Viral Co-Infection in Bats: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Literature Search Strategy
2.2. Screening and Eligibility Criteria
2.3. Data Extraction
2.4. Study-Level Comparison
2.5. Individual-Level Comparison of Pairwise Co-Detection across Viral and Host Families
2.6. Network Analysis of Co-Infection Themes of Discussion
3. Results
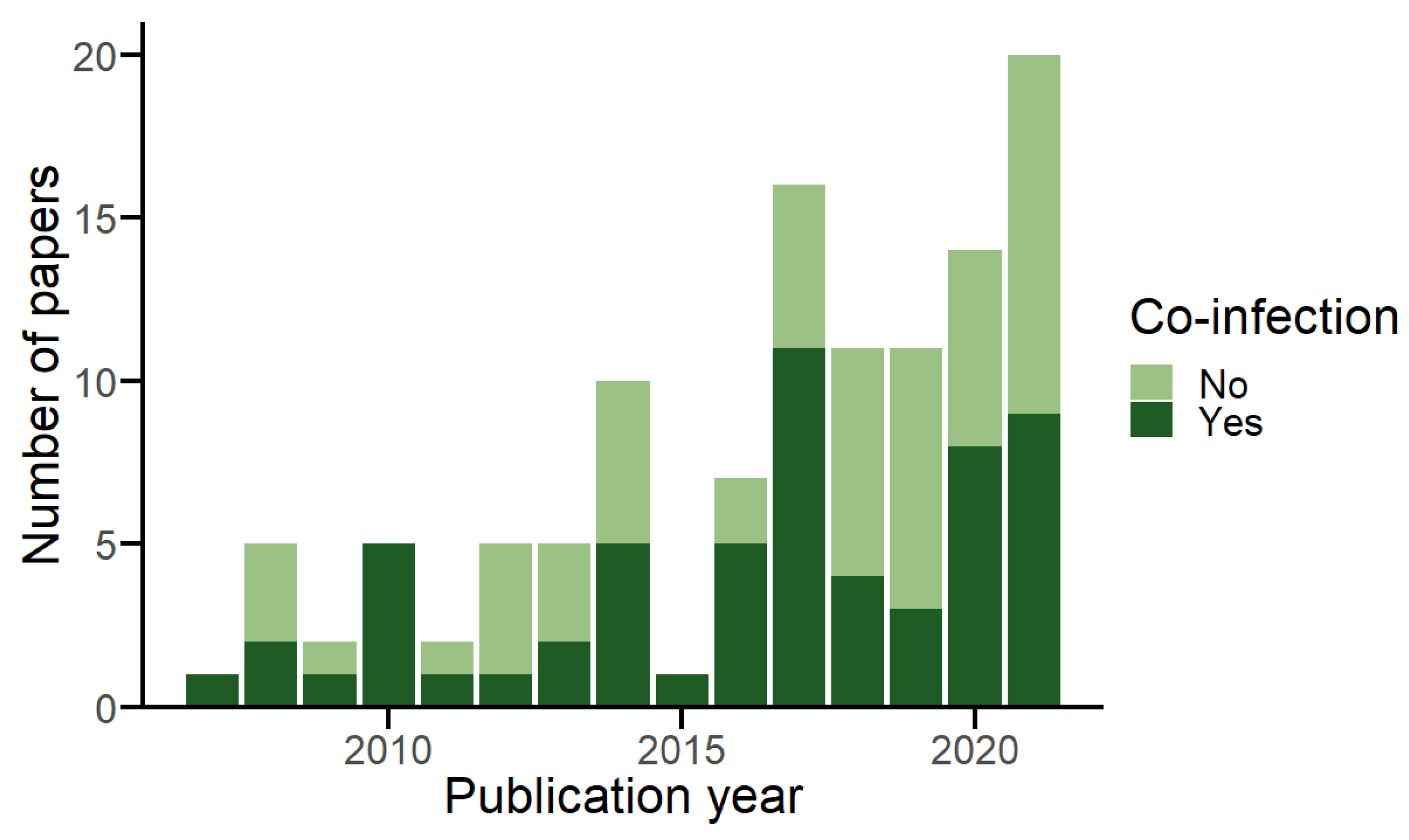
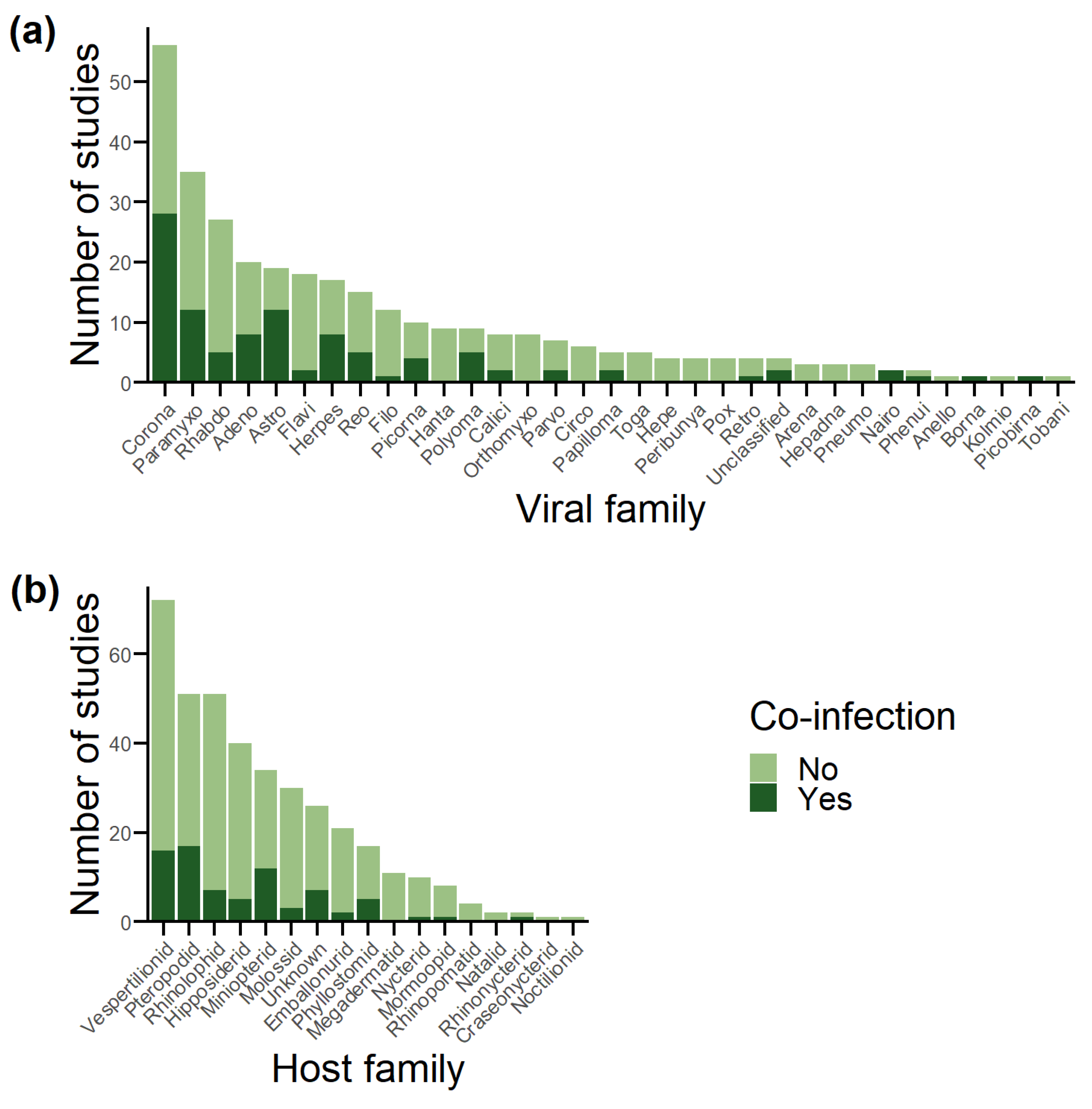
3.1. Database Overview
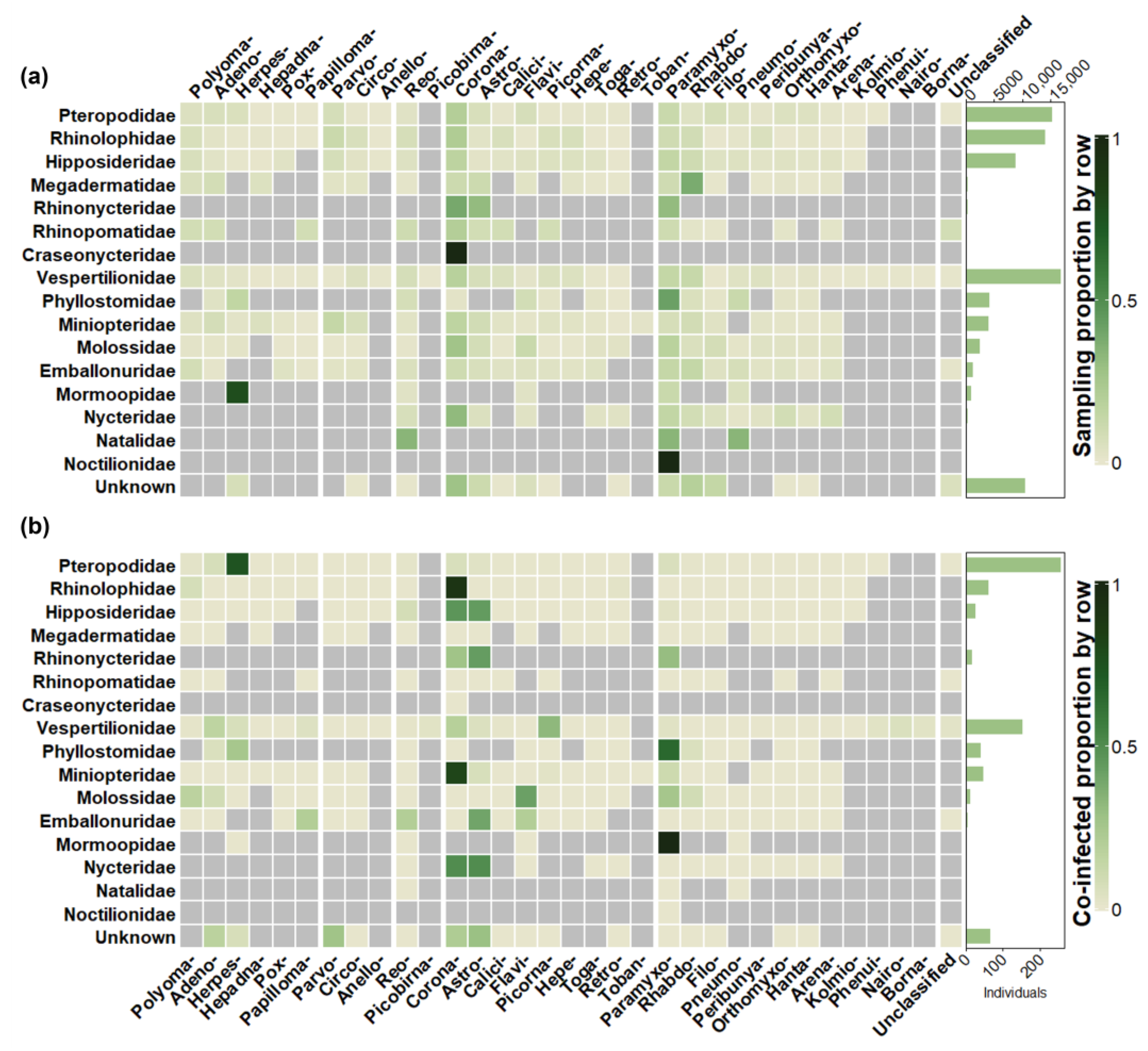
3.2. Sampling Effort vs. Detection
3.3. Co-Infected Proportion
3.4. Pairwise Detections
3.5. Measures of Association
3.6. Terminology and Themes
3.7. Co-Infection of Viral Families with High Emergence Potential
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hoarau, A.O.G.; Mavingui, P.; Lebarbenchon, C. Coinfections in wildlife: Focus on a neglected aspect of infectious disease epidemiology. PLoS Pathog. 2020, 16, e1008790. [Google Scholar] [CrossRef] [PubMed]
- Viney, M.E.; Graham, A.L. Chapter Five-Patterns and Processes in Parasite Co-Infection. In Advances in Parasitology; Rollinson, D., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 82, pp. 321–369. [Google Scholar]
- Pantin-Jackwood, M.J.; Costa-Hurtado, M.; Miller, P.J.; Afonso, C.L.; Spackman, E.; Kapczynski, D.R.; Shepherd, E.; Smith, D.; Swayne, D.E. Experimental co-infections of domestic ducks with a virulent Newcastle disease virus and low or highly pathogenic avian influenza viruses. Veter. Microbiol. 2015, 177, 7–17. [Google Scholar] [CrossRef]
- Davy, C.M.; Donaldson, M.E.; Subudhi, S.; Rapin, N.; Warnecke, L.; Turner, J.M.; Bollinger, T.K.; Kyle, C.J.; Dorville, N.A.S.-Y.; Kunkel, E.L.; et al. White-nose syndrome is associated with increased replication of a naturally persisting coronaviruses in bats. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Polakovicova, N.; Adji, A.V.; Myhill, L.J.; Williams, A.R. Whipworm Infection in Mice Increases Coinfection of Enteric Pathogens but Promotes Clearance of Ascaris Larvae From the Lungs. J. Infect. Dis. 2023, 227, 1428–1432. [Google Scholar] [CrossRef] [PubMed]
- Telfer, S.; Lambin, X.; Birtles, R.; Beldomenico, P.; Burthe, S.; Paterson, S.; Begon, M. Species Interactions in a Parasite Community Drive Infection Risk in a Wildlife Population. Science 2010, 330, 243–246. [Google Scholar] [CrossRef] [PubMed]
- George, P.J.; Anuradha, R.; Kumaran, P.P.; Chandrasekaran, V.; Nutman, T.B.; Babu, S. Modulation of Mycobacterial-Specific Th1 and Th17 Cells in Latent Tuberculosis by Coincident Hookworm Infection. J. Immunol. 2013, 190, 5161–5168. [Google Scholar] [CrossRef]
- Vaumourin, E.; Vourc’h, G.; Gasqui, P.; Vayssier-Taussat, M. The importance of multiparasitism: Examining the consequences of co-infections for human and animal health. Parasites Vectors 2015, 8, 1–13. [Google Scholar] [CrossRef]
- Rigaud, T.; Perrot-Minnot, M.-J.; Brown, M.J.F. Parasite and host assemblages: Embracing the reality will improve our knowledge of parasite transmission and virulence. Proc. R. Soc. B Boil. Sci. 2010, 277, 3693–3702. [Google Scholar] [CrossRef]
- Bordes, F.; Morand, S. The impact of multiple infections on wild animal hosts: A review. Infect. Ecol. Epidemiol. 2011, 1. [Google Scholar] [CrossRef]
- Serrano, E.; Millán, J. What is the price of neglecting parasite groups when assessing the cost of co-infection? Epidemiol. Infect. 2014, 142, 1533–1540. [Google Scholar] [CrossRef]
- Letko, M.; Seifert, S.N.; Olival, K.J.; Plowright, R.K.; Munster, V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 2020, 18, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.-F. Lessons from the host defences of bats, a unique viral reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef]
- Rasche, A.; de Carvalho Dominguez Souza, B.F.; Drexler, J.F. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr. Opin. Virol. 2016, 16, 86–94. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Müller, M.A.; Maganga, G.D.; Vallo, P.; Binger, T.; Gloza-Rausch, F.; Cottontail, V.M.; Rasche, A.; Yordanov, S.; et al. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012, 3, 796. [Google Scholar] [CrossRef]
- Gentles, A.D.; Guth, S.; Rozins, C.; Brook, C.E. A review of mechanistic models of viral dynamics in bat reservoirs for zoonotic disease. Ann. Trop. Med. Parasitol. 2020, 114, 407–425. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Yang, X.; Anderson, D.E.; Wang, L.-F. Bat virome research: The past, the present and the future. Curr. Opin. Virol. 2021, 49, 68–80. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- Mollentze, N.; Streicker, D.G. Predicting zoonotic potential of viruses: Where are we? Curr. Opin. Virol. 2023, 61, 101346. [Google Scholar] [CrossRef]
- Wang, J.; Pan, Y.-F.; Yang, L.-F.; Yang, W.-H.; Lv, K.; Luo, C.-M.; Wang, J.; Kuang, G.-P.; Wu, W.-C.; Gou, Q.-Y.; et al. Individual bat virome analysis reveals co-infection and spillover among bats and virus zoonotic potential. Nat. Commun. 2023, 14, 4079. [Google Scholar] [CrossRef] [PubMed]
- Willoughby, A.R.; Phelps, K.L.; PREDICT Consortium; Olival, K. A Comparative Analysis of Viral Richness and Viral Sharing in Cave-Roosting Bats. Diversity 2017, 9, 35. [Google Scholar] [CrossRef]
- The EndNote Team. EndNote; EndNote 20; Clarivate: Philadelphia, PA, USA, 2013. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Posit Team. RStudio: Integrated Development Environment for R; Posit Software, PBC: Boston, MA, USA, 2023. [Google Scholar]
- Pedersen, T.L. ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. 2022. Available online: https://rdrr.io/cran/ggraph/ (accessed on 6 August 2023).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Pedersen, T.L. tidygraph: A Tidy API for Graph Manipulation. 2023. Available online: https://cran.r-project.org/web/packages/tidygraph/tidygraph.pdf (accessed on 6 August 2023).
- Mills, B.R. MetBrewer: Color Palettes Inspired by Works at the Metropolitan Museum of Art. 2022. Available online: https://cran.rstudio.com/web/packages/MetBrewer/MetBrewer.pdf (accessed on 6 August 2023).
- Lau, S.K.; Woo, P.C.; Li, K.S.; Huang, Y.; Wang, M.; Lam, C.S.; Xu, H.; Guo, R.; Chan, K.-H.; Zheng, B.-J.; et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 2007, 367, 428–439. [Google Scholar] [CrossRef]
- Chu, D.K.W.; Peiris, J.S.M.; Chen, H.; Guan, Y.; Poon, L.L.M. Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J. Gen. Virol. 2008, 89, 1282–1287. [Google Scholar] [CrossRef]
- Chu, D.K.W.; Poon, L.L.M.; Guan, Y.; Peiris, J.S.M. Novel Astroviruses in Insectivorous Bats. J. Virol. 2008, 82, 9107–9114. [Google Scholar] [CrossRef]
- Tong, S.; Conrardy, C.; Ruone, S.; Kuzmin, I.V.; Guo, X.; Tao, Y.; Niezgoda, M.; Haynes, L.; Agwanda, B.; Breiman, R.F.; et al. Detection of Novel SARS-like and Other Coronaviruses in Bats from Kenya. Emerg. Infect. Dis. 2009, 15, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Gloza-Rausch, F.; Glende, J.; Corman, V.M.; Muth, D.; Goettsche, M.; Seebens, A.; Niedrig, M.; Pfefferle, S.; Yordanov, S.; et al. Genomic Characterization of Severe Acute Respiratory Syndrome-Related Coronavirus in European Bats and Classification of Coronaviruses Based on Partial RNA-Dependent RNA Polymerase Gene Sequences. J. Virol. 2010, 84, 11336–11349. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Li, K.S.M.; Huang, Y.; Shek, C.T.; Tse, H.; Wang, M.; Choi, G.K.Y.; Xu, H.F.; Lam, C.S.F.; Guo, R.T.; et al. Ecoepidemiology and Complete Genome Comparison of Different Strains of Severe Acute Respiratory Syndrome-Related Rhinolophus Bat Coronavirus in China Reveal Bats as a Reservoir for Acute, Self-Limiting Infection That Allows Recombination Events. J. Virol. 2010, 84, 2808–2819. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Poon, R.W.S.; Wong, B.H.L.; Wang, M.; Huang, Y.; Xu, H.; Guo, R.; Li, K.S.M.; Gao, K.; Chan, K.-H.; et al. Coexistence of Different Genotypes in the Same Bat and Serological Characterization of Rousettus Bat Coronavirus HKU9 Belonging to a Novel Betacoronavirus Subgroup. J. Virol. 2010, 84, 11385–11394. [Google Scholar] [CrossRef]
- Li, Y.; Ge, X.; Hon, C.-C.; Zhang, H.; Zhou, P.; Zhang, Y.; Wu, Y.; Wang, L.-F.; Shi, Z. Prevalence and genetic diversity of adeno-associated viruses in bats from China. J. Gen. Virol. 2010, 91, 2601–2609. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, C.M.; Cummins, D.M.; Yu, M.; Lunt, R.; Pritchard, L.I.; Hansson, E.; Crameri, S.; Hyatt, A.; Wang, L.-F. Broome virus, a new fusogenic Orthoreovirus species isolated from an Australian fruit bat. Virology 2010, 402, 26–40. [Google Scholar] [CrossRef]
- Mühldorfer, K.; Speck, S.; Kurth, A.; Lesnik, R.; Freuling, C.; Müller, T.; Kramer-Schadt, S.; Wibbelt, G. Diseases and Causes of Death in European Bats: Dynamics in Disease Susceptibility and Infection Rates. PLoS ONE 2011, 6, e29773. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.A.; Temmam, S.; Lebarbenchon, C.; Lagadec, E.; Chotte, J.; Guillebaud, J.; Ramasindrazana, B.; Héraud, J.-M.; de Lamballerie, X.; Goodman, S.M.; et al. Identification of novel paramyxoviruses in insectivorous bats of the Southwest Indian Ocean. Virus Res. 2012, 170, 159–163. [Google Scholar] [CrossRef]
- Anthony, S.J.; Epstein, J.H.; Murray, K.A.; Navarrete-Macias, I.; Zambrana-Torrelio, C.M.; Solovyov, A.; Ojeda-Flores, R.; Arrigo, N.C.; Islam, A.; Khan, S.A.; et al. A Strategy to Estimate Unknown Viral Diversity in Mammals. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.-L.; Firth, C.; Conte, J.M.; Williams, S.H.; Zambrana-Torrelio, C.M.; Anthony, S.J.; Ellison, J.A.; Gilbert, A.T.; Kuzmin, I.V.; Niezgoda, M.; et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. USA 2013, 110, 8194. [Google Scholar] [CrossRef]
- Conrardy, C.; Tao, Y.; Kuzmin, I.V.; Niezgoda, M.; Agwanda, B.; Breiman, R.F.; Anderson, L.J.; Rupprecht, C.E.; Tong, S. Molecular Detection of Adenoviruses, Rhabdoviruses, and Paramyxoviruses in Bats from Kenya. Am. J. Trop. Med. Hyg. 2014, 91, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Dacheux, L.; Cervantes-Gonzalez, M.; Guigon, G.; Thiberge, J.-M.; Vandenbogaert, M.; Maufrais, C.; Caro, V.; Bourhy, H. A Preliminary Study of Viral Metagenomics of French Bat Species in Contact with Humans: Identification of New Mammalian Viruses. PLoS ONE 2014, 9, e87194. [Google Scholar] [CrossRef] [PubMed]
- García-Pérez, R.; Ibáñez, C.; Godínez, J.M.; Aréchiga, N.; Garin, I.; Pérez-Suárez, G.; de Paz, O.; Juste, J.; Echevarría, J.E.; Bravo, I.G. Novel papillomaviruses in free-ranging Iberian bats: No virus-host co-evolution, no strict host specificity, and hints for recombination. Genome Biol. Evol. 2014, 6, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Chmura, A.A.; Li, J.; Zhu, G.; Desmond, J.S.; Zhang, Y.; Zhang, W.; Epstein, J.H.; Daszak, P.; Shi, Z. Detection of diverse novel astroviruses from small mammals in China. J. Gen. Virol. 2014, 95, 2442–2449. [Google Scholar] [CrossRef]
- Kemenesi, G.; Dallos, B.; Görföl, T.; Boldogh, S.; Estók, P.; Kurucz, K.; Kutas, A.; Földes, F.; Oldal, M.; Németh, V.; et al. Molecular Survey of RNA Viruses in Hungarian Bats: Discovering Novel Astroviruses, Coronaviruses, and Caliciviruses. Vector-Borne Zoonotic Dis. 2014, 14, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Wacharapluesadee, S.; Duengkae, P.; Rodpan, A.; Kaewpom, T.; Maneeorn, P.; Kanchanasaka, B.; Yingsakmongkon, S.; Sittidetboripat, N.; Chareesaen, C.; Khlangsap, N.; et al. Diversity of coronavirus in bats from Eastern Thailand. Virol. J. 2015, 12, 1–7. [Google Scholar] [CrossRef]
- Du, J.; Yang, L.; Ren, X.; Zhang, J.; Dong, J.; Sun, L.; Zhu, Y.; Yang, F.; Zhang, S.; Wu, Z.; et al. Genetic diversity of coronaviruses in Miniopterus fuliginosus bats. Sci. China Life Sci. 2016, 59, 604–614. [Google Scholar] [CrossRef]
- Ge, X.-Y.; Wang, N.; Zhang, W.; Hu, B.; Li, B.; Zhang, Y.-Z.; Zhou, J.-H.; Luo, C.-M.; Yang, X.-L.; Wu, L.-J.; et al. Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virol. Sin. 2016, 31, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Pozo, F.; Juste, J.; Vázquez-Morón, S.; Aznar-López, C.; Ibáñez, C.; Garin, I.; Aihartza, J.; Casas, I.; Tenorio, A.; Echevarría, J.E. Identification of Novel Betaherpesviruses in Iberian Bats Reveals Parallel Evolution. PLoS ONE 2016, 11, e0169153. [Google Scholar] [CrossRef]
- Wray, A.K.; Olival, K.J.; Morán, D.; Lopez, M.R.; Alvarez, D.; Navarrete-Macias, I.; Liang, E.; Simmons, N.B.; Lipkin, W.I.; Daszak, P.; et al. Viral Diversity, Prey Preference, and Bartonella Prevalence in Desmodus rotundus in Guatemala. EcoHealth 2016, 13, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, L.; Ren, X.; He, G.; Zhang, J.; Yang, J.; Qian, Z.; Dong, J.; Sun, L.; Zhu, Y.; et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2015, 10, 609–620. [Google Scholar] [CrossRef]
- Han, H.-J.; Wen, H.-L.; Zhao, L.; Liu, J.-W.; Luo, L.-M.; Zhou, C.-M.; Qin, X.-R.; Zhu, Y.-L.; Liu, M.-M.; Qi, R.; et al. Novel coronaviruses, astroviruses, adenoviruses and circoviruses in insectivorous bats from northern China. Zoonoses Public Heal. 2017, 64, 636–646. [Google Scholar] [CrossRef]
- Hu, B.; Zeng, L.-P.; Yang, X.-L.; Ge, X.-Y.; Zhang, W.; Li, B.; Xie, J.-Z.; Shen, X.-R.; Zhang, Y.-Z.; Wang, N.; et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017, 13, e1006698. [Google Scholar] [CrossRef]
- Lacroix, A.; Duong, V.; Hul, V.; San, S.; Davun, H.; Omaliss, K.; Chea, S.; Hassanin, A.; Theppangna, W.; Silithammavong, S.; et al. Diversity of bat astroviruses in Lao PDR and Cambodia. Infect. Genet. Evol. 2017, 47, 41–50. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Ahmed, S.S.; Tsoi, H.-W.; Yeung, H.C.; Li, K.S.M.; Fan, R.Y.Y.; Zhao, P.S.H.; Lau, C.C.C.; Lam, C.S.F.; Choi, K.K.F.; et al. Bats host diverse parvoviruses as possible origin of mammalian dependoparvoviruses and source for bat–swine interspecies transmission. J. Gen. Virol. 2017, 98, 3046–3059. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Yang, X.-L.; Li, B.; Liu, Q.; Zhang, Q.; Liu, H.; Kan, H.-P.; Wong, K.-C.; Chek, S.-N.; He, X.; et al. Detection of diverse viruses in alimentary specimens of bats in Macau. Virol. Sin. 2017, 32, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Munnink, B.B.O.; Phan, M.V.; Simmonds, P.; Koopmans, M.P.; Kellam, P.; van der Hoek, L.; Cotten, M. The VIZIONS Consortium Characterization of Posa and Posa-like virus genomes in fecal samples from humans, pigs, rats, and bats collected from a single location in Vietnam. Virus Evol. 2017, 3, vex022. [Google Scholar] [CrossRef]
- Rizzo, F.; Edenborough, K.M.; Toffoli, R.; Culasso, P.; Zoppi, S.; Dondo, A.; Robetto, S.; Rosati, S.; Lander, A.; Kurth, A.; et al. Coronavirus and paramyxovirus in bats from Northwest Italy. BMC Veter.- Res. 2017, 13, 1–11. [Google Scholar] [CrossRef]
- Seltmann, A.; Corman, V.M.; Rasche, A.; Drosten, C.; Czirják, G.; Bernard, H.; Struebig, M.J.; Voigt, C.C. Seasonal Fluctuations of Astrovirus, But Not Coronavirus Shedding in Bats Inhabiting Human-Modified Tropical Forests. EcoHealth 2017, 14, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Santos, A.; Moreira-Soto, A.; Soto-Garita, C.; Chaverri, L.G.; Chaves, A.; Drexler, J.F.; Morales, J.A.; Alfaro-Alarcón, A.; Rodríguez-Herrera, B.; Corrales-Aguilar, E. Neotropical bats that co-habit with humans function as dead-end hosts for dengue virus. PLoS Negl. Trop. Dis. 2017, 11, e0005537. [Google Scholar] [CrossRef] [PubMed]
- Waruhiu, C.; Ommeh, S.; Obanda, V.; Agwanda, B.; Gakuya, F.; Ge, X.-Y.; Yang, X.-L.; Wu, L.-J.; Zohaib, A.; Hu, B.; et al. Molecular detection of viruses in Kenyan bats and discovery of novel astroviruses, caliciviruses and rotaviruses. Virol. Sin. 2017, 32, 101–114. [Google Scholar] [CrossRef]
- Yang, X.L.; Zhang, Y.Z.; Jiang, R.D.; Guo, H.; Zhang, W.; Li, B.; Wang, N.; Wang, L.; Waruhiu, C.; Zhou, J.H.; et al. Genetically Diverse Filoviruses in Rousettus and Eonycteris spp. Bats, China, 2009 and 2015. Emerg. Infect. Dis. 2017, 23, 482–486. [Google Scholar] [CrossRef]
- Phan, M.V.T.; Tri, T.N.; Anh, P.H.; Baker, S.; Kellam, P.; Cotten, M. Identification and characterization of Coronaviridae genomes from Vietnamese bats and rats based on conserved protein domains. Virus Evol. 2018, 4, vey035. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Duengkae, P.; Chaiyes, A.; Kaewpom, T.; Rodpan, A.; Yingsakmongkon, S.; Petcharat, S.; Phengsakul, P.; Maneeorn, P.; Hemachudha, T. Longitudinal study of age-specific pattern of coronavirus infection in Lyle’s flying fox (Pteropus lylei) in Thailand. Virol. J. 2018, 15, 1–10. [Google Scholar] [CrossRef]
- Wada, Y.; Sasaki, M.; Setiyono, A.; Handharyani, E.; Rahmadani, I.; Taha, S.; Adiani, S.; Latief, M.; Kholilullah, Z.A.; Subangkit, M.; et al. Detection of novel gammaherpesviruses from fruit bats in Indonesia. J. Med. Microbiol. 2018, 67, 415–422. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Son, K.-D.; Yong-Sik, K.; Wang, S.-J.; Kim, Y.-K.; Jheong, W.-H.; Oem, J.-K. Genetic diversity and phylogenetic analysis of newly discovered bat astroviruses in Korea. Arch. Virol. 2018, 163, 3065–3072. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Fagbo, S.F.; Alagaili, A.N.; Nitido, A.; Williams, S.H.; Ng, J.; Lee, B.; Durosinlorun, A.; Garcia, J.A.; Jain, K.; et al. A viral metagenomic survey identifies known and novel mammalian viruses in bats from Saudi Arabia. PLoS ONE 2019, 14, e0214227. [Google Scholar] [CrossRef]
- Mortlock, M.; Dietrich, M.; Weyer, J.; Paweska, J.T.; Markotter, W. Co-Circulation and Excretion Dynamics of Diverse Rubula- and Related Viruses in Egyptian Rousette Bats from South Africa. Viruses 2019, 11, 37. [Google Scholar] [CrossRef]
- Prada, D.; Boyd, V.; Baker, M.L.; O’dea, M.; Jackson, B. Viral Diversity of Microbats within the South West Botanical Province of Western Australia. Viruses 2019, 11, 1157. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, M.E.; Bergner, L.M.; Broos, A.; Meza, D.K.; Filipe, A.d.S.; Davison, A.; Tello, C.; Becker, D.J.; Streicker, D.G. Epidemiology and biology of a herpesvirus in rabies endemic vampire bat populations. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- James, S.; Donato, D.; de Thoisy, B.; Lavergne, A.; Lacoste, V. Novel herpesviruses in neotropical bats and their relationship with other members of the Herpesviridae family. Infect. Genet. Evol. 2020, 84, 104367. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, A.; Vidal, N.; Keita, A.K.; Thaurignac, G.; Esteban, A.; De Nys, H.; Diallo, R.; Toure, A.; Goumou, S.; Soumah, A.K.; et al. Wide Diversity of Coronaviruses in Frugivorous and Insectivorous Bat Species: A Pilot Study in Guinea, West Africa. Viruses 2020, 12, 855. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Chung, C.-U.; Park, J.S.; Oem, J.-K. Novel viruses detected in bats in the Republic of Korea. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Tan, Z.; Gonzalez, G.; Sheng, J.; Wu, J.; Zhang, F.; Xu, L.; Zhang, P.; Zhu, A.; Qu, Y.; Tu, C.; et al. Extensive Genetic Diversity of Polyomaviruses in Sympatric Bat Communities: Host Switching versus Coevolution. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Valitutto, M.T.; Aung, O.; Tun, K.Y.N.; Vodzak, M.E.; Zimmerman, D.; Yu, J.H.; Win, Y.T.; Maw, M.T.; Thein, W.Z.; Win, H.H.; et al. Detection of novel coronaviruses in bats in Myanmar. PLoS ONE 2020, 15, e0230802. [Google Scholar] [CrossRef]
- Mourya, D.; Yadav, P.; Shete-Aich, A.; Nyayanit, D.; Pardeshi, P.; Majumdar, T.; Balasubramanian, R.; Ullas, P.; Mohandas, S.; Dighe, H.; et al. Detection of coronaviruses in Pteropus & Rousettus species of bats from different States of India. Indian J. Med. Res. 2020, 151, 226–235. [Google Scholar] [CrossRef]
- Zeus, V.M.; Köhler, A.; Reusch, C.; Fischer, K.; Balkema-Buschmann, A.; Kerth, G. Analysis of astrovirus transmission pathways in a free-ranging fission-fusion colony of Natterer’s bats (Myotis nattereri). Behav. Ecol. Sociobiol. 2020, 74. [Google Scholar] [CrossRef]
- Darcissac, E.; Donato, D.; de Thoisy, B.; Lacoste, V.; Lavergne, A. Paramyxovirus circulation in bat species from French Guiana. Infect. Genet. Evol. 2021, 90, 104769. [Google Scholar] [CrossRef]
- Hoarau, A.O.G.; Goodman, S.M.; Al Halabi, D.; Ramasindrazana, B.; Lagadec, E.; Le Minter, G.; Köster, M.; Dos Santos, A.; Schoeman, M.C.; Gudo, E.S.; et al. Investigation of astrovirus, coronavirus and paramyxovirus co-infections in bats in the western Indian Ocean. Virol. J. 2021, 18, 1–8. [Google Scholar] [CrossRef]
- Kohl, C.; Brinkmann, A.; Radonić, A.; Dabrowski, P.W.; Mühldorfer, K.; Nitsche, A.; Wibbelt, G.; Kurth, A. The virome of German bats: Comparing virus discovery approaches. Sci. Rep. 2021, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kumakamba, C.; Niama, F.R.; Muyembe, F.; Mombouli, J.-V.; Kingebeni, P.M.; Nina, R.A.; Lukusa, I.N.; Bounga, G.; N’kawa, F.; Nkoua, C.G.; et al. Coronavirus surveillance in wildlife from two Congo basin countries detects RNA of multiple species circulating in bats and rodents. PLoS ONE 2021, 16, e0236971. [Google Scholar] [CrossRef] [PubMed]
- Lazov, C.M.; Belsham, G.J.; Bøtner, A.; Rasmussen, T.B. Full-Genome Sequences of Alphacoronaviruses and Astroviruses from Myotis and Pipistrelle Bats in Denmark. Viruses 2021, 13, 1073. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Ashworth, J.; Nguyen, D.; Li, K.; Smith, D.B.; Woolhouse, M.; on behalf of the VIZIONS Consortium. No Exchange of Picornaviruses in Vietnam between Humans and Animals in a High-Risk Cohort with Close Contact despite High Prevalence and Diversity. Viruses 2021, 13, 1709. [Google Scholar] [CrossRef] [PubMed]
- Ntumvi, N.F.; Valantine Ngum, N.; Gillis, A.; Joseph Le Doux, D.; Tamoufe, U.; Jean-Michel, T.; Moctar, M.M.M.; Nwobegahay, J.; Lebreton, M.; Rimoin, A.W.; et al. Wildlife in Cameroon harbor diverse coronaviruses including many isolates closely related to human coronavirus 229E. Viruses Evol. 2022, 8, veab110. [Google Scholar] [CrossRef] [PubMed]
- Orłowska, A.; Smreczak, M.; Potyrało, P.; Bomba, A.; Trębas, P.; Rola, J. First Detection of Bat Astroviruses (BtAstVs) among Bats in Poland: The Genetic BtAstVs Diversity Reveals Multiple Co-Infection of Bats with Different Strains. Viruses 2021, 13, 158. [Google Scholar] [CrossRef]
- Simsek, C.; Corman, V.M.; Everling, H.U.; Lukashev, A.N.; Rasche, A.; Maganga, G.D.; Binger, T.; Jansen, D.; Beller, L.; Deboutte, W.; et al. At Least Seven Distinct Rotavirus Genotype Constellations in Bats with Evidence of Reassortment and Zoonotic Transmissions. mBio 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Anderson, D.E.; Halpin, K.; Hong, X.; Chen, H.; Walker, S.; Valdeter, S.; van der Heide, B.; Neave, M.J.; Bingham, J.; et al. A new Hendra virus genotype found in Australian flying foxes. Virol. J. 2021, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wells, H.L.; Loh, E.; Nava, A.; Mei Ho, L.; Lee, J.; Jum Ra, S.; Navarrete-Macias, I.; Liang, E.; Firth, C.; Epstein, J.; et al. Taxonomic classification methods reveal a new subgenus in the paramyxovirus subfamily Orthoparamyxovirinae. BioRxiv 2021. [Google Scholar] [CrossRef]
- Torres-Castro, M.; Noh-Pech, H.; Hernández-Betancourt, S.; Peláez-Sánchez, R.; Lugo-Caballero, C.; Puerto, F.I. West Nile and Zika viruses in bats from a suburban area of Merida, Yucatan, Mexico. Zoonoses Public Health 2021, 68, 834–841. [Google Scholar] [CrossRef]
- Ramos-Nino, M.E.; Fitzpatrick, D.M.; Eckstrom, K.M.; Tighe, S.; Dragon, J.A.; Cheetham, S. The Kidney-Associated Microbiome of Wild-Caught Artibeus spp. in Grenada, West Indies. Animals 2021, 11, 1571. [Google Scholar] [CrossRef]
- Marrero, L.M.; Nuñez, G.B.; Malta, L.; Delfraro, A.; Frabasile, S. Ecological and Conservation Significance of Herpesvirus Infection in Neotropical Bats. EcoHealth 2021, 18, 123–133. [Google Scholar] [CrossRef]
- Luo, D.-S.; Li, B.; Shen, X.-R.; Jiang, R.-D.; Zhu, Y.; Wu, J.; Fan, Y.; Bourhy, H.; Hu, B.; Ge, X.-Y.; et al. Characterization of Novel Rhabdoviruses in Chinese Bats. Viruses 2021, 13, 64. [Google Scholar] [CrossRef]
- Luo, D.; Serra-Cobo, J.; Harazim, M.; Maufrais, C.; Bonas, S.; Martinkova, N.; Lalis, A.; Nakouné, E.; Adjogoua, E.; Kadjo, B.; et al. Novel bat vesiculovirus in the Mediterranean region. Virologie 2021, 25 (Suppl. S1). [Google Scholar] [CrossRef]
- Lacroix, A.; Kingebeni, P.M.; Kumugo, S.P.N.; Lempu, G.; Butel, C.; Serrano, L.; Vidal, N.; Thaurignac, G.; Esteban, A.; Bamuleka, D.M.; et al. Investigating the Circulation of Ebola Viruses in Bats during the Ebola Virus Disease Outbreaks in the Equateur and North Kivu Provinces of the Democratic Republic of Congo from 2018. Pathogens 2021, 10, 557. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.; Calvelage, S.; Schlottau, K.; Hoffmann, B.; Eggerbauer, E.; Müller, T.; Freuling, C.M. Retrospective Enhanced Bat Lyssavirus Surveillance in Germany between 2018–2020. Viruses 2021, 13, 1538. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Gao, Y.; Wen, Y.; Ke, X.; Ou, Z.; Li, Y.; He, H.; Chen, Q. Detection of Virus-Related Sequences Associated With Potential Etiologies of Hepatitis in Liver Tissue Samples From Rats, Mice, Shrews, and Bats. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Delaune, D.; Hul, V.; Karlsson, E.A.; Hassanin, A.; Ou, T.P.; Baidaliuk, A.; Gámbaro, F.; Prot, M.; Tu, V.T.; Chea, S.; et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat. Commun. 2021, 12, 6563. [Google Scholar] [CrossRef]
- de Souza, W.M.; Fumagalli, M.J.; Carrera, J.P.; de Araujo, J.; Cardoso, J.F.; de Carvalho, C.; Durigon, E.L.; Queiroz, L.H.; Faria, N.R.; Murcia, P.R.; et al. Paramyxoviruses from neotropical bats suggest a novel genus and nephrotropism. Infect. Genet. Evol. 2021, 95, 105041. [Google Scholar] [CrossRef]
- Schulz, J.E.; Seifert, S.N.; Thompson, J.T.; Avanzato, V.; Sterling, S.L.; Lianying, Y.; Letko, M.C.; Matson, M.J.; Fischer, R.J.; Tremeau-Bravard, A.; et al. Serological Evidence for Henipa-like and Filo-like Viruses in Trinidad Bats. J. Infect. Dis. 2020, 221, S375–S382. [Google Scholar] [CrossRef] [PubMed]
- Nziza, J.; Goldstein, T.; Cranfield, M.; Webala, P.; Nsengimana, O.; Nyatanyi, T.; Mudakikwa, A.; Tremeau-Bravard, A.; Byarugaba, D.; Tumushime, J.C.; et al. Coronaviruses Detected in Bats in Close Contact with Humans in Rwanda. EcoHealth 2019, 17, 152–159. [Google Scholar] [CrossRef]
- Kivistö, I.; Tidenberg, E.-M.; Lilley, T.; Suominen, K.; Forbes, K.M.; Vapalahti, O.; Huovilainen, A.; Sironen, T. First Report of Coronaviruses in Northern European Bats. Vector-Borne Zoonotic Dis. 2020, 20, 155–158. [Google Scholar] [CrossRef]
- Islam, A.; Hossain, M.E.; Rostal, M.K.; Ferdous, J.; Islam, A.; Hasan, R.; Miah, M.; Rahman, M.; Rahman, M.Z.; Daszak, P.; et al. Epidemiology and Molecular Characterization of Rotavirus A in Fruit Bats in Bangladesh. EcoHealth 2020, 17, 398–405. [Google Scholar] [CrossRef]
- Amman, B.R.; Bird, B.H.; Bakarr, I.A.; Bangura, J.; Schuh, A.J.; Johnny, J.; Sealy, T.K.; Conteh, I.; Koroma, A.H.; Foday, I.; et al. Isolation of Angola-like Marburg virus from Egyptian rousette bats from West Africa. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Sjodin, A.R.; Willig, M.R.; Anthony, S.J. Quantitative delineation of herpesviruses in bats for use in ecological studies. BioRxiv 2019. [Google Scholar] [CrossRef]
- Markotter, W.; Geldenhuys, M.; van Vuren, P.J.; Kemp, A.; Mortlock, M.; Mudakikwa, A.; Nel, L.; Nziza, J.; Paweska, J.; Weyer, J. Paramyxo- and Coronaviruses in Rwandan Bats. Trop. Med. Infect. Dis. 2019, 4, 99. [Google Scholar] [CrossRef]
- Lim, X.F.; Lee, C.B.; Pascoe, S.M.; How, C.B.; Chan, S.; Tan, J.H.; Yang, X.; Zhou, P.; Shi, Z.; Sessions, O.M.; et al. Detection and characterization of a novel bat-borne coronavirus in Singapore using multiple molecular approaches. J. Gen. Virol. 2019, 100, 1363–1374. [Google Scholar] [CrossRef]
- Lecis, R.; Mucedda, M.; Pidinchedda, E.; Pittau, M.; Alberti, A. Molecular identification of Betacoronavirus in bats from Sardinia (Italy): First detection and phylogeny. Virus Genes 2018, 55, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Matsugo, H.; Maruyama, J.; Kamiki, H.; Takada, A.; Maeda, K.; Takenaka-Uema, A.; Tohya, Y.; Murakami, S.; Horimoto, T. Characterization of a novel species of adenovirus from Japanese microbat and role of CXADR as its entry factor. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Kikuchi, F.; Bawm, S.; Sơn, N.T.; Lin, K.S.; Tú, V.T.; Aoki, K.; Tsuchiya, K.; Tanaka-Taya, K.; Morikawa, S.; et al. Molecular Phylogeny of Mobatviruses (Hantaviridae) in Myanmar and Vietnam. Viruses 2019, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ren, X.; Xu, Z.; Fu, S.; Li, X.; Zhang, H.; Yang, W.; Zhang, Y.; Liang, G. Genetic diversity of the Yokose virus, XYBX1332, isolated from bats (Myotis daubentonii) in China. Virol. J. 2019, 16, 1–8. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Q.; Xu, L.; Tu, C.; Huang, X.; He, B. Detection and Characterization of a Novel Norovirus in Bats, China. Virol. Sin. 2018, 33, 100–103. [Google Scholar] [CrossRef]
- Van Nguyen, D.; Van Nguyen, C.; Bonsall, D.; Ngo, T.T.; Carrique-Mas, J.; Pham, A.H.; Bryant, J.E.; Thwaites, G.; Baker, S.; Woolhouse, M.; et al. Detection and Characterization of Homologues of Human Hepatitis Viruses and Pegiviruses in Rodents and Bats in Vietnam. Viruses 2018, 10, 102. [Google Scholar] [CrossRef]
- Holz, P.H.; Lumsden, L.F.; Druce, J.; Legione, A.R.; Vaz, P.; Devlin, J.M.; Hufschmid, J. Virus survey in populations of two subspecies of bent-winged bats (Miniopterus orianae bassanii and oceanensis) in south-eastern Australia reveals a high prevalence of diverse herpesviruses. PLoS ONE 2018, 13, e0197625. [Google Scholar] [CrossRef]
- Goldstein, T.; Anthony, S.J.; Gbakima, A.; Bird, B.H.; Bangura, J.; Tremeau-Bravard, A.; Belaganahalli, M.N.; Wells, H.L.; Dhanota, J.K.; Liang, E.; et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat. Microbiol. 2018, 3, 1084–1089. [Google Scholar] [CrossRef]
- Amoroso, M.G.; Russo, D.; Lanave, G.; Cistrone, L.; Pratelli, A.; Martella, V.; Galiero, G.; Decaro, N.; Fusco, G. Detection and phylogenetic characterization of astroviruses in insectivorous bats from Central-Southern Italy. Zoonoses Public Health 2018, 65, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, X.-L.; Li, W.; Zhu, Y.; Ge, X.-Y.; Zhang, L.-B.; Zhang, Y.-Z.; Bock, C.-T.; Shi, Z.-L. Detection and genome characterization of four novel bat hepadnaviruses and a hepevirus in China. Virol. J. 2017, 14, 1–10. [Google Scholar] [CrossRef]
- Pauly, M.; Pir, J.B.; Loesch, C.; Sausy, A.; Snoeck, C.J.; Hübschen, J.M.; Muller, C.P. Novel Alphacoronaviruses and Paramyxoviruses Cocirculate with Type 1 and Severe Acute Respiratory System (SARS)-Related Betacoronaviruses in Synanthropic Bats of Luxembourg. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Noh, J.Y.; Yoon, S.-W.; Kim, D.-J.; Lee, M.-S.; Kim, J.-H.; Na, W.; Song, D.; Jeong, D.G.; Kim, H.K. Simultaneous detection of severe acute respiratory syndrome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch. Virol. 2017, 162, 1617–1623. [Google Scholar] [CrossRef]
- Hu, D.; Zhu, C.; Wang, Y.; Ai, L.; Yang, L.; Ye, F.; Ding, C.; Chen, J.; He, B.; Zhu, J.; et al. Virome analysis for identification of novel mammalian viruses in bats from Southeast China. Sci. Rep. 2017, 7, 10917. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Yoon, S.-W.; Kim, D.-J.; Koo, B.-S.; Noh, J.Y.; Kim, J.H.; Choi, Y.G.; Na, W.; Chang, K.-T.; Song, D.; et al. Detection of Severe Acute Respiratory Syndrome-Like, Middle East Respiratory Syndrome-Like Bat Coronaviruses and Group H Rotavirus in Faeces of Korean Bats. Transbound. Emerg. Dis. 2016, 63, 365–372. [Google Scholar] [CrossRef]
- Fischer, K.; Zeus, V.; Kwasnitschka, L.; Kerth, G.; Haase, M.; Groschup, M.H.; Balkema-Buschmann, A. Insectivorous bats carry host specific astroviruses and coronaviruses across different regions in Germany. Infect. Genet. Evol. 2016, 37, 108–116. [Google Scholar] [CrossRef]
- Wilkinson, D.A.; Mélade, J.; Dietrich, M.; Ramasindrazana, B.; Soarimalala, V.; Lagadec, E.; le Minter, G.; Tortosa, P.; Heraud, J.-M.; de Lamballerie, X.; et al. Highly Diverse Morbillivirus-Related Paramyxoviruses in Wild Fauna of the Southwestern Indian Ocean Islands: Evidence of Exchange between Introduced and Endemic Small Mammals. J. Virol. 2014, 88, 8268–8277. [Google Scholar] [CrossRef] [PubMed]
- Maganga, G.D.; Bourgarel, M.; Vallo, P.; Dallo, T.D.; Ngoagouni, C.; Drexler, J.F.; Drosten, C.; Nakouné, E.R.; Leroy, E.M.; Morand, S. Bat Distribution Size or Shape as Determinant of Viral Richness in African Bats. PLoS ONE 2014, 9, e100172. [Google Scholar] [CrossRef]
- Maganga, G.D.; Bourgarel, M.; Nkoghe, J.O.; N’Dilimabaka, N.; Drosten, C.; Paupy, C.; Morand, S.; Drexler, J.F.; Leroy, E.M. Identification of an Unclassified Paramyxovirus in Coleura afra: A Potential Case of Host Specificity. PLoS ONE 2014, 9, e115588. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhang, Y.; Xu, L.; Yang, W.; Yang, F.; Feng, Y.; Xia, L.; Zhou, J.; Zhen, W.; Feng, Y.; et al. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J. Virol. 2014, 88, 7070–7082. [Google Scholar] [CrossRef] [PubMed]
- Cadar, D.; Becker, N.; de Mendonca Campos, R.; Börstler, J.; Jöst, H.; Schmidt-Chanasit, J. Usutu virus in bats, Germany, 2013. Emerg. Infect. Dis. 2014, 20, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.F.; Kapoor, V.; Epstein, J.H.; Alhakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Li, K.S.M.; Tsang, A.K.L.; Lam, C.S.F.; Ahmed, S.; Chen, H.; Chan, K.-H.; Woo, P.C.Y.; Yuen, K.-Y. Genetic Characterization of Betacoronavirus Lineage C Viruses in Bats Reveals Marked Sequence Divergence in the Spike Protein of Pipistrellus Bat Coronavirus HKU5 in Japanese Pipistrelle: Implications for the Origin of the Novel Middle East Respiratory Syndrome Coronavirus. J. Virol. 2013, 87, 8638–8650. [Google Scholar] [CrossRef] [PubMed]
- Raut, C.; Yadav, P.; Towner, J.; Amman, B.; Erickson, B.; Cannon, D.; Sivaram, A.; Basu, A.; Nichol, S.; Mishra, A.; et al. Isolation of a Novel Adenovirus from Rousettus leschenaultii Bats from India. Intervirology 2012, 55, 488–490. [Google Scholar] [CrossRef]
- Kurth, A.; Kohl, C.; Brinkmann, A.; Ebinger, A.; Harper, J.A.; Wang, L.-F.; Mühldorfer, K.; Wibbelt, G. Novel Paramyxoviruses in Free-Ranging European Bats. PLoS ONE 2012, 7, e38688. [Google Scholar] [CrossRef]
- Crockett, R.J.; Gilbert, A.; Kityo, R.; Ledermann, J.; Borland, E.; Powers, A.; Panella, N.; Crabtree, M.; Nakayiki, T.; Kuzmin, I.; et al. Arbovirus surveillance and virus isolations from bats in Uganda, Kenya, and the democratic republic of the Congo. Am. J. Trop. Med. Hyg. 2012, 87, 170. [Google Scholar]
- Negredo, A.; Palacios, G.; Vázquez-Morón, S.; González, F.; Dopazo, H.; Molero, F.; Juste, J.; Quetglas, J.; Savji, N.; de la Cruz Martínez, M.; et al. Discovery of an Ebolavirus-Like Filovirus in Europe. PLoS Pathog. 2011, 7, e1002304. [Google Scholar] [CrossRef]
- Misra, V.; Dumonceaux, T.; Dubois, J.; Willis, C.; Nadin-Davis, S.; Severini, A.; Wandeler, A.; Lindsay, R.; Artsob, H. Detection of polyoma and corona viruses in bats of Canada. J. Gen. Virol. 2009, 90, 2015–2022. [Google Scholar] [CrossRef]
- Molnár, V.; Jánoska, M.; Harrach, B.; Glávits, R.; Pálmai, N.; Rigó, D.; Sós, E.; Liptovszky, M. Detection of a novel bat gammaherpesvirus in Hungary. Acta Veter. Hung. 2008, 56, 529–538. [Google Scholar] [CrossRef]
- Aguilar-Setién, Á.; Romero-Almaraz, M.L.; Sánchez-Hernández, C.; Figueroa, R.; Juárez-Palma, L.P.; García-Flores, M.M.; Vázquez-Salinas, C.; Salas-Rojas, M.; Hidalgo-Martínez, A.C.; Pierlé, S.A.; et al. Dengue virus in Mexican bats. Epidemiol. Infect. 2008, 136, 1678–1683. [Google Scholar] [CrossRef]
- Vázquez-Morón, S.; Juste, J.; Ibáñez, C.; Aznar, C.; Ruiz-Villamor, E.; Echevarría, J.E. Asymptomatic rhabdovirus infection in meridional serotine bats (Eptesicus isabellinus) from Spain. In Proceedings of the Joint OIE/WHO/EU International Conference on Towards the Elimination of Rabies in Eurasia, Paris, France, 27–30 May 2007; pp. 311–316. [Google Scholar]
- Kia, G.S.N.; Tao, Y.; Umoh, J.U.; Kwaga, J.K.P.; Tong, S. Identification of Coronaviruses, Paramyxoviruses, Reoviruses, and Rotaviruses among Bats in Nigeria. Am. J. Trop. Med. Hyg. 2021, 104, 1106. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Nelson, E.; Christopher-Hennings, J. Novel and Diverse Non-Rabies Rhabdoviruses Identified in Bats with Human Exposure, South Dakota, USA. Viruses 2020, 12, 1408. [Google Scholar] [CrossRef] [PubMed]
- Finoketti, F.; dos Santos, R.N.; Campos, A.A.S.; Zani, A.L.d.S.; Barboza, C.M.; Fernandes, M.E.S.; Souza, T.d.C.P.d.; dos Santos, D.D.; Bortolanza, G.W.; Filho, H.O.; et al. Detection of adenovirus, papillomavirus and parvovirus in Brazilian bats of the species Artibeus lituratus and Sturnira lilium. Arch. Virol. 2019, 164, 1015–1025. [Google Scholar] [CrossRef]
- Kemenesi, G.; Kurucz, K.; Zana, B.; Földes, F.; Urbán, P.; Vlaschenko, A.; Kravchenko, K.; Budinski, I.; Szodoray-Parádi, F.; Bücs, S.; et al. Diverse replication-associated protein encoding circular DNA viruses in guano samples of Central-Eastern European bats. Arch. Virol. 2017, 163, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Afelt, A.; Lacroix, A.; Zawadzka-Pawlewska, U.; Pokojski, W.; Buchy, P.; Frutos, R. Distribution of bat-borne viruses and environment patterns. Infect. Genet. Evol. 2017, 58, 181–191. [Google Scholar] [CrossRef]
- Mourya, D.T.; Shete, A.M.; Yadav, P.; Kumar, V.; Nikam, T.; Mehershahi, K.; Kokate, P.; Patil, D. Development of polymerase chain reaction-based diagnostic tests for detection of Malsoor virus & adenovirus isolated from Rousettus species of bats in Maharashtra, India. Indian J. Med. Res. 2017, 145, 90–96. [Google Scholar] [CrossRef]
- He, B.; Li, Z.; Yang, F.; Zheng, J.; Ye, F.; Guo, H.; Li, Y.; Wang, Y.; Su, N.; Zhang, F.; et al. Virome Profiling of Bats from Myanmar by Metagenomic Analysis of Tissue Samples Reveals More Novel Mammalian Viruses. PLoS ONE 2013, 8, e61950. [Google Scholar]
- Simmons, N.B.; Cirranello, A.L. Bat Species of the World: A Taxonomic and Geographic Database. 2022. Available online: https://batnames.org/ (accessed on 27 April 2023).
- Wang, L.-F.; Anderson, D.E. Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 2019, 34, 79–89. [Google Scholar] [CrossRef]
- Hellard, E.; Fouchet, D.; Vavre, F.; Pontier, D. Parasite–Parasite Interactions in the Wild: How To Detect Them? Trends Parasitol. 2015, 31, 640–652. [Google Scholar] [CrossRef]
- Alizon, S. Co-infection and super-infection models in evolutionary epidemiology. Interface Focus 2013, 3, 20130031. [Google Scholar] [CrossRef]
- Young, C.C.W.; Olival, K.J. Optimizing Viral Discovery in Bats. PLoS ONE 2016, 11, e0149237. [Google Scholar] [CrossRef]
- Hayman, D.T.S. Bats as Viral Reservoirs. Ann. Rev. Virol. 2016, 3, 77–99. [Google Scholar] [CrossRef]
- Van Brussel, K.; Holmes, E.C. Zoonotic disease and virome diversity in bats. Curr. Opin. Virol. 2022, 52, 192–202. [Google Scholar] [CrossRef]
- Singh, R.K.; Dhama, K.; Chakraborty, S.; Tiwari, R.; Natesan, S.; Khandia, R.; Munjal, A.; Vora, K.S.; Latheef, S.K.; Karthik, K.; et al. Nipah virus: Epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies—A comprehensive review. Veter. Quart. 2019, 39, 26–55. [Google Scholar] [CrossRef]
- Chinese SARS Molecular Epidemiology Consortium. Molecular Evolution of the SARS Coronavirus During the Course of the SARS Epidemic in China. Science 2004, 303, 1666–1669. [Google Scholar] [CrossRef]
- DaPalma, T.; Doonan, B.; Trager, N.; Kasman, L. A systematic approach to virus–virus interactions. Virus Res. 2010, 149, 1–9. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, T.J.; Cryan, P.M.; Hayman, D.T.; Plowright, R.K.; Streicker, D.G. Multiple mortality events in bats: A global review. Mammal Rev. 2016, 46, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Munson, L.; Terio, K.A.; Kock, R.; Mlengeya, T.; Roelke, M.E.; Dubovi, E.; Summers, B.; Sinclair, A.R.E.; Packer, C. Climate Extremes Promote Fatal Co-Infections during Canine Distemper Epidemics in African Lions. PLoS ONE 2008, 3, e2545. [Google Scholar] [CrossRef] [PubMed]
- Petney, T.N.; Andrews, R.H. Multiparasite communities in animals and humans: Frequency, structure and pathogenic significance. Int. J. Parasitol. 1998, 28, 377–393. [Google Scholar] [CrossRef]
- Davis, A.; Gordy, P.; Rudd, R.; Jarvis, J.A.; Bowen, R.A.; O’Shea, T.J.; Cryan, P.M.; Hayman, D.T.; Plowright, R.K.; Streicker, D.G.; et al. Naturally Acquired Rabies Virus Infections in Wild-Caught Bats. Vector-Borne Zoonotic Dis. 2012, 12, 55–60. [Google Scholar] [CrossRef]
- Banerjee, A.; Misra, V.; Schountz, T.; Baker, M.L. Tools to study pathogen-host interactions in bats. Virus Res. 2018, 248, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Maljkovic Berry, I.; Melendrez, M.C.; Bishop-Lilly, K.A.; Rutvisuttinunt, W.; Pollett, S.; Talundzic, E.; Morton, L.; Jarman, R.G. Next Generation Sequencing and Bioinformatics Methodologies for Infectious Disease Research and Public Health: Approaches, Applications, and Considerations for Development of Laboratory Capacity. J. Infect. Dis. 2019, 221, S292–S307. [Google Scholar] [CrossRef] [PubMed]
- Belak, S.; Karlsson, O.; Leijon, M.; Granberg, F. High-throughput sequencing in veterinary infection biology and diagnostics. Rev. Sci. Et Techol. l’OIE 2013, 32, 893–915. [Google Scholar] [CrossRef]
- Edson, D.; Field, H.; McMichael, L.; Vidgen, M.; Goldspink, L.; Broos, A.; Melville, D.; Kristoffersen, J.; de Jong, C.; McLaughlin, A.; et al. Routes of Hendra Virus Excretion in Naturally-Infected Flying-Foxes: Implications for Viral Transmission and Spillover Risk. PLoS ONE 2015, 10, e0140670. [Google Scholar] [CrossRef]
- Essaidi-Laziosi, M.; Geiser, J.; Huang, S.; Constant, S.; Kaiser, L.; Tapparel, C. Interferon-Dependent and Respiratory Virus-Specific Interference in Dual Infections of Airway Epithelia. Sci. Rep. 2020, 10, 10246. [Google Scholar] [CrossRef] [PubMed]
- Pepin, K.M.; Lambeth, K.; Hanley, K.A. Asymmetric competitive suppression between strains of dengue virus. BMC Microbiol. 2008, 8, 28. [Google Scholar] [CrossRef]
- Hao, X.; Li, Y.; Chen, H.; Chen, B.; Liu, R.; Wu, Y.; Xiao, X.; Zhou, P.; Li, S. Canine Circovirus Suppresses the Type I Interferon Response and Protein Expression but Promotes CPV-2 Replication. Int. J. Mol. Sci. 2022, 23, 6382. [Google Scholar] [CrossRef]
- Du, X.; Zhou, D.; Zhou, J.; Xue, J.; Cheng, Z. Marek’s Disease Virus and Reticuloendotheliosis Virus Coinfection Enhances Viral Replication and Alters Cellular Protein Profiles. Front. Vet. Sci. 2022, 9, 854007. [Google Scholar] [CrossRef]
- Fenton, A.; Knowles, S.C.; Petchey, O.L.; Pedersen, A.B. The reliability of observational approaches for detecting interspecific parasite interactions: Comparison with experimental results. Int. J. Parasitol. 2014, 44, 437–445. [Google Scholar] [CrossRef]
- Hamelin, F.M.; Allen, L.J.S.; Bokil, V.A.; Gross, L.J.; Hilker, F.M.; Jeger, M.J.; Manore, C.A.; Power, A.G.; Rúa, M.A.; Cunniffe, N.J. Coinfections by noninteracting pathogens are not independent and require new tests of interaction. PLoS Biol. 2019, 17, e3000551. [Google Scholar] [CrossRef] [PubMed]
- Clark, N.J.; Wells, K.; Lindberg, O. Unravelling changing interspecific interactions across environmental gradients using Markov random fields. Ecology 2018, 99, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, A.B.; Fenton, A. Emphasizing the ecology in parasite community ecology. Trends Ecol. Evol. 2007, 22, 133–139. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jones, B.D.; Kaufman, E.J.; Peel, A.J. Viral Co-Infection in Bats: A Systematic Review. Viruses 2023, 15, 1860. https://doi.org/10.3390/v15091860
Jones BD, Kaufman EJ, Peel AJ. Viral Co-Infection in Bats: A Systematic Review. Viruses. 2023; 15(9):1860. https://doi.org/10.3390/v15091860
Chicago/Turabian StyleJones, Brent D., Eli J. Kaufman, and Alison J. Peel. 2023. "Viral Co-Infection in Bats: A Systematic Review" Viruses 15, no. 9: 1860. https://doi.org/10.3390/v15091860






