The Human Cytomegalovirus Latency-Associated Gene Product Latency Unique Natural Antigen Regulates Latent Gene Expression
Abstract
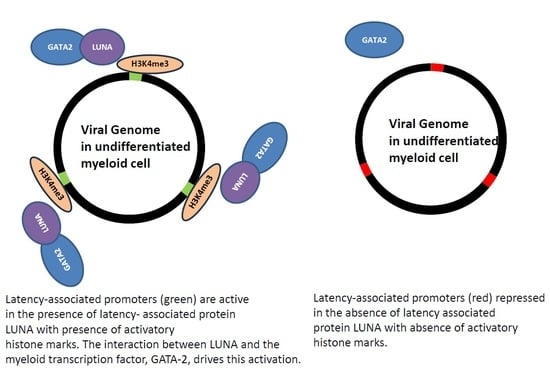
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Chromatin Immunoprecipitation
2.3. Immunoprecipitation (IP)
2.4. Drug Treatments
2.5. qRT-PCR
2.6. Droplet PCR
3. Results
3.1. LUNA Expression Promotes an Open Chromatin Conformation at Latent Promoters and Increased Latent Viral Gene Expression
3.2. LUNA deSUMOylase Motif Is Important for Binding to GATA-2 and Promoting Changes in Chromatin Structure
3.3. LUNA Expression Contributes to an Extended Lifespan in Infected CD14+ Monocytes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poole, E.; Wills, M.; Sinclair, J. Human Cytomegalovirus Latency: Targeting Differences in the Latently Infected Cell with a view to Clearing Latent Infection. New J. Sci. 2014, 2014, 10. [Google Scholar] [CrossRef]
- Sinclair, J.; Poole, E. Human cytomegalovirus latency and reactivation in and beyond the myeloid lineage. Future Virol. 2014, 6, 7. [Google Scholar] [CrossRef]
- Belzile, J.-P.; Stark, T.J.; Yeo, G.W.; Spector, D.H. Infection of primitive neural stem and progenitor cells by HCMV is restricted at several steps. J. Virol. 2014, 88, 4021–4039. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.; Stern-Ginossar, N. Rethinking human cytomegalovirus latency reservoir. Ann. N. Y. Acad. Sci. 2023, 1524, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, M.A.; Nelson, J.A. Human cytomegalovirus persistence and latency in endothelial cells and macrophages. Curr. Opin. Microbiol. 2002, 5, 403–407. [Google Scholar] [CrossRef]
- Sinclair, J.; Sissons, P. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 2006, 87, 1763–1779. [Google Scholar] [CrossRef]
- Poole, E.; Carlan da Silva, M.C.; Huang, C.; Perera, M.; Jackson, S.; Groves, I.J.; Wills, M.; Rana, A.; Sinclair, J. A BMPR2/YY1 Signaling Axis Is Required for Human Cytomegalovirus Latency in Undifferentiated Myeloid Cells. mBio 2021, 12, e0022721. [Google Scholar] [CrossRef]
- Dooley, A.L.; O’Connor, C.M. Regulation of the MIE Locus during HCMV Latency and Reactivation. Pathogens 2020, 9, 869. [Google Scholar] [CrossRef]
- Krishna, B.A.; Poole, E.L.; Jackson, S.E.; Smit, M.J.; Wills, M.R.; Sinclair, J.H. Latency-Associated Expression of Human Cytomegalovirus US28 Attenuates Cell Signaling Pathways To Maintain Latent Infection. mBio 2017, 8, 10–1128. [Google Scholar] [CrossRef]
- Weekes, M.P.; Tan, S.Y.; Poole, E.; Talbot, S.; Antrobus, R.; Smith, D.L.; Montag, C.; Gygi, S.P.; Sinclair, J.H.; Lehner, P.J. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 2013, 340, 199–202. [Google Scholar] [CrossRef]
- Goodrum, F.; Reeves, M.; Sinclair, J.; High, K.; Shenk, T. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 2007, 110, 937–945. [Google Scholar] [CrossRef]
- Poole, E.; Lau, J.C.; Sinclair, J. Latent infection of myeloid progenitors by human cytomegalovirus protects cells from FAS-mediated apoptosis through the cellular IL-10/PEA-15 pathway. J. Gen. Virol. 2015, 96, 2355–2359. [Google Scholar] [CrossRef]
- Poole, E.; Avdic, S.; Hodkinson, J.; Jackson, S.; Wills, M.; Slobedman, B.; Sinclair, J. Latency-associated viral interleukin-10 (IL-10) encoded by human cytomegalovirus modulates cellular IL-10 and CCL8 Secretion during latent infection through changes in the cellular microRNA hsa-miR-92a. J. Virol. 2014, 88, 13947–13955. [Google Scholar] [CrossRef] [PubMed]
- Poole, E.; McGregor Dallas, S.R.; Colston, J.; Joseph, R.S.; Sinclair, J. Virally induced changes in cellular microRNAs maintain latency of human cytomegalovirus in CD34 progenitors. J. Gen. Virol. 2011, 92, 1539–1549. [Google Scholar] [CrossRef]
- Pantaleao, S.Q.; Camillo, L.M.B.; Neves, T.C.; Menezes, I.G.; Stangherlin, L.M.; Batista, H.; Poole, E.; Nevels, M.; Philot, E.A.; Scott, A.L.; et al. Molecular modelling of the HCMV IL-10 protein isoforms and analysis of their interaction with the human IL-10 receptor. PLoS ONE 2022, 17, e0277953. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.K.; Abendroth, A.; Cunningham, A.L.; Slobedman, B. Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood 2006, 108, 3691–3699. [Google Scholar] [CrossRef] [PubMed]
- Beisser, P.S.; Laurent, L.; Virelizier, J.L.; Michelson, S. Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes. J. Virol. 2001, 75, 5949–5957. [Google Scholar] [CrossRef]
- Kotenko, S.V.; Saccani, S.; Izotova, L.S.; Mirochnitchenko, O.V.; Pestka, S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA 2000, 97, 1695–1700. [Google Scholar] [CrossRef]
- Mlera, L.; Moy, M.; Maness, K.; Tran, L.N.; Goodrum, F.D. The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation. Viruses 2020, 12, 714. [Google Scholar] [CrossRef]
- Bego, M.; Maciejewski, J.; Khaiboullina, S.; Pari, G.; St Jeor, S. Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J. Virol. 2005, 79, 11022–11034. [Google Scholar] [CrossRef]
- Keyes, L.R.; Hargett, D.; Soland, M.; Bego, M.G.; Rossetto, C.C.; Almeida-Porada, G.; St Jeor, S. HCMV protein LUNA is required for viral reactivation from latently infected primary CD14(+) cells. PLoS ONE 2013, 7, e52827. [Google Scholar] [CrossRef]
- Poole, E.L.; Kew, V.G.; Lau, J.C.H.; Murray, M.J.; Stamminger, T.; Sinclair, J.H.; Reeves, M.B. A Virally Encoded DeSUMOylase Activity Is Required for Cytomegalovirus Reactivation from Latency. Cell Rep. 2018, 24, 594–606. [Google Scholar] [CrossRef]
- Poole, E.; Walther, A.; Raven, K.; Benedict, C.A.; Mason, G.M.; Sinclair, J. The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner. J. Virol. 2013, 87, 4261–4271. [Google Scholar] [CrossRef]
- Reeves, M.; Woodhall, D.; Compton, T.; Sinclair, J. Human cytomegalovirus IE72 protein interacts with the transcriptional repressor hDaxx to regulate LUNA gene expression during lytic infection. J. Virol. 2010, 84, 7185–7194. [Google Scholar] [CrossRef] [PubMed]
- Poole, E.; Groves, I.; Jackson, S.; Wills, M.; Sinclair, J. Using Primary Human Cells to Analyze Human Cytomegalovirus Biology. Methods Mol. Biol. 2021, 2244, 51–81. [Google Scholar] [CrossRef]
- Aslam, Y.; Williamson, J.; Romashova, V.; Elder, E.; Krishna, B.; Wills, M.; Lehner, P.; Sinclair, J.; Poole, E. Human Cytomegalovirus Upregulates Expression of HCLS1 Resulting in Increased Cell Motility and Transendothelial Migration during Latency. iScience 2019, 20, 60–72. [Google Scholar] [CrossRef]
- Elder, E.; Krishna, B.; Williamson, J.; Aslam, Y.; Farahi, N.; Wood, A.; Romashova, V.; Roche, K.; Murphy, E.; Chilvers, E.; et al. Monocytes latently infected with human cytomegalovirus evade neutrophil killing. iScience 2019, 12, 13–26. [Google Scholar] [CrossRef]
- Poole, E.; King, C.; Sinclair, J.; Alcami, A. The UL144 gene product of human cytomegalovirus activates NF-kB via a TRAF6-dependent mechanism. EMBO J. 2006, 25, 4390–4399. [Google Scholar] [CrossRef]
- Jackson, S.E.; Sedikides, G.X.; Okecha, G.; Poole, E.L.; Sinclair, J.H.; Wills, M.R. Latent Cytomegalovirus (CMV) Infection Does Not Detrimentally Alter T Cell Responses in the Healthy Old, But Increased Latent CMV Carriage Is Related to Expanded CMV-Specific T Cells. Front. Immunol. 2017, 8, 733. [Google Scholar] [CrossRef] [PubMed]
- Poole, E.; Sinclair, J. Latency-associated upregulation of SERBP1 is important for the recruitment of transcriptional repressors to the viral major immediate early promoter of human cytomegalovirus during latent carriage. Front. Microbiol. 2022, 13, 999290. [Google Scholar] [CrossRef] [PubMed]
- Avdic, S.; McSharry, B.P.; Steain, M.; Poole, E.; Sinclair, J.; Abendroth, A.; Slobedman, B. Human Cytomegalovirus-Encoded Human Interleukin-10 (IL-10) Homolog Amplifies Its Immunomodulatory Potential by Upregulating Human IL-10 in Monocytes. J. Virol. 2016, 90, 3819–3827. [Google Scholar] [CrossRef] [PubMed]
- Nachtwey, J.; Spencer, J.V. HCMV IL-10 suppresses cytokine expression in monocytes through inhibition of nuclear factor-kappaB. Viral Immunol. 2008, 21, 477–482. [Google Scholar] [CrossRef]
- Jenkins, C.; Garcia, W.; Godwin, M.J.; Spencer, J.V.; Stern, J.L.; Abendroth, A.; Slobedman, B. Immunomodulatory properties of a viral homolog of human interleukin-10 expressed by human cytomegalovirus during the latent phase of infection. J. Virol. 2008, 82, 3736–3750. [Google Scholar] [CrossRef]
- Mason, G.M.; Jackson, S.; Okecha, G.; Poole, E.; Sissons, J.G.; Sinclair, J.; Wills, M.R. Human Cytomegalovirus Latency-Associated Proteins Elicit Immune-Suppressive IL-10 Producing CD4(+) T Cells. PLoS Pathog. 2013, 9, e1003635. [Google Scholar] [CrossRef] [PubMed]
- Mason, G.M.; Poole, E.; Sissons, J.G.; Wills, M.R.; Sinclair, J.H. Human cytomegalovirus latency alters the cellular secretome, inducing cluster of differentiation (CD)4+ T-cell migration and suppression of effector function. Proc. Natl. Acad. Sci. USA 2012, 109, 14538–14543. [Google Scholar] [CrossRef] [PubMed]
- Poole, E.; Neves, T.C.; Oliveira, M.T.; Sinclair, J.; da Silva, M.C.C. Human Cytomegalovirus Interleukin 10 Homologs: Facing the Immune System. Front. Cell Infect. Microbiol. 2020, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Buck, I.; Morceau, F.; Cristofanon, S.; Heintz, C.; Chateauvieux, S.; Reuter, S.; Dicato, M.; Diederich, M. Tumor necrosis factor alpha inhibits erythroid differentiation in human erythropoietin-dependent cells involving p38 MAPK pathway, GATA-1 and FOG-1 downregulation and GATA-2 upregulation. Biochem. Pharmacol. 2008, 76, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Svensson, E.C.; Huggins, G.S.; Dardik, F.B.; Polk, C.E.; Leiden, J.M. A functionally conserved N-terminal domain of the friend of GATA-2 (FOG-2) protein represses GATA4-dependent transcription. J. Biol. Chem. 2000, 275, 20762–20769. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.H.; Itoh, H.; Subramanian, L.; Iniguez-Lluhi, J.A.; Nakao, K. Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. Circ. Res. 2003, 92, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Everett, R.D.; Chelbi-Alix, M.K. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie 2007, 89, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Tavalai, N.; Stamminger, T. Interplay between Herpesvirus Infection and Host Defense by PML Nuclear Bodies. Viruses 2009, 1, 1240–1264. [Google Scholar] [CrossRef]
- Wagenknecht, N.; Reuter, N.; Scherer, M.; Reichel, A.; Muller, R.; Stamminger, T. Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1. Viruses 2015, 7, 2884–2907. [Google Scholar] [CrossRef]
- Saffert, R.T.; Penkert, R.R.; Kalejta, R.F. Cellular and viral control over the initial events of human cytomegalovirus experimental latency in CD34+ cells. J. Virol. 2013, 84, 5594–5604. [Google Scholar] [CrossRef]
- Soderberg-Naucler, C.; Fish, K.N.; Nelson, J.A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 1997, 91, 119–126. [Google Scholar] [CrossRef]
- Taylor-Wiedeman, J.; Sissons, P.; Sinclair, J. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 1994, 68, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Hahn, G.; Jores, R.; Mocarski, E.S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 1998, 95, 3937–3942. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.B.; Compton, T. Inhibition of inflammatory interleukin-6 activity via extracellular signal-regulated kinase-mitogen-activated protein kinase signaling antagonizes human cytomegalovirus reactivation from dendritic cells. J. Virol. 2011, 85, 12750–12758. [Google Scholar] [CrossRef]
- Hargett, D.; Shenk, T.E. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc. Natl. Acad. Sci. USA 2010, 107, 20039–20044. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poole, E.; Lau, J.; Groves, I.; Roche, K.; Murphy, E.; Carlan da Silva, M.; Reeves, M.; Sinclair, J. The Human Cytomegalovirus Latency-Associated Gene Product Latency Unique Natural Antigen Regulates Latent Gene Expression. Viruses 2023, 15, 1875. https://doi.org/10.3390/v15091875
Poole E, Lau J, Groves I, Roche K, Murphy E, Carlan da Silva M, Reeves M, Sinclair J. The Human Cytomegalovirus Latency-Associated Gene Product Latency Unique Natural Antigen Regulates Latent Gene Expression. Viruses. 2023; 15(9):1875. https://doi.org/10.3390/v15091875
Chicago/Turabian StylePoole, Emma, Jonathan Lau, Ian Groves, Kate Roche, Eain Murphy, Maria Carlan da Silva, Matthew Reeves, and John Sinclair. 2023. "The Human Cytomegalovirus Latency-Associated Gene Product Latency Unique Natural Antigen Regulates Latent Gene Expression" Viruses 15, no. 9: 1875. https://doi.org/10.3390/v15091875
APA StylePoole, E., Lau, J., Groves, I., Roche, K., Murphy, E., Carlan da Silva, M., Reeves, M., & Sinclair, J. (2023). The Human Cytomegalovirus Latency-Associated Gene Product Latency Unique Natural Antigen Regulates Latent Gene Expression. Viruses, 15(9), 1875. https://doi.org/10.3390/v15091875