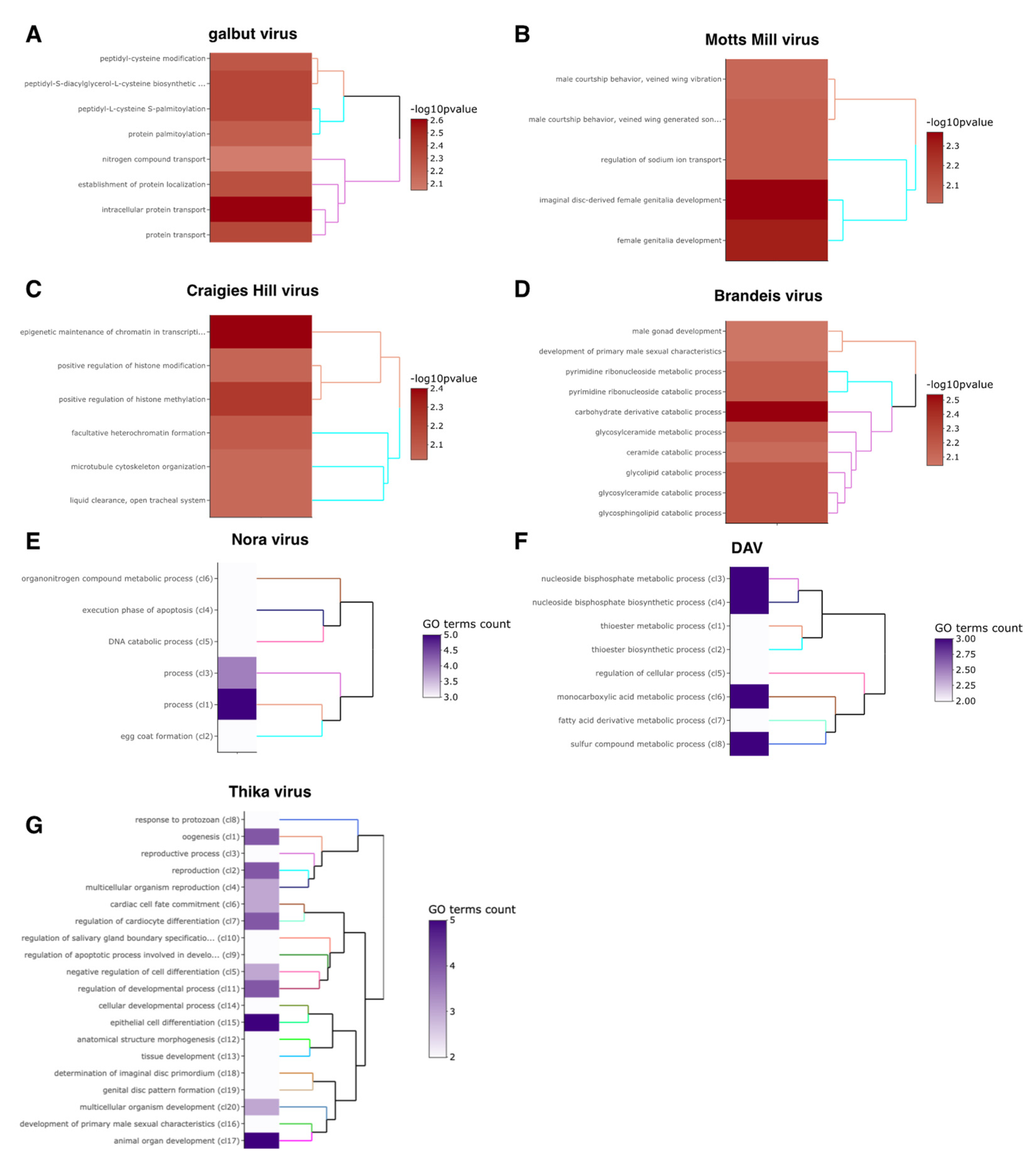
3.4. Motts Mill Virus and Craigies Hill Virus Affected the Expression of Genes Related to Development
3.5. Brandeis Virus and Thika Virus Affected the Expression of Genes Related to Reproduction and Immunity
3.6. The Enteric Viruses Nora Virus and DAV May Trigger Host Innate Immune Response and Gut Epithelium Repair
3.7. Sex–Virus Interaction
3.8. Changes in Gene Expression Were Positively Correlated between Flies Infected with Galbut Virus, Nora Virus, and Motts Mill Virus
3.9. Significantly Enriched GO Terms
4. Discussion
4.1. Prevalence
4.2. Changes in Gene Expression in Virus-Infected Flies
4.3. Potential Virus-Induced Pathologies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mussabekova, A.; Daeffler, L.; Imler, J.-L. Innate and intrinsic antiviral immunity in Drosophila. Cell Mol. Life Sci. 2017, 74, 2039–2054. [Google Scholar] [CrossRef] [PubMed]
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.-L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953. [Google Scholar] [CrossRef]
- Holleufer, A.; Winther, K.G.; Gad, H.H.; Ai, X.; Chen, Y.; Li, L.; Wei, Z.; Deng, H.; Liu, J.; Frederiksen, N.A.; et al. Two cGAS-like receptors induce antiviral immunity in Drosophila. Nature 2021, 597, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kausar, S.; Tang, Y.; Huang, W.; Tang, B.; Abbas, M.N.; Dai, L. The Emerging Role of STING in Insect Innate Immune Responses and Pathogen Evasion Strategies. Front. Immunol. 2022, 13, 874605. [Google Scholar] [CrossRef] [PubMed]
- Palmer, W.H.; Joosten, J.; Overheul, G.J.; Jansen, P.W.; Vermeulen, M.; Obbard, D.J.; Van Rij, R.P. Induction and Suppression of NF-κB Signalling by a DNA Virus of Drosophila. J. Virol. 2019, 93, e01443-18. [Google Scholar] [CrossRef]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy Is an Essential Component of Drosophila Immunity against Vesicular Stomatitis Virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-H.; Aliyari, R.; Li, W.-X.; Li, H.-W.; Kim, K.; Carthew, R.; Atkinson, P.; Ding, S.-W. RNA Interference Directs Innate Immunity Against Viruses in Adult Drosophila. Science 2006, 312, 452–454. [Google Scholar] [CrossRef]
- Zambon, R.A.; Nandakumar, M.; Vakharia, V.N.; Wu, L.P. The Toll Pathway Is Important for an Antiviral Response in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 7257–7262. [Google Scholar] [CrossRef]
- Kemp, C.; Mueller, S.; Goto, A.; Barbier, V.; Paro, S.; Bonnay, F.; Dostert, C.; Troxler, L.; Hetru, C.; Meignin, C.; et al. Broad RNA Interference–Mediated Antiviral Immunity and Virus-Specific Inducible Responses in Drosophila. J. Immunol. 2013, 190, 650–658. [Google Scholar] [CrossRef]
- Hedges, L.M.; Johnson, K.N. Induction of host defence responses by Drosophila C virus. J. Gen. Virol. 2008, 89, 1497–1501. [Google Scholar] [CrossRef]
- Palmer, W.H.; Medd, N.C.; Beard, P.M.; Obbard, D.J. Isolation of a natural DNA virus of Drosophila melanogaster, and characterisation of host resistance and immune responses. PLoS Pathog. 2018, 14, e1007050. [Google Scholar] [CrossRef] [PubMed]
- Cordes, E.J.; Licking-Murray, K.D.; Carlson, K.A. Differential gene expression related to Nora virus infection of Drosophila melanogaster. Virus Res. 2013, 175, 95–100. [Google Scholar] [CrossRef]
- Merkling, S.H.; Overheul, G.J.; van Mierlo, J.T.; Arends, D.; Gilissen, C.; van Rij, R.P. The heat shock response restricts virus infection in Drosophila. Sci. Rep. 2015, 5, 12758. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Ding, H.; Zhu, B. Transcriptional profiling of Drosophila S2 cells in early response to Drosophila C virus. Virol. J. 2013, 10, 210–219. [Google Scholar] [CrossRef]
- Gupta, V.; Vasanthakrishnan, R.B.; Siva-Jothy, J.; Monteith, K.M.; Brown, S.P.; Vale, P.F. The route of infection determines Wolbachia antibacterial protection in Drosophila. Proc. R. Soc. B Boil. Sci. 2017, 284, 20170809. [Google Scholar] [CrossRef]
- Mondotte, J.A.; Gausson, V.; Frangeul, L.; Blanc, H.; Lambrechts, L.; Saleh, M.-C. Immune priming and clearance of orally acquired RNA viruses in Drosophila. Nat. Microbiol. 2018, 3, 1394–1403. [Google Scholar] [CrossRef]
- Mondotte, J.A.; Saleh, M.-C. Antiviral Immune Response and the Route of Infection in Drosophila melanogaster. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2018; Volume 100, pp. 247–278. [Google Scholar] [CrossRef]
- Ferreira, Á.G.; Naylor, H.; Esteves, S.S.; Pais, I.S.; Martins, N.E.; Teixeira, L. The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in Drosophila. PLoS Pathog. 2014, 10, e1004507. [Google Scholar] [CrossRef]
- Wong, Z.S.; Brownlie, J.C.; Johnson, K.N. Impact of ERK activation on fly survival and Wolbachia-mediated protection during virus infection. J. Gen. Virol. 2016, 97, 1446–1452. [Google Scholar] [CrossRef] [PubMed]
- Longdon, B.; Cao, C.; Martinez, J.; Jiggins, F.M. Previous Exposure to an RNA Virus Does Not Protect against Subsequent Infection in Drosophila melanogaster. PLoS ONE 2013, 8, e73833. [Google Scholar] [CrossRef]
- Tafesh-Edwards, G.; Eleftherianos, I. Drosophila immunity against natural and nonnatural viral pathogens. Virology 2020, 540, 165–171. [Google Scholar] [CrossRef]
- van Mierlo, J.T.; Overheul, G.J.; Obadia, B.; van Cleef, K.W.R.; Webster, C.L.; Saleh, M.-C.; Obbard, D.J.; van Rij, R.P. Novel Drosophila Viruses Encode Host-Specific Suppressors of RNAi. PLoS Pathog. 2014, 10, e1004256. [Google Scholar] [CrossRef] [PubMed]
- Avadhanula, V.; Weasner, B.P.; Hardy, G.G.; Kumar, J.P.; Hardy, R.W. A Novel System for the Launch of Alphavirus RNA Synthesis Reveals a Role for the Imd Pathway in Arthropod Antiviral Response. PLoS Pathog. 2009, 5, e1000582. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.W.; van Cleef, K.W.R.; Vodovar, N.; İnce, İ.A.; Blanc, H.; Vlak, J.M.; Saleh, M.-C.; van Rij, R.P. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. USA 2012, 109, E3604–E3613. [Google Scholar] [CrossRef]
- Nayak, A.; Berry, B.; Tassetto, M.; Kunitomi, M.; Acevedo, A.; Deng, C.; Krutchinsky, A.; Gross, J.; Antoniewski, C.; Andino, R. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Medd, N.C.; Fellous, S.; Waldron, F.M.; Xuéreb, A.; Nakai, M.; Cross, J.V.; Obbard, D.J. The virome of Drosophila suzukii, an invasive pest of soft fruit. Virus Evol. 2018, 4, vey009. [Google Scholar] [CrossRef]
- Shi, M.; White, V.L.; Schlub, T.; Eden, J.-S.; Hoffmann, A.A.; Holmes, E.C. No detectable effect of Wolbachia w Mel on the prevalence and abundance of the RNA virome of Drosophila melanogaster. Proc. R. Soc. B Boil. Sci. 2018, 285, 20181165. [Google Scholar] [CrossRef] [PubMed]
- Wallace, M.A.; Coffman, K.A.; Gilbert, C.; Ravindran, S.; Albery, G.F.; Abbott, J.; Argyridou, E.; Bellosta, P.; Betancourt, A.J.; Colinet, H.; et al. The Discovery, Distribution, and Diversity of DNA Viruses Associated with Drosophila Melanogaster in Europe. Virus Evol. 2021, 7, veab031. [Google Scholar] [CrossRef]
- Webster, C.L.; Waldron, F.M.; Robertson, S.; Crowson, D.; Ferrari, G.; Quintana, J.F.; Brouqui, J.-M.; Bayne, E.H.; Longdon, B.; Buck, A.H.; et al. The Discovery, Distribution, and Evolution of Viruses Associated with Drosophila melanogaster. PLoS Biol. 2015, 13, e1002210. [Google Scholar] [CrossRef]
- Webster, C.L.; Longdon, B.; Lewis, S.H.; Obbard, D.J. Twenty-Five New Viruses Associated with the Drosophilidae (Diptera). Evol. Bioinform. 2016, 12s2, EBO.S39454. [Google Scholar] [CrossRef]
- Habayeb, M.S.; Ekengren, S.K.; Hultmark, D. Nora virus, a persistent virus in Drosophila, defines a new picorna-like virus family. J. Gen. Virol. 2006, 87, 3045–3051. [Google Scholar] [CrossRef]
- Jousset, F.X.; Plus, N.; Croizier, G.; Thomas, M. Existence in Drosophila of 2 groups of picornavirus with different biological and serological properties. Comptes Rendus Hebd. Des Seances L’Academie Des Sci. Ser. D Sci. Nat. 1972, 275, 3043–3046. [Google Scholar]
- Teninges, D.; Ohanessian, A.; Richard-Molard, C.; Contamine, D. Isolation and Biological Properties of Drosophila X Virus. J. Gen. Virol. 1979, 42, 241–254. [Google Scholar] [CrossRef]
- Bruner-Montero, G.; Luque, C.M.; Cesar, C.S.; Ding, S.D.; Day, J.P.; Jiggins, F.M. Hunting Drosophila viruses from wild populations: A novel isolation approach and characterisation of viruses. PLoS Pathog. 2023, 19, e1010883. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.T.; Maertens, B.L.; Dunham, T.J.; Rodgers, C.P.; Brehm, A.L.; Miller, M.R.; Williams, A.M.; Foy, B.D.; Stenglein, M.D. Partitiviruses Infecting Drosophila melanogaster and Aedes aegypti Exhibit Efficient Biparental Vertical Transmission. J. Virol. 2020, 94, e01070-20. [Google Scholar] [CrossRef]
- Echalier, G. Drosophila Viruses and Other Infections of Cultured Cells. In Drosophila Cells in Culture; Academic Press: Cambridge, MA, USA, 1997; pp. 555–595. [Google Scholar] [CrossRef]
- Brosh, O.; Fabian, D.K.; Cogni, R.; Tolosana, I.; Day, J.P.; Olivieri, F.; Merckx, M.; Akilli, N.; Szkuta, P.; Jiggins, F.M. A novel transposable element-mediated mechanism causes antiviral resistance in Drosophila through truncating the Veneno protein. Proc. Natl. Acad. Sci. USA 2022, 119, e2122026119. [Google Scholar] [CrossRef]
- Gomariz-Zilber, E.; Thomas-Orillard, M. Drosophila C virus and Drosophila hosts: A good association in various environments. J. Evol. Biol. 1993, 6, 677–689. [Google Scholar] [CrossRef]
- Gupta, V.; Stewart, C.O.; Rund, S.S.C.; Monteith, K.; Vale, P.F. Costs and benefits of sublethal Drosophila C virus infection. J. Evol. Biol. 2017, 30, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Brun, G.; Sigot, A. Etude de la sensibilité héréditaire au gaz carbonique chez la Drosophile. II. —Installation du virus σ dans la lignée germinale à la suite d’une inoculation. Ann. Inst. Pasteur 1955, 88, 488–513. [Google Scholar]
- Rogers, A.; Towery, L.; McCown, A.; Carlson, K.A. Impaired Geotaxis as a Novel Phenotype of Nora Virus Infection of Drosophila melanogaster. Scientifica 2020, 2020, 1804510. [Google Scholar] [CrossRef]
- European Nucleotide Archive. 2022. Available online: https://www.ebi.ac.uk/ena/browser/home (accessed on 16 August 2023).
- Lin, Y.; Golovnina, K.; Chen, Z.-X.; Lee, H.N.; Negron, Y.L.S.; Sultana, H.; Oliver, B.; Harbison, S.T. Comparison of normalization and differential expression analyses using RNA-Seq data from 726 individual Drosophila melanogaster. BMC Genom. 2016, 17, 28. [Google Scholar] [CrossRef]
- Everett, L.J.; Huang, W.; Zhou, S.; Carbone, M.A.; Lyman, R.F.; Arya, G.H.; Geisz, M.S.; Ma, J.; Morgante, F.; Armour, G.S.; et al. Gene expression networks in the Drosophila Genetic Reference Panel. Genome Res. 2020, 30, 485–496. [Google Scholar] [CrossRef]
- Litovchenko, M.; Meireles-Filho, A.C.A.; Frochaux, M.V.; Bevers, R.P.J.; Prunotto, A.; Anduaga, A.M.; Hollis, B.; Gardeux, V.; Braman, V.S.; Russeil, J.M.C.; et al. Extensive tissue-specific expression variation and novel regulators underlying circadian behavior. Sci. Adv. 2021, 7, eabc3781. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Gurdziel, K.; Pique-Regi, R.; Ruden, D.M. Identification of Splicing Quantitative Trait Loci (sQTL) in Drosophila melanogaster with Developmental Lead (Pb2+) Exposure. Front. Genet. 2017, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Ingleby, F.C.; Webster, C.L.; Pennell, T.M.; Flis, I.; Morrow, E.H. Sex-biased gene expression in Drosophila melanogaster is constrained by ontogeny and genetic architecture. Genetics, 2016; preprint. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, Z.-X.; Oliver, B.; Harbison, S.T. Microenvironmental Gene Expression Plasticity Among Individual Drosophila melanogaster. G3 Genes Genomes Genet. 2016, 6, 4197–4210. [Google Scholar] [CrossRef]
- Lee, H.; Cho, D.-Y.; Whitworth, C.; Eisman, R.; Phelps, M.; Roote, J.; Kaufman, T.; Cook, K.; Russell, S.; Przytycka, T.; et al. Effects of Gene Dose, Chromatin, and Network Topology on Expression in Drosophila melanogaster. PLoS Genet. 2016, 12, e1006295. [Google Scholar] [CrossRef]
- Lee, H.; Cho, D.-Y.; Wojtowicz, D.; Harbison, S.T.; Russell, S.; Oliver, B.; Przytycka, T.M. Dosage-Dependent Expression Variation Suppressed on the Drosophila Male X Chromosome. G3 Genes Genomes Genet. 2018, 8, 587–598. [Google Scholar] [CrossRef]
- Fear, J.M.; León-Novelo, L.G.; Morse, A.M.; Gerken, A.R.; Van Lehmann, K.; Tower, J.; Nuzhdin, S.V.; McIntyre, L.M. Buffering of Genetic Regulatory Networks in Drosophila Melanogaster. Genetics 2016, 203, 1177–1190. [Google Scholar] [CrossRef] [PubMed]
- Kurmangaliyev, Y.Z.; Ali, S.; Nuzhdin, S.V. Genetic Determinants of RNA Editing Levels of ADAR Targets in Drosophila melanogaster. G3 Genes Genomes Genet. 2016, 6, 391–396. [Google Scholar] [CrossRef]
- Djordjevic, J.; Dumas, Z.; Robinson-Rechavi, M.; Schwander, T.; Parker, D.J. Dynamics of sex-biased gene expression during development in the stick insect Timema californicum. Heredity 2022, 129, 113–122. [Google Scholar] [CrossRef]
- Qu, W.; Gurdziel, K.; Pique-Regi, R.; Ruden, D.M. Lead Modulates trans- and cis-Expression Quantitative Trait Loci (eQTLs) in Drosophila melanogaster Heads. Front. Genet. 2018, 9, 395. [Google Scholar] [CrossRef]
- Kondo, S.; Vedanayagam, J.; Mohammed, J.; Eizadshenass, S.; Kan, L.; Pang, N.; Aradhya, R.; Siepel, A.; Steinhauer, J.; Lai, E.C. New genes often acquire male-specific functions but rarely become essential in Drosophila. Genes Dev. 2017, 31, 1841–1846. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, C.; Robichon, A. Temporal and sequential order of nonoverlapping gene networks unraveled in mated female Drosophila. Life Sci. Alliance 2022, 5, e202101119. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Frochaux, M.; Gardeux, V.; Deplancke, B.; Robinson-Rechavi, M. Inter-embryo gene expression variability recapitulates the hourglass pattern of evo-devo. BMC Biol. 2020, 18, 129. [Google Scholar] [CrossRef] [PubMed]
- Wallace, M.A. Virus Discovery, Dynamics, and Disease in a Wild Drosophila Community. Ph.D. Thesis, The University of Edinburgh, Edinburgh, UK, 2021. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Griffiths, J.A.; Richard, A.C.; Bach, K.; Lun, A.T.L.; Marioni, J.C. Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun. 2018, 9, 2667. [Google Scholar] [CrossRef]
- The Flybase Consortium. FlyBase: The Drosophila database. Nucleic Acids Res. 1996, 24, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.r-project.org/ (accessed on 16 August 2023).
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Roussos, P. Dream: Powerful differential expression analysis for repeated measures designs. Bioinformatics 2021, 37, 192–201. [Google Scholar] [CrossRef]
- Brionne, A.; Juanchich, A.; Hennequet-Antier, C. ViSEAGO: A Bioconductor package for clustering biological functions using Gene Ontology and semantic similarity. BioData Min. 2019, 12, 1–13. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. TopGO: Enrichment Analysis for Gene Ontology, R Package Version 2.26.0; 2016.
- Harrel, F.E., Jr. Hmisc: Harrell Miscellaneous 2022
- Habayeb, M.S.; Cantera, R.; Casanova, G.; Ekström, J.-O.; Albright, S.; Hultmark, D. The Drosophila Nora virus is an enteric virus, transmitted via feces. J. Invertebr. Pathol. 2009, 101, 29–33. [Google Scholar] [CrossRef]
- Contamine, D. Sigma Rhabdoviruses. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2008; pp. 576–581. [Google Scholar] [CrossRef]
- Kanamori, Y.; Saito, A.; Hagiwara-Komoda, Y.; Tanaka, D.; Mitsumasu, K.; Kikuta, S.; Watanabe, M.; Cornette, R.; Kikawada, T.; Okuda, T. The trehalose transporter 1 gene sequence is conserved in insects and encodes proteins with different kinetic properties involved in trehalose import into peripheral tissues. Insect Biochem. Mol. Biol. 2010, 40, 30–37. [Google Scholar] [CrossRef]
- Carrillo, R.A.; Özkan, E.; Menon, K.P.; Nagarkar-Jaiswal, S.; Lee, P.-T.; Jeon, M.; Birnbaum, M.E.; Bellen, H.J.; Garcia, K.C.; Zinn, K. Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins. Cell 2015, 163, 1770–1782. [Google Scholar] [CrossRef]
- Bannan, B.A.; Van Etten, J.; Kohler, J.A.; Tsoi, Y.; Hansen, N.M.; Sigmon, S.; Fowler, E.; Buff, H.; Williams, T.S.; Ault, J.G.; et al. The Drosophila protein palmitoylome: Characterizing palmitoyl-thioesterases and DHHC palmitoyl-transferases. Fly 2008, 2, 198–214. [Google Scholar] [CrossRef] [PubMed]
- Boeynaems, S.; Bogaert, E.; Michiels, E.; Gijselinck, I.; Sieben, A.; Jovičić, A.; De Baets, G.; Scheveneels, W.; Steyaert, J.; Cuijt, I.; et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 2016, 6, 20877. [Google Scholar] [CrossRef] [PubMed]
- Lollies, A.; Krsmanovic, T.; Jussen, L.C.; Behr, M. Wurst-Mediated Airway Clearance is Required for Postembryonic Development. J. Allergy Ther. 2012, 1, 1–6. [Google Scholar] [CrossRef]
- Wang, L.; Jahren, N.; Vargas, M.L.; Andersen, E.F.; Benes, J.; Zhang, J.; Miller, E.L.; Jones, R.S.; Simon, J.A. Alternative ESC and ESC-Like Subunits of a Polycomb Group Histone Methyltransferase Complex Are Differentially Deployed during Drosophila Development. Mol. Cell Biol. 2006, 26, 2637–2647. [Google Scholar] [CrossRef] [PubMed]
- Findlay, G.D.; Yi, X.; MacCoss, M.J.; Swanson, W.J. Proteomics Reveals Novel Drosophila Seminal Fluid Proteins Transferred at Mating. PLoS Biol. 2008, 6, e178. [Google Scholar] [CrossRef]
- Dobbelaere, J.; Josué, F.; Suijkerbuijk, S.; Baum, B.; Tapon, N.; Raff, J. A Genome-Wide RNAi Screen to Dissect Centriole Duplication and Centrosome Maturation in Drosophila. PLoS Biol. 2008, 6, e224. [Google Scholar] [CrossRef]
- Burmester, T.; Antoniewski, C.; Lepesant, J.-A. Ecdysone-regulation of synthesis and processing of fat body protein 1, the larval serum protein receptor of Drosophila melanogaster. JBIC J. Biol. Inorg. Chem. 1999, 262, 49–55. [Google Scholar] [CrossRef]
- Wolfe, J.; Akam, M.E.; Roberts, D.B. Biochemical and Immunological Studies on Larval Serum Protein 1, the Major Haemolymph Protein of Drosophila melanogaster Third-Instar Larvae. Eur. J. Biochem. 1977, 79, 47–53. [Google Scholar] [CrossRef]
- Feng, G.; Deak, P.; Chopra, M.; Hall, L.M. Cloning and functional analysis of tipE, a novel membrane protein that enhances drosophila para sodium channel function. Cell 1995, 82, 1001–1011. [Google Scholar] [CrossRef]
- Sitnik, J.L.; Gligorov, D.; Maeda, R.K.; Karch, F.; Wolfner, M.F. The Female Post-Mating Response Requires Genes Expressed in the Secondary Cells of the Male Accessory Gland in Drosophila melanogaster. Genetics 2016, 202, 1029–1041. [Google Scholar] [CrossRef]
- Shukla, A.K.; Spurrier, J.; Kuzina, I.; Giniger, E. Hyperactive Innate Immunity Causes Degeneration of Dopamine Neurons upon Altering Activity of Cdk5. Cell Rep. 2019, 26, 131–144.e4. [Google Scholar] [CrossRef] [PubMed]
- Dweck, H.K.; Talross, G.J.; Luo, Y.; Ebrahim, S.A.; Carlson, J.R. Ir56b is an atypical ionotropic receptor that underlies appetitive salt response in Drosophila. Curr. Biol. 2022, 32, 1776–1787.e4. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Gao, B.; Rodriguez, M.d.C.; Lanz-Mendoza, H.; Ma, B.; Zhu, S. Gene expression, antiparasitic activity, and functional evolution of the drosomycin family. Mol. Immunol. 2008, 45, 3909–3916. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.W.; Biteau, B. A Sox Transcription Factor Is a Critical Regulator of Adult Stem Cell Proliferation in the Drosophila Intestine. Cell Rep. 2015, 13, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Tootle, T.L.; Williams, D.; Hubb, A.; Frederick, R.; Spradling, A. Drosophila Eggshell Production: Identification of New Genes and Coordination by Pxt. PLoS ONE 2011, 6, e19943. [Google Scholar] [CrossRef]
- Gligorov, D.; Sitnik, J.L.; Maeda, R.K.; Wolfner, M.F.; Karch, F. A Novel Function for the Hox Gene Abd-B in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in Drosophila. PLoS Genet. 2013, 9, e1003395. [Google Scholar] [CrossRef]
- Ross, J.; Jiang, H.; Kanost, M.R.; Wang, Y. Serine proteases and their homologs in the Drosophila melanogaster genome: An initial analysis of sequence conservation and phylogenetic relationships. Gene 2003, 304, 117–131. [Google Scholar] [CrossRef]
- Okumura, T.; Takeda, K.; Kuchiki, M.; Akaishi, M.; Taniguchi, K.; Adachi-Yamada, T. GATAe regulates intestinal stem cell maintenance and differentiation in Drosophila adult midgut. Dev. Biol. 2016, 410, 24–35. [Google Scholar] [CrossRef]
- Vorbrüggen, G.; Constien, R.; Zilian, O.; A Wimmer, E.; Dowe, G.; Taubert, H.; Noll, M.; Jäckle, H. Embryonic expression and characterization of a Ptx1 homolog in Drosophila. Mech. Dev. 1997, 68, 139–147. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, W.; Gao, X.; Li, W.; Qi, S.; Guo, D.; Ajayi, O.E.; Ding, S.-W.; Wu, Q. lncRNA Sensing of a Viral Suppressor of RNAi Activates Non-canonical Innate Immune Signaling in Drosophila. Cell Host Microbe 2020, 27, 115–128.e8. [Google Scholar] [CrossRef]
- Lee, J.H.; Cho, K.S.; Lee, J.; Yoo, J.; Lee, J.; Chung, J. Diptericin-like protein: An immune response gene regulated by the anti-bacterial gene induction pathway in Drosophila. Gene 2001, 271, 233–238. [Google Scholar] [CrossRef]
- Lemaitre, B.; Reichhart, J.-M.; Hoffmann, J.A. Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 1997, 94, 14614–14619. [Google Scholar] [CrossRef]
- Levashina, E.A.; Ohresser, S.; Bulet, P.; Reichhart, J.-M.; Hetru, C.; Hoffmann, J.A. Metchnikowin, a Novel Immune-Inducible Proline-Rich Peptide from Drosophila with Antibacterial and Antifungal Properties. Eur. J. Biochem. 1995, 233, 694–700. [Google Scholar] [CrossRef]
- Cornman, R.S. Molecular Evolution of Drosophila Cuticular Protein Genes. PLoS ONE 2009, 4, e8345. [Google Scholar] [CrossRef]
- Karouzou, M.V.; Spyropoulos, Y.; Iconomidou, V.A.; Cornman, R.; Hamodrakas, S.J.; Willis, J.H. Drosophila cuticular proteins with the R&R Consensus: Annotation and classification with a new tool for discriminating RR-1 and RR-2 sequences. Insect Biochem. Mol. Biol. 2007, 37, 754–760. [Google Scholar] [CrossRef]
- Zuber, R.; Wang, Y.; Gehring, N.; Bartoszewski, S.; Moussian, B. Tweedle proteins form extracellular two-dimensional structures defining body and cell shape in Drosophila melanogaster. Open Biol. 2020, 10, 200214. [Google Scholar] [CrossRef]
- Hales, K.G.; Fuller, M.T. Developmentally Regulated Mitochondrial Fusion Mediated by a Conserved, Novel, Predicted GTPase. Cell 1997, 90, 121–129. [Google Scholar] [CrossRef]
- Meng, H.; Yamashita, C.; Shiba-Fukushima, K.; Inoshita, T.; Funayama, M.; Sato, S.; Hatta, T.; Natsume, T.; Umitsu, M.; Takagi, J.; et al. Loss of Parkinson’s disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat. Commun. 2017, 8, 15500. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef]
- Zhong, W.; McClure, C.D.; Evans, C.R.; Mlynski, D.T.; Immonen, E.; Ritchie, M.G.; Priest, N.K. Immune anticipation of mating in Drosophila: Turandot M promotes immunity against sexually transmitted fungal infections. Proc. R. Soc. B Boil. Sci. 2013, 280, 20132018. [Google Scholar] [CrossRef]
- Subramanian, M.; Metya, S.K.; Sadaf, S.; Kumar, S.; Schwudke, D.; Hasan, G. Altered lipid homeostasis in Drosophila InsP3 receptor mutants leads to obesity and hyperphagia. Dis. Model. Mech. 2013, 6, 734–744. [Google Scholar] [CrossRef]
- Watkins, P.A.; Maiguel, D.; Jia, Z.; Pevsner, J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J. Lipid Res. 2007, 48, 2736–2750. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-A.; Cho, K.-C.; Kim, B.; Jang, I.-H.; Nam, K.; Kwon, Y.E.; Kim, M.; Hyeon, D.Y.; Hwang, D.; Seol, J.-H.; et al. Inflammation-Modulated Metabolic Reprogramming Is Required for DUOX-Dependent Gut Immunity in Drosophila. Cell Host Microbe 2018, 23, 338–352.e5. [Google Scholar] [CrossRef]
- Wijffels, G.; Eisemann, C.; Riding, G.; Pearson, R.; Jones, A.; Willadsen, P.; Tellam, R. A Novel Family of Chitin-binding Proteins from Insect Type 2 Peritrophic Matrix. J. Biol. Chem. 2001, 276, 15527–15536. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, Z. Notch signaling downstream target E(spl)mbeta is dispensable for adult midgut homeostasis in Drosophila. Gene 2015, 560, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Voßfeldt, H.; Butzlaff, M.; Prüßing, K.; Chárthaigh, R.-A.N.; Karsten, P.; Lankes, A.; Hamm, S.; Simons, M.; Adryan, B.; Schulz, J.B.; et al. Large-Scale Screen for Modifiers of Ataxin-3-Derived Polyglutamine-Induced Toxicity in Drosophila. PLoS ONE 2012, 7, e47452. [Google Scholar] [CrossRef]
- Neely, G.G.; Hess, A.; Costigan, M.; Keene, A.C.; Goulas, S.; Langeslag, M.; Griffin, R.S.; Belfer, I.; Dai, F.; Smith, S.B.; et al. A Genome-wide Drosophila Screen for Heat Nociception Identifies α2δ3 as an Evolutionarily Conserved Pain Gene. Cell 2010, 143, 628–638. [Google Scholar] [CrossRef]
- Hedengren, M.; Borge, K.; Hultmark, D. Expression and Evolution of the Drosophila Attacin/Diptericin Gene Family. Biochem. Biophys. Res. Commun. 2000, 279, 574–581. [Google Scholar] [CrossRef]
- Vig, M.; Peinelt, C.; Beck, A.; Koomoa, D.L.; Rabah, D.; Koblan-Huberson, M.; Kraft, S.; Turner, H.; Fleig, A.; Penner, R.; et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 2006, 312, 1220–1223. [Google Scholar] [CrossRef]
- Kongton, K.; McCall, K.; Phongdara, A. Identification of gamma-interferon-inducible lysosomal thiol reductase (GILT) homologues in the fruit fly Drosophila melanogaster. Dev. Comp. Immunol. 2014, 44, 389–396. [Google Scholar] [CrossRef]
- Verleyen, P.; Baggerman, G.; D’hertog, W.; Vierstraete, E.; Husson, S.J.; Schoofs, L. Identification of new immune induced molecules in the haemolymph of Drosophila melanogaster by 2D-nanoLC MS/MS. J. Insect Physiol. 2006, 52, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Ertürk-Hasdemir, D.; Broemer, M.; Leulier, F.; Lane, W.S.; Paquette, N.; Hwang, D.; Kim, C.-H.; Stöven, S.; Meier, P.; Silverman, N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc. Natl. Acad. Sci. USA 2009, 106, 9779–9784. [Google Scholar] [CrossRef] [PubMed]
- Linnemannstöns, K.; Ripp, C.; Honemann-Capito, M.; Brechtel-Curth, K.; Hedderich, M.; Wodarz, A. The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Drosophila Wnt2 Required for Male Fertility. PLoS Genet. 2014, 10, e1004443. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Qiang, K.M.; Beckingham, K.M. Failure to Burrow and Tunnel Reveals Roles for jim lovell in the Growth and Endoreplication of the Drosophila Larval Tracheae. PLoS ONE 2016, 11, e0160233. [Google Scholar] [CrossRef]
- Lerch, S.; Zuber, R.; Gehring, N.; Wang, Y.; Eckel, B.; Klass, K.-D.; Lehmann, F.-O.; Moussian, B. Resilin matrix distribution, variability and function in Drosophila. BMC Biol. 2020, 18, 195. [Google Scholar] [CrossRef]
- Syed, Z.A.; Härd, T.; Uv, A.; van Dijk-Härd, I.F. A Potential Role for Drosophila Mucins in Development and Physiology. PLoS ONE 2008, 3, e3041. [Google Scholar] [CrossRef]
- Lung, O.; Wolfner, M. Identification and characterization of the major Drosophila melanogaster mating plug protein. Insect Biochem. Mol. Biol. 2001, 31, 543–551. [Google Scholar] [CrossRef]
- Campos, I.; A Geiger, J.; Santos, A.C.; Carlos, V.; Jacinto, A. Genetic Screen in Drosophila melanogaster Uncovers a Novel Set of Genes Required for Embryonic Epithelial Repair. Genetics 2010, 184, 129–140. [Google Scholar] [CrossRef]
- Schnakenberg, S.L.; Matias, W.R.; Siegal, M.L. Sperm-Storage Defects and Live Birth in Drosophila Females Lacking Spermathecal Secretory Cells. PLoS Biol. 2011, 9, e1001192. [Google Scholar] [CrossRef]
- Lake, C.M.; Nielsen, R.J.; Bonner, A.M.; Eche, S.; White-Brown, S.; McKim, K.S.; Hawley, R.S. Narya, a RING finger domain-containing protein, is required for meiotic DNA double-strand break formation and crossover maturation in Drosophila melanogaster. PLoS Genet. 2019, 15, e1007886. [Google Scholar] [CrossRef]
- Paine-Saunders, S.; Fristrom, D.; Fristrom, J.W. The Drosophila IMP-E2 gene encodes an apically secreted protein expressed during imaginal disc morphogenesis. Dev. Biol. 1990, 140, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Akhmetova, K.; Balasov, M.; Chesnokov, I. Drosophila STING protein has a role in lipid metabolism. eLife 2021, 10, e67358. [Google Scholar] [CrossRef] [PubMed]
- Goto, A.; Okado, K.; Martins, N.; Cai, H.; Barbier, V.; Lamiable, O.; Troxler, L.; Santiago, E.; Kuhn, L.; Paik, D.; et al. The Kinase IKKβ Regulates a STING- and NF-κB-Dependent Antiviral Response Pathway in Drosophila. Immunity 2018, 49, 225–234.e4. [Google Scholar] [CrossRef]
- Sadanandan, S.A.; Ekström, J.-O.; Hultmark, D. Drosophila Melanogaster Transcriptional Response to Nora Virus Infection. 2016. Available online: https://umu.diva-portal.org/smash/get/diva2:1045375/FULLTEXT01.pdf (accessed on 16 August 2023).
- Carpenter, J.; Hutter, S.; Baines, J.F.; Roller, J.; Saminadin-Peter, S.S.; Parsch, J.; Jiggins, F.M. The Transcriptional Response of Drosophila melanogaster to Infection with the Sigma Virus (Rhabdoviridae). PLoS ONE 2009, 4, e6838. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.-F.; Wu, C.-P.; Tang, C.-K.; Tsai, C.-W.; Rouhová, L. Identification of Regulatory Host Genes Involved in Sigma Virus Replication Using RNAi Knockdown in Drosophila. Insects 2019, 10, 339. [Google Scholar] [CrossRef]
- Chtarbanova, S.; Lamiable, O.; Lee, K.-Z.; Galiana, D.; Troxler, L.; Meignin, C.; Hetru, C.; Hoffmann, J.A.; Daeffler, L.; Imler, J.-L. Drosophila C Virus Systemic Infection Leads to Intestinal Obstruction. J. Virol. 2014, 88, 14057–14069. [Google Scholar] [CrossRef]
- Cohen, L.; Moran, Y.; Sharon, A.; Segal, D.; Gordon, D.; Gurevitz, M. Drosomycin, an Innate Immunity Peptide of Drosophila melanogaster, Interacts with the Fly Voltage-gated Sodium Channel. J. Biol. Chem. 2009, 284, 23558–23563. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Dudzic, J.P.; Li, X.; Collas, E.J.; Boquete, J.-P.; Lemaitre, B. Remote Control of Intestinal Stem Cell Activity by Haemocytes in Drosophila. PLoS Genet. 2016, 12, e1006089. [Google Scholar] [CrossRef]
- A Callus, B.; Mathey-Prevot, B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene 2002, 21, 4812–4821. [Google Scholar] [CrossRef]
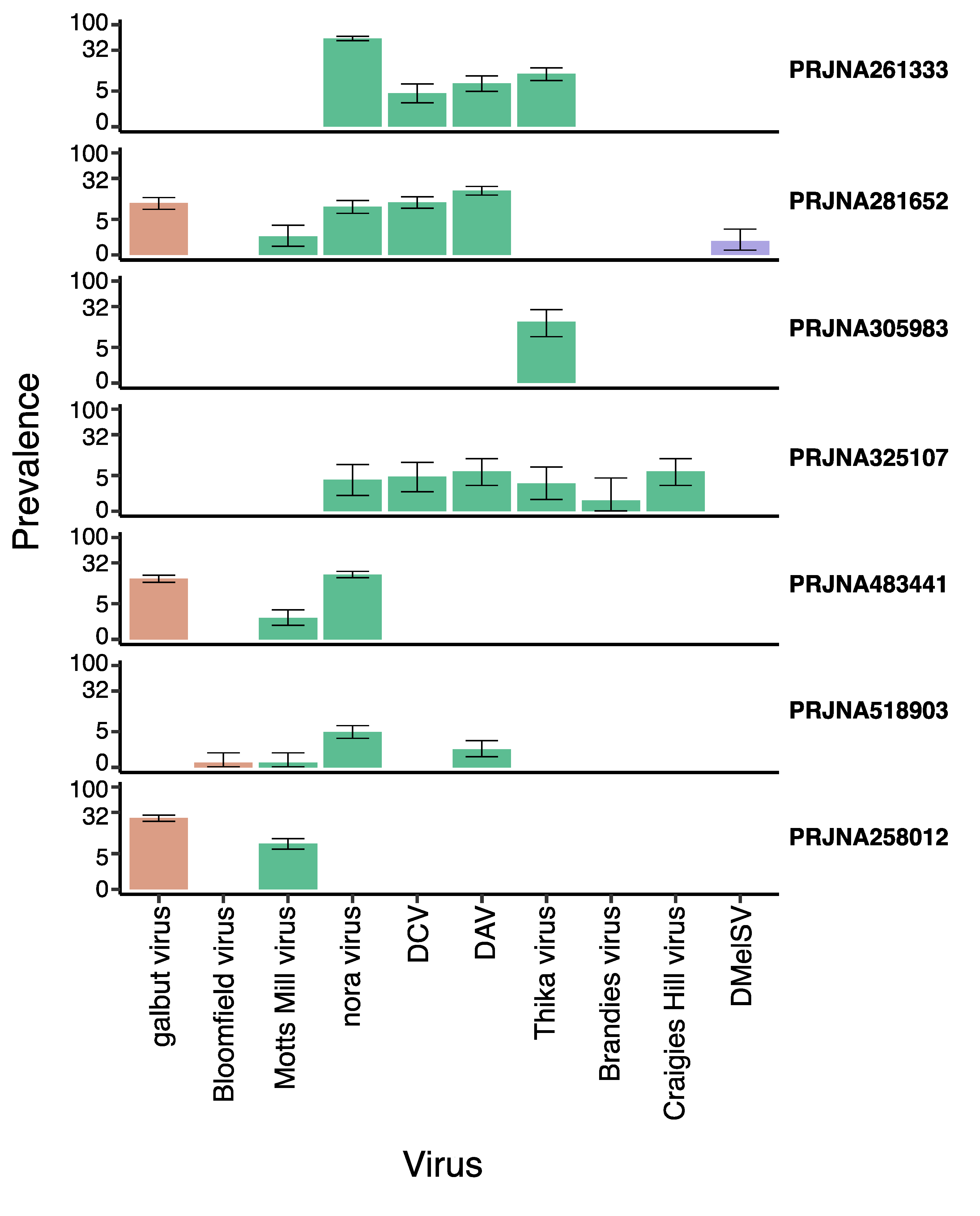


| Project Code | Number of Libraries | Data | Genotype | Sex | Number of Reads | Viruses Found | Reference |
|---|---|---|---|---|---|---|---|
| PRJNA258012 | 768 | Single virgin adult flies | 16 DGRP | Males and females | 7.74 billion | Galbut virus, Motts Mill virus | [43,48] |
| PRJNA261333 | 396 | Single, 3–5-day post-eclosion adult | 99 DrosDel lines | Male and females | 2.94 billion | DAV, DCV nora virus, Thika virus | [49,50] |
| PRJNA281652 | 919 | Single virgin and mated adult fly heads | F1 of Female DGRP and Male w118 crosses | Females | 2.27 billion | DAV, DCV, galbut virus, Motts Mill virus, nora virus, DmelSV | [51,52] |
| PRJNA305983 | 179 | Single whole larvae, pupae & virgin adults | 10 different Drosophila genotypes | Males and females | 6.26 billion | Thika virus | [47,53] |
| PRJNA325107 | 158 | Control and lead-exposed single adults | Adult w1118 | Males | 5.18 billion | Brandeis virus, Craigie’s Hill virus, DAV, DCV, Motts Mill virus, nora virus, | [46,54] |
| PRJNA483441 | 800 | Pooled and mated adult lies | 200 DGRP | Males and females | 15.53 billion | Galbut virus, Motts Mill viurus, nora virus | [44] |
| PRJNA518903 | 778 | Brain, abdominal fat body, whole gut without the crop, and malpighian tubes of mated flies | w1118 and DGRP | Males | 1.54 billion | Bloomfield, DAV, Motts Mill, nora | [45] |
| PRJNA75285 | 699 | Mixture of cell culture and adult flies | Not mentioned | Cell culture and Mixed-sex pools | 22.94 billion | None | [55,56] |
| PRJNA527284 | 288 | Single embryos | Fly lines (w1118) | N/A | 894 million | None | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuyateh, O.; Obbard, D.J. Viruses in Laboratory Drosophila and Their Impact on Host Gene Expression. Viruses 2023, 15, 1849. https://doi.org/10.3390/v15091849
Kuyateh O, Obbard DJ. Viruses in Laboratory Drosophila and Their Impact on Host Gene Expression. Viruses. 2023; 15(9):1849. https://doi.org/10.3390/v15091849
Chicago/Turabian StyleKuyateh, Oumie, and Darren J. Obbard. 2023. "Viruses in Laboratory Drosophila and Their Impact on Host Gene Expression" Viruses 15, no. 9: 1849. https://doi.org/10.3390/v15091849
APA StyleKuyateh, O., & Obbard, D. J. (2023). Viruses in Laboratory Drosophila and Their Impact on Host Gene Expression. Viruses, 15(9), 1849. https://doi.org/10.3390/v15091849






