Colorimetric RT-LAMP Detection of Multiple SARS-CoV-2 Variants and Lineages of Concern Direct from Nasopharyngeal Swab Samples without RNA Isolation
Abstract
:1. Introduction
2. Methods
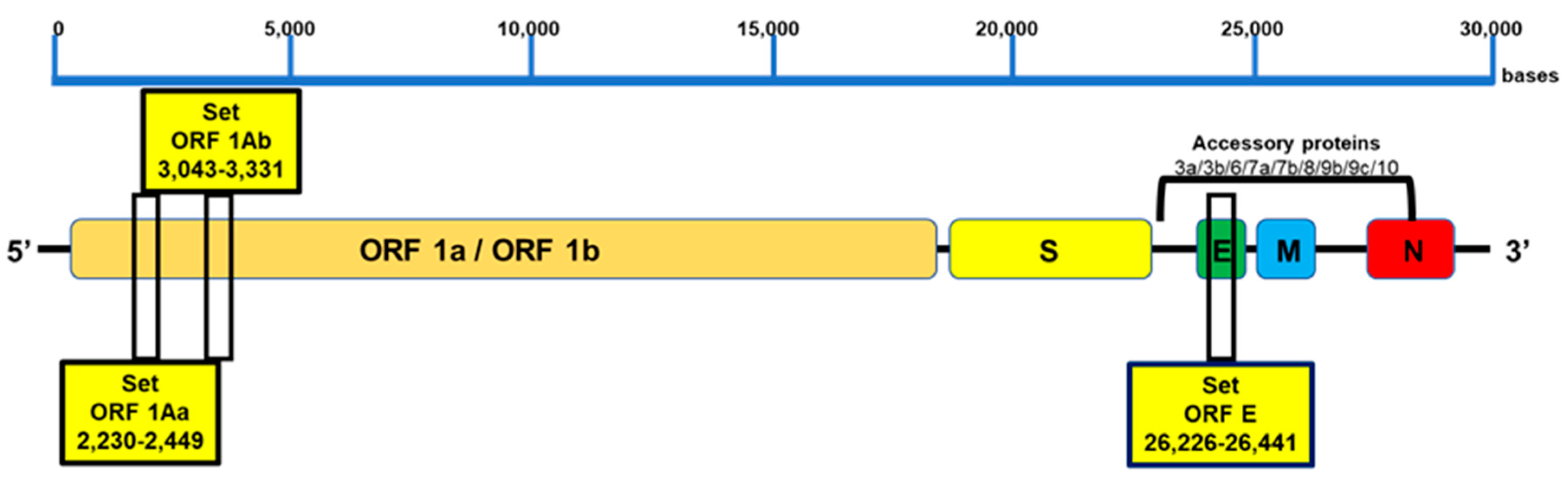
2.1. Design of RT-LAMP Primers for SARS-CoV-2 Genome Detection
2.2. Sample Inactivation and Treatment for Direct Assay
2.3. RT-LAMP Assay
2.4. Primer Cross-Reactivity (In Silico Sequence Analysis)
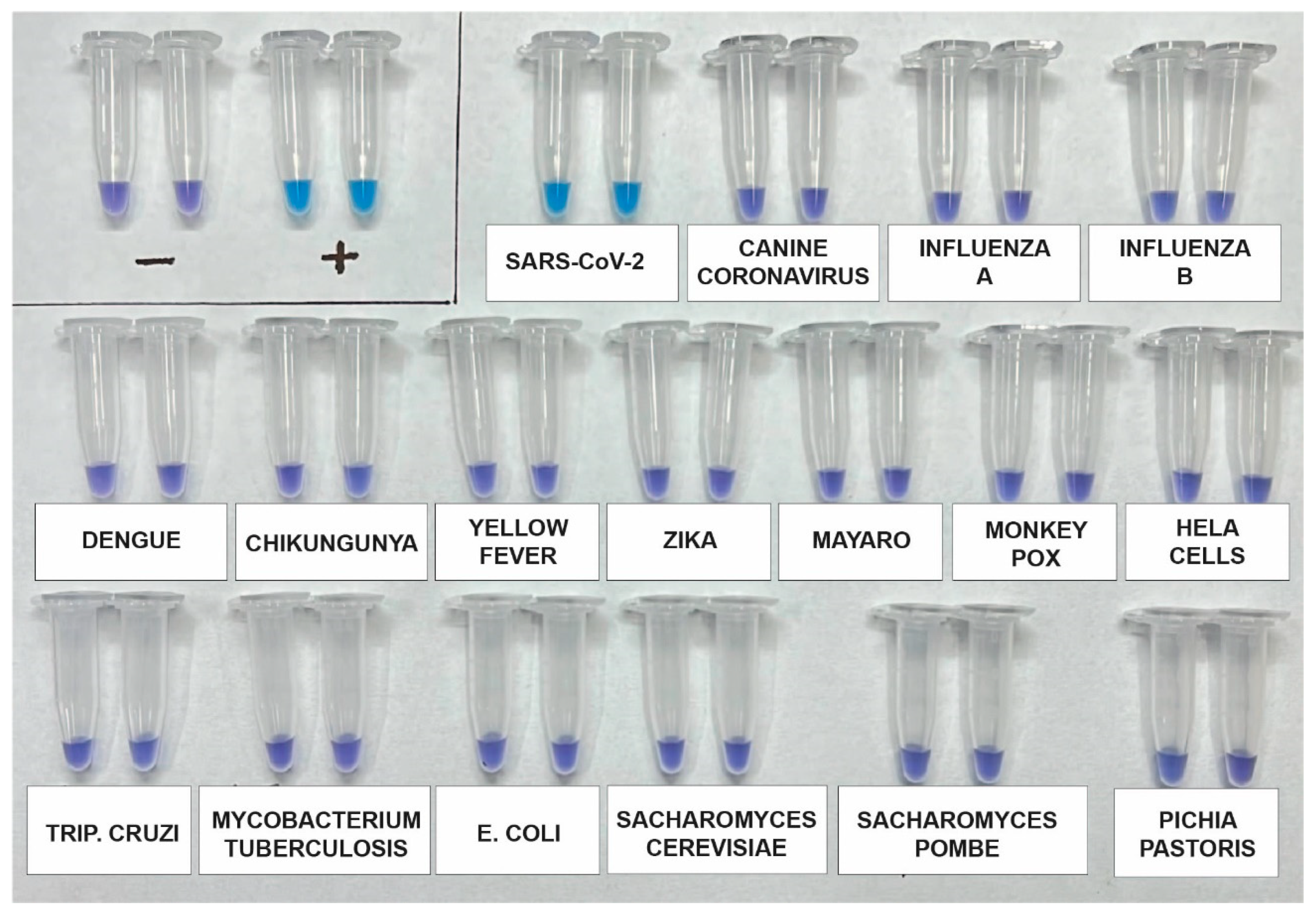
2.5. Analytical Specificity of the RT-LAMP Assay
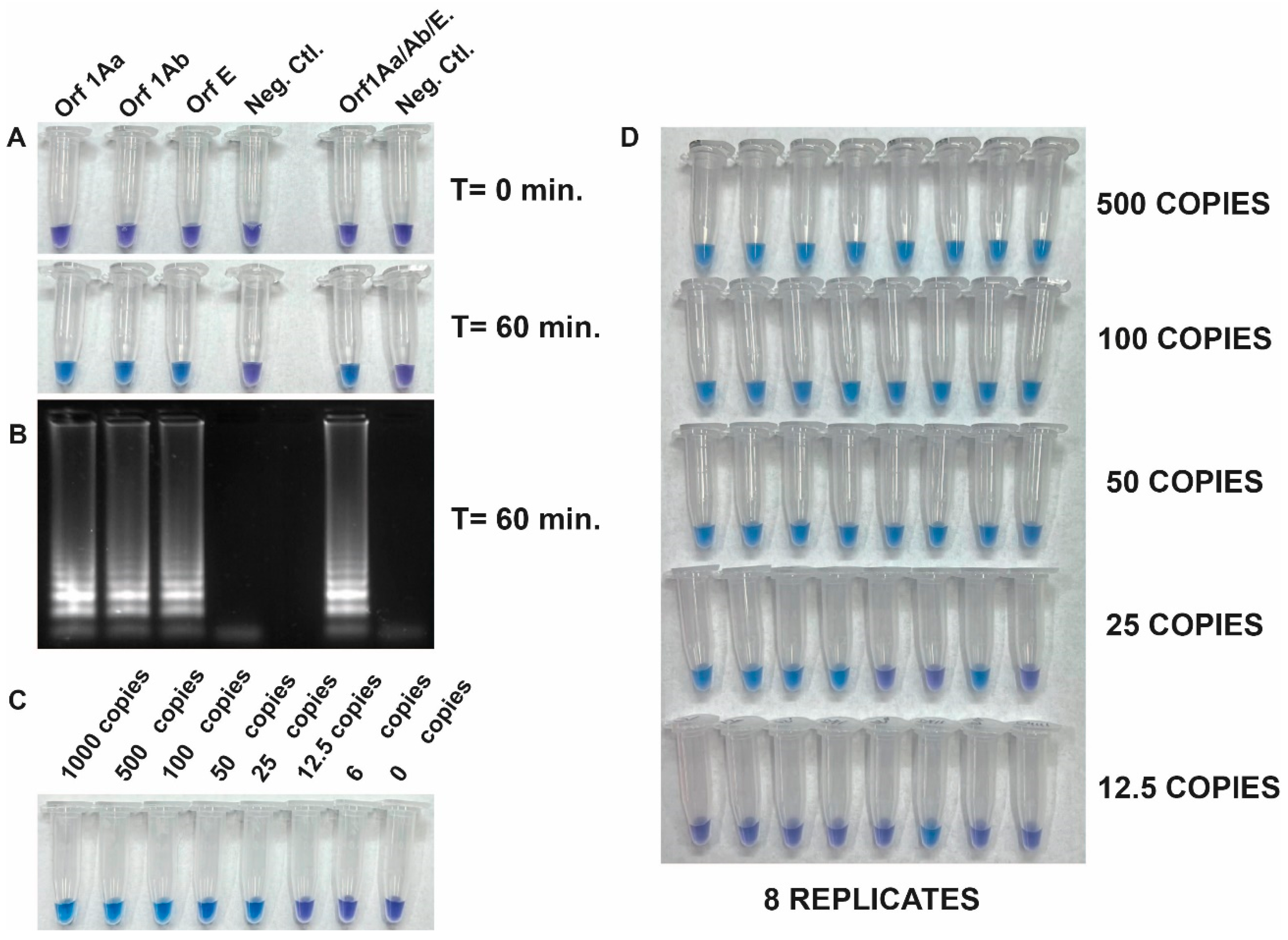
2.6. Repeatability, Reproducibility, and Detection Limit of the RT-LAMP Assay
2.7. Clinical Evaluation of the RT-LAMP Assay for SARS-CoV-2 Detection
3. Results
3.1. Design and Specificity of the ORF1aa, ORF1ab, and E Gene Primers for SARS-CoV-2 Target Sequence Identification
3.2. Evaluation of Primer Annealing with SARS-CoV-2 Variants and Mutations
3.3. Development of the RT-LAMP Assay and Evaluation of Its Analytical Sensitivity
3.4. Analytical Specificity
3.5. Clinical Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Approval
References
- Drosten, C.; Göttig, S.; Schilling, S.; Asper, M.; Panning, M.; Schmitz, H.; Günther, S. Rapid Detection and Quantification of RNA of Ebola and Marburg Viruses, Lassa Virus, Crimean-Congo Hemorrhagic Fever Virus, Rift Valley Fever Virus, Dengue Virus, and Yellow Fever Virus by Real-Time Reverse Transcription-PCR. J. Clin. Microbiol. 2002, 40, 2323–2330. [Google Scholar] [CrossRef]
- Espy, M.J.; Uhl, J.R.; Sloan, L.M.; Buckwalter, S.P.; Jones, M.F.; Vetter, E.A.; Yao, J.D.C.; Wengenack, N.L.; Rosenblatt, J.E.; Cockerill, F.R.; et al. Real-Time PCR in Clinical Microbiology: Applications for Routine Laboratory Testing. Clin. Microbiol. Rev. 2006, 19, 165–256. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Survey and summary survey and summary real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- Boehme, C.C.; Nabeta, P.; Henostroza, G.; Raqib, R.; Rahim, Z.; Gerhardt, M.; Sanga, E.; Hoelscher, M.; Notomi, T.; Hase, T.; et al. Operational Feasibility of Using Loop-Mediated Isothermal Amplification for Diagnosis of Pulmonary Tuberculosis in Microscopy Centers of Developing Countries. J. Clin. Microbiol. 2007, 45, 1936–1940. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-Mediated Isothermal Amplification (LAMP) of Gene Sequences and Simple Visual Detection of Products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Mori, Y.; Notomi, T. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Accurate, and Cost-Effective Diagnostic Method for Infectious Diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef]
- Law, J.W.F.; Mutalib, N.S.A.; Chan, K.G.; Lee, L.H. Rapid Metho Ds for the Detection of Foodborne Bacterial Pathogens: Principles, Applications, Advantages and Limitations. Front. Microbiol. 2014, 5, 770. [Google Scholar] [CrossRef]
- Thai, H.T.C.; Le, M.Q.; Vuong, C.D.; Parida, M.; Minekawa, H.; Notomi, T.; Hasebe, F.; Morita, K. Development and Evaluation of a Novel Loop-Mediated Isothermal Amplification Method for Rapid Detection of Severe Acute Respiratory Syndrome Coronavirus. J. Clin. Microbiol. 2004, 42, 1956–1961. [Google Scholar] [CrossRef]
- Reboud, J.; Xu, G.; Garrett, A.; Adriko, M.; Yang, Z.; Tukahebwa, E.M.; Rowell, C.; Cooper, J.M. Paper-based Microfluidics for DNA Diagnostics of Malaria in Low Resource Under Served Rural Communities. Proc. Natl. Acad. Sci. USA 2019, 116, 4834–4842. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated Reaction by Loop-Mediated Isothermal Amplification Using Loop Primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A Novel Coronavirus Outbreak of Global Health Concern Published. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Lu, Y.; Meng, Y.; Ye, Z.; Wang, L.; Wang, R.; Zheng, Q.; Wu, H.; Wu, J. Field Detection of Citrus Huanglongbing Associated with ‘ Candidatus Liberibacter Asiaticus’ by Recombinese Polymerase Amplification within 15 Min. J. Agric. Food Chem. 2018, 66, 5473–5480. [Google Scholar] [CrossRef]
- Zhao, Y.; Lu, H.; Thai, S.; Li, X.; Hui, J.; Tang, H.; Zhai, S.; Sun, L.; Wang, T. Development and Validation of an Algorithm to Identify Drug-Induced Anaphylaxis in the Beijing Pharmacovigilance Database. Int. J. Clin. Pharm. 2018, 40, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W. Principles and Applications of Loop-Mediated Isothermal Amplification to Point-of-Care Tests. Biosensors 2022, 12, 857. [Google Scholar] [CrossRef]
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y.; et al. Rapid and Visual Detection of 2019 Novel Coronavirus (SARS-CoV-2) by a Reverse Transcription Loop-Mediated Isothermal Amplification Assay. Clin. Microbiol. Infect. 2020, 26, 773–779. [Google Scholar] [CrossRef]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid Detection of Novel Coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Reverse Transcription-Loop-Mediated Isothermal Amplification. PLoS ONE 2020, 15, e0234682. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Qiu, Y.; Zheng, L.; Chen, J.; Yang, J. Development of a Visual Loop-Mediated Isothermal Amplification Method for Rapid Detection of the Bacterial Pathogen Pseudomonas putida of the Large Yellow Croaker (Pseudosciaena crocea). J. Microbiol. Methods 2012, 89, 179–184. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, Y.; Zhang, Z.; Chen, M.; Su, Y.; Yuan, Y.; Alam, M.J.; Yan, H.; Shi, L. Rapid Detection of Food-Borne Listeria Monocytogenes by Real-Time Quantitative Loop-Mediated Isothermal Amplification. Food Sci. Biotechnol. 2012, 21, 101–106. [Google Scholar] [CrossRef]
- Goto, M.; Honda, E.; Ogura, A.; Nomoto, A.; Hanaki, K.I. Colorimetric Detection of Loop-Mediated Isothermal Amplification Reaction by Using Hydroxy Naphthol Blue. Biotechniques 2009, 46, 167–172. [Google Scholar] [CrossRef]
- Prakash, S.; Priyatma; Aasarey, R.; Pandey, P.K.; Mathur, P.; Arulselvi, S. An Inexpensive and Rapid Diagnostic Method for Detection of SARS-CoV-2 RNA by Loop-Mediated Isothermal Amplification (LAMP). MethodsX 2023, 10, 102011. [Google Scholar] [CrossRef]
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M.; et al. RT-LAMP for Rapid Diagnosis of Coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Jiang, F.; Zhang, Y.; Li, Y.; Pang, B.; Liang, H.; Kou, Z.; Jiang, X.; Wen, H.; Xu, Y. SARS-CoV-2 Variant of Concern 202 012/01 (B.1.1.7) in a Traveller from the UK to China. J. Travel Med. 2021, 28, taab032. [Google Scholar] [CrossRef]
- Iacobucci, G. Covid-19: New UK Variant May Be Linked to Increased Death Rate, Early Data Indicate. BMJ 2021, 372, n230. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Emergence and Rapid Spread of a New Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-2) Lineage with Multiple Spike Mutations in South Africa. medRxiv 2020, 2. [Google Scholar] [CrossRef]
- Voloch, C.M.; da Silva Francisco, R., Jr.; de Almeida, L.G.P.; Cardoso, C.C.; Brustolini, O.J.; Gerber, A.L.; de C Guimarães, A.P.; Mariani, D.; da Costa, R.M.; Ferreira, O.C., Jr.; et al. Genomic Characterization of a Novel SARS-CoV-2 Lineage from Rio de Janeiro, Brazil. J. Virol. 2021, 95, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Long, S.W.; Olsen, R.J.; Christensen, P.A.; Subedi, S.; Olson, R.; Davis, J.J. Short Communication Sequence Analysis of 20,453 Severe Acute Respiratory Syndrome Coronavirus 2 Genomes from the Houston Metropolitan Area Identi Fi Es the Emergence and Widespread Distribution of Multiple Isolates of All Major Variants of Concern. Am. J. Pathol. 2021, 191, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Sun, Y.; Xu, H.; Ye, Q. The Emergence and Epidemic Characteristics of the Highly Mutated SARS-CoV-2 Omicron Variant. J. Med. Virol. 2022, 94, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bivins, A.; Smith, W.J.M.; Metcalfe, S.; Stephens, M.; Jennison, A.V.; Moore, F.A.J.; Bourke, J.; Schlebusch, S.; McMahon, J.; et al. Detection of the Omicron (B.1.1.529) Variant of SARS-CoV-2 in Aircraft Wastewater. Sci. Total Environ. 2022, 820, 153171. [Google Scholar] [CrossRef]
- Lyke, K.E.; Atmar, R.L.; Dominguez Islas, C.; Posavad, C.M.; Szydlo, D.; PaulChourdhury, R.; Deming, M.E.; Eaton, A.; Jackson, L.A.; Branche, A.R.; et al. Rapid Decline in Vaccine-Boosted Neutralizing Antibodies Against SARS-CoV-2 Omicron Variant. Cell Rep. Med. 2022, 3, 100679. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Huang, Y.; Lau, S.K.P.; Yuen, K.Y. Coronavirus Genomics and Bioinformatics Analysis. Viruses 2010, 2, 1805–1820. [Google Scholar] [CrossRef] [PubMed]
| Target | Primer | Sequence |
|---|---|---|
| ORF1aa | R1Aa-2F3 | TGCTTGTGAAATTGTCGGT |
| R1Aa-2B3 | GCCAGTTTCTTCTCTGGAT | |
| R1Aa-2FIP | TCAGCACACAAAGCCAAAAATTTAT-AAATTGTCACCTGTGCAAAG | |
| R1Aa-2BIP | TATTGGTGGAGCTAAACTTAAAGCC-ACACTTTCTGTACAATCCCTT | |
| R1Aa-2LP | CTTAAAGAATGTCTGAACACTCTCC | |
| R1Aa-2LB | GAATTTAGGTGAAACATTTGTCACG | |
| ORF1ab | R1Ab-1F3(R1AB) | TCCAGATGAGGATGAAGAAGA |
| R1Ab-1B3 | AGTCTGAACAACTGGTGTAAG | |
| R1Ab-1FIP | AGAGCAGCAGAAGTGGCACAGGTGATTGTGAAGAAGAAGAG | |
| R1Ab-1BIP | TCAACCTGAAGAAGAGCAAGAACTGATTGTCCTCACTGCC | |
| R1Ab-1LF | CTCATATTGAGTTGATGGCTCA | |
| R1Ab-1LB | ACAAACTGTTGGTCAACAAGAC | |
| ORF E | pE-3F3(PE3) | AGCTGATGAGTACGAACTT |
| pE-3B3 | TTCAGATTTTTAACACGAGAGT | |
| pE-3FIP | ACCACGAAAGCAAGAAAAAGAAGTATTCGTTTCGGAAGAGACAG | |
| pE-3BIP | TTGCTAGTTACACTAGCCATCCTTAGGTTTTACAAGACTCACGT | |
| pE-3LF | ACGCTATTAACTATTAACGTAC | |
| pE-3LB | CTGCGCTTCGATTGTGTGCGT |
| Reference Technique RT-PCR | Total | ||
|---|---|---|---|
| Positive Samples | Negative Samples | ||
| NEOKIT positives | 126 | 0 | 126 |
| NEOKIT negatives | 13 | 53 | 66 |
| TOTAL | 139 | 53 | 192 |
| True Positives (TP) = 126 (NEOKIT positives that were also positive in RT-qPCR). False Positives (FP) = 0 (NEOKIT positives that were negative in RT-qPCR). True Negatives (TN) = 53 (NEOKIT negatives that were also negative in RT-qPCR). False Negatives (FN) = 13 (NEOKIT negatives that were positive in RT-qPCR). PPV = TP/(TP + FP) = 126/(126 + 0) = 1.000 = 100%NPV = TN/(TN + FN) = 53/(53 + 13) = 0.803 = 80.3%Sensitivity = TP)/TP + FN = 126/(126 + 13) = 0.9067 Specificity = TN/TN + FP = 53/(53 + 0) = 1 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Werbajh, S.; Larocca, L.; Carrillo, C.; Stolowicz, F.; Ogas, L.; Pallotto, S.; Cassará, S.; Mammana, L.; Zapiola, I.; Bouzas, M.B.; et al. Colorimetric RT-LAMP Detection of Multiple SARS-CoV-2 Variants and Lineages of Concern Direct from Nasopharyngeal Swab Samples without RNA Isolation. Viruses 2023, 15, 1910. https://doi.org/10.3390/v15091910
Werbajh S, Larocca L, Carrillo C, Stolowicz F, Ogas L, Pallotto S, Cassará S, Mammana L, Zapiola I, Bouzas MB, et al. Colorimetric RT-LAMP Detection of Multiple SARS-CoV-2 Variants and Lineages of Concern Direct from Nasopharyngeal Swab Samples without RNA Isolation. Viruses. 2023; 15(9):1910. https://doi.org/10.3390/v15091910
Chicago/Turabian StyleWerbajh, Santiago, Luciana Larocca, Carolina Carrillo, Fabiana Stolowicz, Lorena Ogas, Sergio Pallotto, Solange Cassará, Liliana Mammana, Inés Zapiola, María Belén Bouzas, and et al. 2023. "Colorimetric RT-LAMP Detection of Multiple SARS-CoV-2 Variants and Lineages of Concern Direct from Nasopharyngeal Swab Samples without RNA Isolation" Viruses 15, no. 9: 1910. https://doi.org/10.3390/v15091910
APA StyleWerbajh, S., Larocca, L., Carrillo, C., Stolowicz, F., Ogas, L., Pallotto, S., Cassará, S., Mammana, L., Zapiola, I., Bouzas, M. B., & Vojnov, A. A. (2023). Colorimetric RT-LAMP Detection of Multiple SARS-CoV-2 Variants and Lineages of Concern Direct from Nasopharyngeal Swab Samples without RNA Isolation. Viruses, 15(9), 1910. https://doi.org/10.3390/v15091910





