Characterization and Comparative Genomic Analysis of a Deep-Sea Bacillus Phage Reveal a Novel Genus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phage Isolation and Purification
2.2. Transmission Electron Microscopy (TEM) Observation
2.3. One-Step Growth Curve
2.4. Phage Genomic DNA Extraction and Genome Sequencing
2.5. Phage Genome Assembly, Annotation, and Analysis
3. Results and Discussion
3.1. Phage Isolation and Characterization
3.2. Genomic Analysis of vB_BteM-A9Y
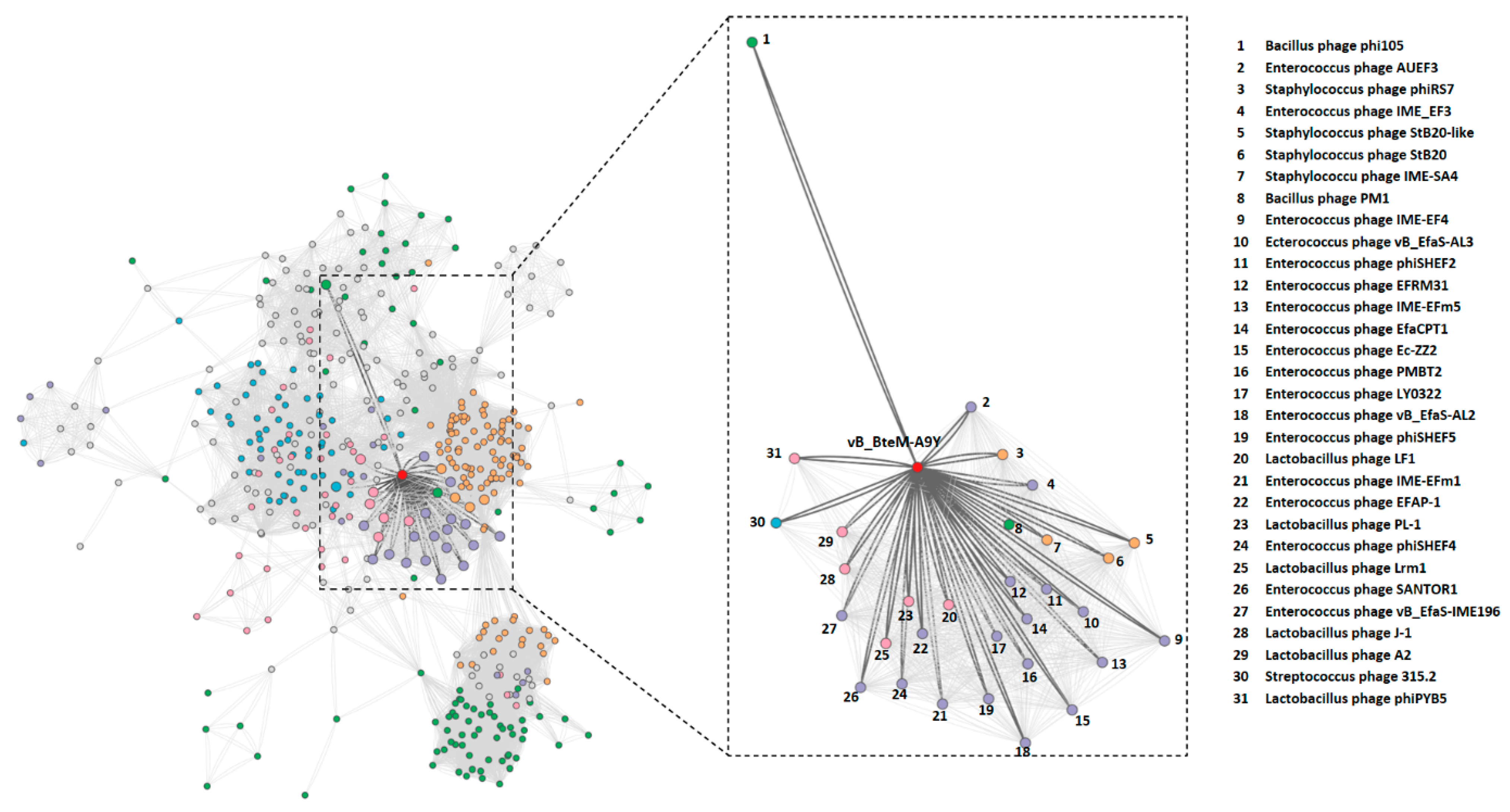
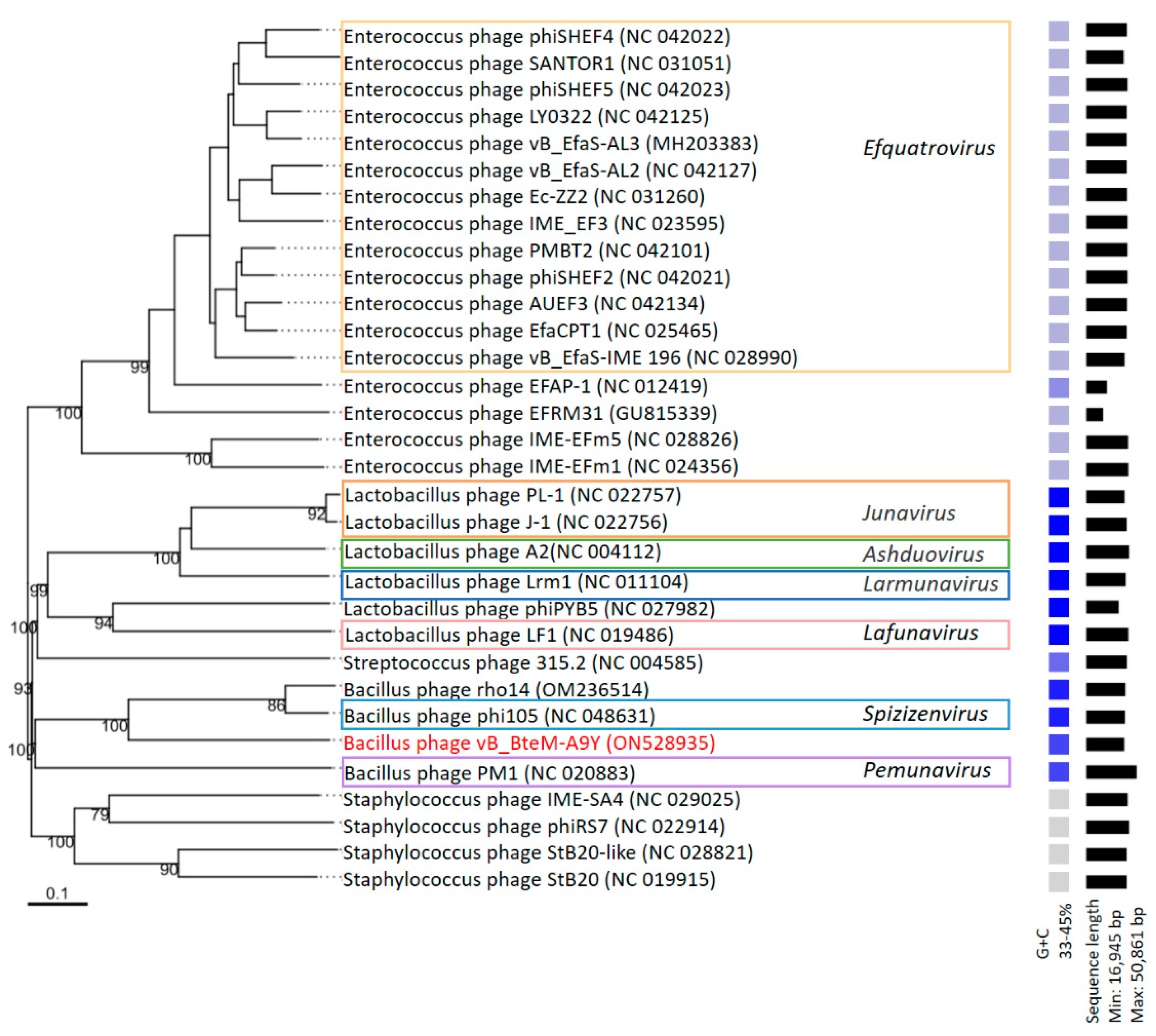
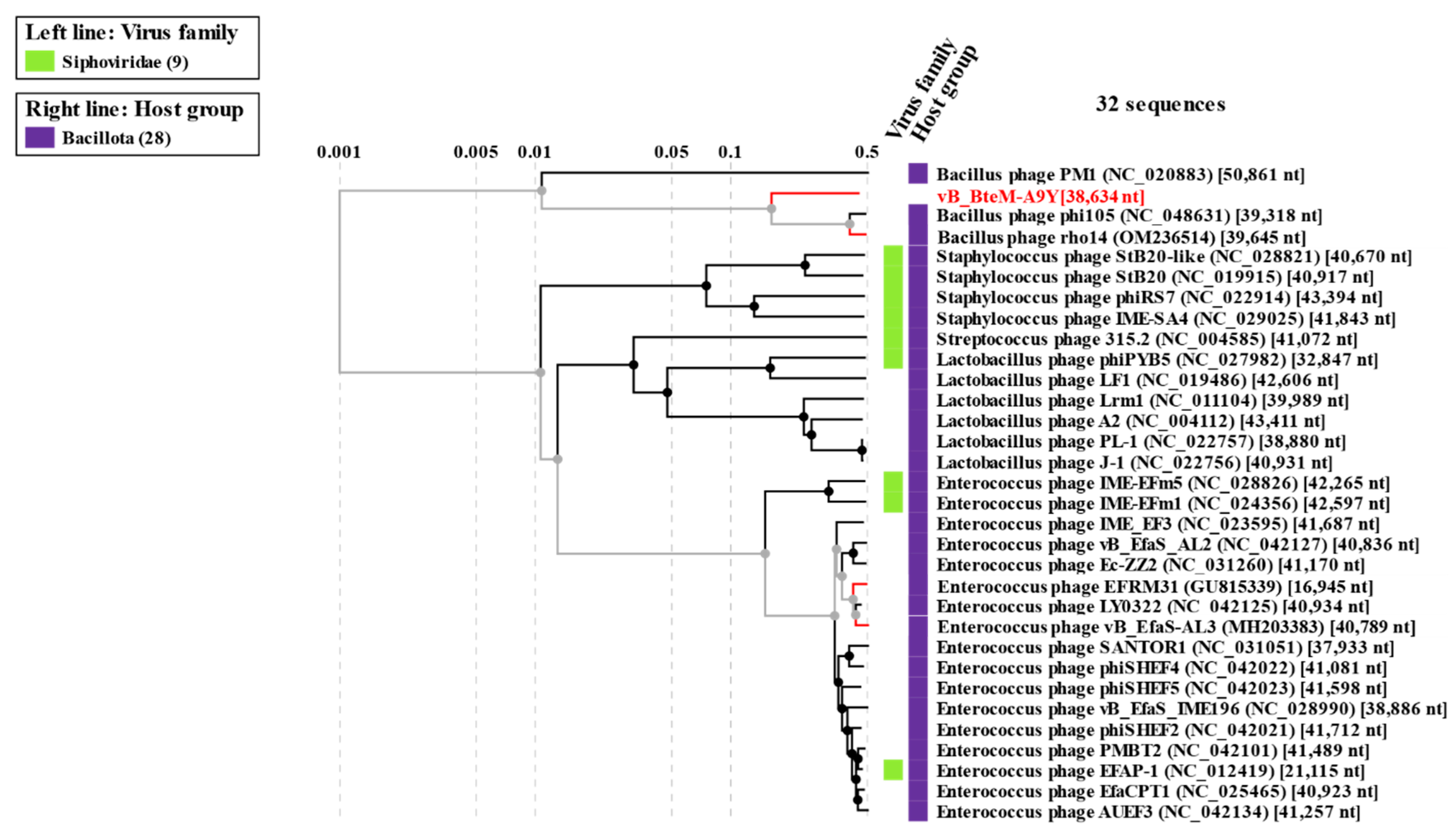
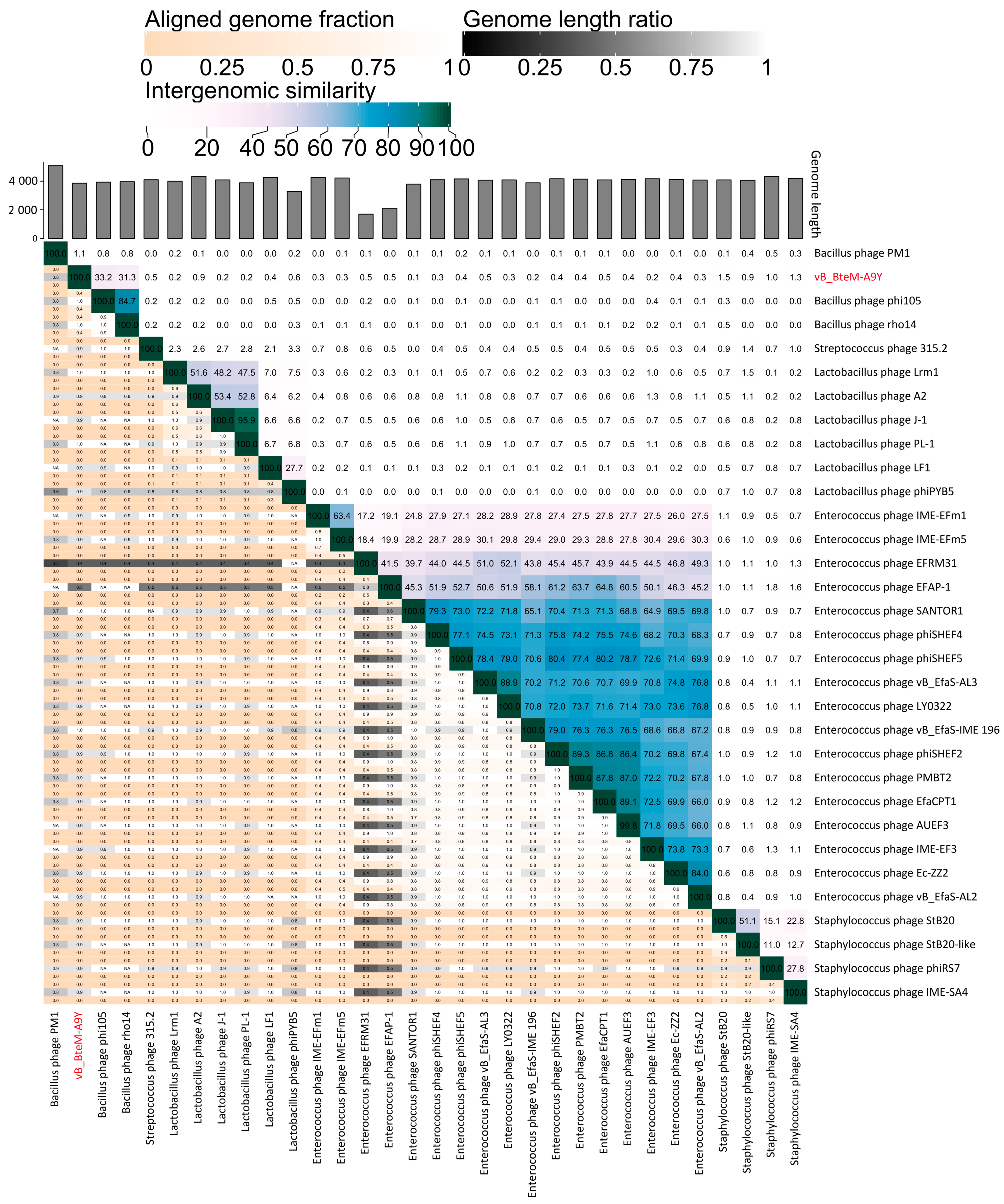
3.3. vB_BteM-A9Y Belongs to a Novel Genus under Caudoviricetes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breitbart, M. Marine Viruses: Truth or Dare. Annu. Rev. Mar. Sci. 2012, 4, 425–448. [Google Scholar] [CrossRef]
- Suttle, C.A. Viruses in the Sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef]
- Paul, J.H. Use of Hoechst Dyes 33258 and 33342 for Enumeration of Attached and Planktonic Bacteria. Appl. Environ. Microbiol. 1982, 43, 939–944. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Rassoulzadegan, F. Are Viruses Driving Microbial Diversification and Diversity? Environ. Microbiol. 2004, 6, 1–11. [Google Scholar] [CrossRef]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in Aquatic Ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. Available online: https://journals.asm.org/doi/10.1128/mmbr.64.1.69-114.2000 (accessed on 27 June 2023). [CrossRef]
- Suttle, C.A. Marine Viruses—Major Players in the Global Ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Rohwer, F.; Prangishvili, D.; Lindell, D. Roles of Viruses in the Environment. Environ. Microbiol. 2009, 11, 2771–2774. [Google Scholar] [CrossRef]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de Vargas, C.; Gasol, J.M.; et al. Patterns and Ecological Drivers of Ocean Viral Communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef]
- Paez-Espino, D.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Thomas, A.D.; Huntemann, M.; Mikhailova, N.; Rubin, E.; Ivanova, N.N.; Kyrpides, N.C. Uncovering Earth’s Virome. Nature 2016, 536, 425–430. Available online: https://www.nature.com/articles/nature19094 (accessed on 27 June 2023). [CrossRef]
- Roux, S.; Hallam, S.J.; Woyke, T.; Sullivan, M.B. Viral Dark Matter and Virus–Host Interactions Resolved from Publicly Available Microbial Genomes. eLife 2015, 4, e08490. Available online: https://elifesciences.org/articles/08490 (accessed on 27 June 2023). [CrossRef] [PubMed]
- Chen, Y.; Guo, X.; Wu, J.; Jin, M.; Zeng, R. A Novel Deep-Sea Bacteriophage Possesses Features of Wbeta-Like Viruses and Prophages. Arch. Virol. 2020, 165, 1219–1223. Available online: https://link.springer.com/article/10.1007/s00705-020-04579-6 (accessed on 27 June 2023). [CrossRef] [PubMed]
- Gatson, J.W.; Benz, B.F.; Chandrasekaran, C.; Satomi, M.; Venkateswaran, K.; Hart, M.E. Bacillus tequilensis sp. nov., Isolated from a 2000-Year-Old Mexican Shaft-Tomb, Is Closely Related to Bacillus Subtilis. Int. J. Syst. Evol. Microbiol. 2006, 56, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Shafique, M.; Nawaz, H.R.; Jabeen, N.; Naz, S.A. Bacillus Tequilensis ZMS-2: A Novel Source of Alkaline Protease with Antimicrobial, Anti-Coagulant, Fibrinolytic and Dehairing Potentials. Pak. J. Pharm. Sci. 2019, 32, 1913–1918. [Google Scholar] [PubMed]
- Wang, J.; Niu, C.; Liu, X.; Chen, X.; Li, Q. Characterization of a New 1,3-1,4-β-Glucanase Gene from Bacillus Tequilensis CGX5-1. Appl. Biochem. Biotechnol. 2014, 173, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Chiliveri, S.R.; Linga, V.R. A Novel Thermostable, Alkaline Pectate Lyase from Bacillus tequilensis SV11 with Potential in Textile Industry. Carbohydr. Polym. 2014, 111, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Shukla, N.; Mishra, P.; Gaur, R. Enhanced Production and Characterization of a Solvent Stable Amylase from Solvent Tolerant Bacillus tequilensis RG-01: Thermostable and Surfactant Resistant. Sci. World J. 2014, 2014, 972763. Available online: https://www.hindawi.com/journals/tswj/2014/972763/ (accessed on 27 June 2023). [CrossRef]
- Dai, J.; Dong, A.; Xiong, G.; Liu, Y.; Hossain, M.S.; Liu, S.; Gao, N.; Li, S.; Wang, J.; Qiu, D. Production of Highly Active Extracellular Amylase and Cellulase from Bacillus subtilis ZIM3 and a Recombinant Strain with a Potential Application in Tobacco Fermentation. Front. Microbiol. 2020, 11, 1539. Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01539/full (accessed on 27 June 2023). [CrossRef]
- Sondhi, S.; Sharma, P.; George, N.; Chauhan, P.S.; Puri, N.; Gupta, N. An Extracellular Thermo-Alkali-Stable Laccase from Bacillus tequilensis SN4, with a Potential to Biobleach Softwood Pulp. 3 Biotech 2015, 5, 175. [Google Scholar] [CrossRef]
- Rani, R.P.; Anandharaj, M.; Sabhapathy, P.; Ravindran, A.D. Physiochemical and Biological Characterization of Novel Exopolysaccharide Produced by Bacillus tequilensis FR9 Isolated from Chicken. Int. J. Biol. Macromol. 2017, 96, 1–10. [Google Scholar] [CrossRef]
- Pradhan, A.K.; Pradhan, N.; Mall, G.; Panda, H.T.; Sukla, L.B.; Panda, P.K.; Mishra, B.K. Application of Lipopeptide Biosurfactant Isolated from a Halophile: Bacillus tequilensis CH for Inhibition of Biofilm. Appl. Biochem. Biotechnol. 2013, 171, 1362–1375. [Google Scholar] [CrossRef]
- Pradhan, A.K.; Rath, A.; Pradhan, N.; Hazra, R.K.; Nayak, R.R.; Kanjilal, S. Cyclic Lipopeptide Biosurfactant from Bacillus Tequilensis Exhibits Multifarious Activity. 3 Biotech 2018, 8, 261. [Google Scholar] [CrossRef]
- Kropinski, A.M. Practical Advice on the One-Step Growth Curve. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Lavigne, R., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 3, pp. 41–47. ISBN 978-1-4939-7343-9. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z.; et al. Genome Sequencing in Open Microfabricated High Density Picoliter Reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. GigaScience 2012, 1, 18. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.-C.; Bikard, D.; Monot, M. PhageTerm: A Tool for Fast and Accurate Determination of Phage Termini and Packaging Mechanism Using next-Generation Sequencing Data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinforma. Oxf. Engl. 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A Self-Training Method for Prediction of Gene Starts in Microbial Genomes. Implications for Finding Sequence Motifs in Regulatory Regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.; Tavares, P.; Petit, M.-A.; Guérois, R.; Zinn-Justin, S. Automated Classification of Tailed Bacteriophages According to Their Neck Organization. BMC Genom. 2014, 15, 1027. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-Line: Integrating Search and Context for Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Paez-Espino, D.; Chen, I.-M.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Szeto, E.; Pillay, M.; Huang, J.; Markowitz, V.M.; Nielsen, T.; et al. IMG/VR: A Database of Cultured and Uncultured DNA Viruses and Retroviruses. Nucleic Acids Res. 2017, 45, D457–D465. [Google Scholar] [CrossRef]
- Bin Jang, H.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Brister, J.R.; Kropinski, A.M.; Krupovic, M.; Lavigne, R.; et al. Taxonomic Assignment of Uncultivated Prokaryotic Virus Genomes Is Enabled by Gene-Sharing Networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-Based Phylogeny and Classification of Prokaryotic Viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequences (RefSeq): A Curated Non-Redundant Sequence Database of Genomes, Transcripts and Proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef]
- Blakely, G.W. A Genetic Switch, Third Edition, Phage Lambda Revisited. M. Ptashne. Cold Spring Harbor Laboratory Press. 2004. 154 Pages. ISBN 0 87969 716 4. Price $39 (Paperback). Genet. Res. 2004, 84, 193–194. [Google Scholar] [CrossRef]
- Earl, A.M.; Mohundro, M.M.; Mian, I.S.; Battista, J.R. The IrrE Protein of Deinococcus radiodurans R1 Is a Novel Regulator of recA Expression. J. Bacteriol. 2002, 184, 6216–6224. [Google Scholar] [CrossRef] [PubMed]
- Narumi, I.; Satoh, K.; Cui, S.; Funayama, T.; Kitayama, S.; Watanabe, H. PprA: A Novel Protein from Deinococcus radiodurans That Stimulates DNA Ligation. Mol. Microbiol. 2004, 54, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Earl, A.M.; Howell, H.A.; Park, M.-J.; Eisen, J.A.; Peterson, S.N.; Battista, J.R. Analysis of Deinococcus radiodurans’s Transcriptional Response to Ionizing Radiation and Desiccation Reveals Novel Proteins That Contribute to Extreme Radioresistance. Genetics 2004, 168, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Vujičić-Žagar, A.; Dulermo, R.; Le Gorrec, M.; Vannier, F.; Servant, P.; Sommer, S.; de Groot, A.; Serre, L. Crystal Structure of the IrrE Protein, a Central Regulator of DNA Damage Repair in Deinococcaceae. J. Mol. Biol. 2009, 386, 704–716. [Google Scholar] [CrossRef] [PubMed]
- Akroyd, J.E.; Clayson, E.; Higgins, N.P. Purification of the Gam Gene-Product of Bacteriophage Mu and Determination of the Nucleotide Sequence of the Gam Gene. Nucleic Acids Res. 1986, 14, 6901–6914. [Google Scholar] [CrossRef] [PubMed]
- Strack, B.; Lessl, M.; Calendar, R.; Lanka, E. A Common Sequence Motif, -E-G-Y-A-T-A-, Identified within the Primase Domains of Plasmid-Encoded I- and P-Type DNA Primases and the Alpha Protein of the Escherichia Coli Satellite Phage P4. J. Biol. Chem. 1992, 267, 13062–13072. [Google Scholar] [CrossRef]
- Westerveld, A.; Hoeijmakers, J.H.J.; van Duin, M.; de Wit, J.; Odijk, H.; Pastink, A.; Wood, R.D.; Bootsma, D. Molecular Cloning of a Human DNA Repair Gene. Nature 1984, 310, 425–429. [Google Scholar] [CrossRef]
- Busch, D.; Greiner, C.; Lewis, K.; Ford, R.; Adair, G.; Thompson, L. Summary of Complementation Groups of UV-Sensitive CHO Cell Mutants Isolated by Large-Scale Screening. Mutagenesis 1989, 4, 349–354. [Google Scholar] [CrossRef]
- Moodley, S.; Maxwell, K.L.; Kanelis, V. The Protein Gp74 from the Bacteriophage HK97 Functions as a HNH Endonuclease. Protein Sci. Publ. Protein Soc. 2012, 21, 809–818. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, D.; Huang, Y.; Zhu, X.; Rui, M.; Wan, T.; Zheng, X.; Shen, Y.; Chen, X.; Ma, K.; et al. Structural and Functional Characterization of Deep-Sea Thermophilic Bacteriophage GVE2 HNH Endonuclease. Sci. Rep. 2017, 7, 42542. [Google Scholar] [CrossRef]
- Xu, S.; Gupta, Y.K. Natural Zinc Ribbon HNH Endonucleases and Engineered Zinc Finger Nicking Endonuclease. Nucleic Acids Res. 2013, 41, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, B.J.; Hayes, J.A.; Stone, N.P.; Duffy, C.M.; Sankaran, B.; Kelch, B.A. Structure and Mechanism of the ATPase That Powers Viral Genome Packaging. Proc. Natl. Acad. Sci. USA 2015, 112, E3792–E3799. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Bajpai, U. Functional Characterization of a DNA-Dependent AAA ATPase in a F-Cluster Mycobacteriophage. Virus Res. 2022, 323, 198957. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fan, H.; An, X.; Fan, H.; Jiang, H.; Chen, Y.; Tong, Y. Scrutinizing Virus Genome Termini by High-Throughput Sequencing. PLoS ONE 2014, 9, e85806. [Google Scholar] [CrossRef]
- Huang, R.K.; Khayat, R.; Lee, K.K.; Gertsman, I.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E. The Prohead-I Structure of Bacteriophage HK97: Implications for Scaffold-Mediated Control of Particle Assembly and Maturation. J. Mol. Biol. 2011, 408, 541–554. [Google Scholar] [CrossRef]
- Conway, J.F.; Duda, R.L.; Cheng, N.; Hendrix, R.W.; Steven, A.C. Proteolytic and Conformational Control of Virus Capsid Maturation: The Bacteriophage HK97 System. J. Mol. Biol. 1995, 253, 86–99. [Google Scholar] [CrossRef]
- Cardarelli, L.; Lam, R.; Tuite, A.; Baker, L.A.; Sadowski, P.D.; Radford, D.R.; Rubinstein, J.L.; Battaile, K.P.; Chirgadze, N.; Maxwell, K.L.; et al. The Crystal Structure of Bacteriophage HK97 Gp6: Defining a Large Family of Head–Tail Connector Proteins. J. Mol. Biol. 2010, 395, 754–768. [Google Scholar] [CrossRef]
- Lhuillier, S.; Gallopin, M.; Gilquin, B.; Brasilès, S.; Lancelot, N.; Letellier, G.; Gilles, M.; Dethan, G.; Orlova, E.V.; Couprie, J.; et al. Structure of Bacteriophage SPP1 Head-to-Tail Connection Reveals Mechanism for Viral DNA Gating. Proc. Natl. Acad. Sci. USA 2009, 106, 8507–8512. [Google Scholar] [CrossRef]
- Wang, I.-N.; Smith, D.L.; Young, R. Holins: The Protein Clocks of Bacteriophage Infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a Widely Distributed Protein Motif for Binding to (Peptido)Glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef]
- Quiles-Puchalt, N.; Tormo-Más, M.Á.; Campoy, S.; Toledo-Arana, A.; Monedero, V.; Lasa, Í.; Novick, R.P.; Christie, G.E.; Penadés, J.R. A Super-Family of Transcriptional Activators Regulates Bacteriophage Packaging and Lysis in Gram-Positive Bacteria. Nucleic Acids Res. 2013, 41, 7260–7275. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Edwards, R.; Nash, J.H.E.; Mahadevan, P.; Seto, D.; Ackermann, H.-W.; Lavigne, R.; Kropinski, A.M. Integration of Genomic and Proteomic Analyses in the Classification of the Siphoviridae Family. Virology 2015, 477, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef] [PubMed]




| vB_BteM-A9Y | |
|---|---|
| Genome length (bp) | 38,634 |
| GC content (%) | 41.05% |
| Predicted ORF number | 54 |
| ORF total length (bp) | 35,436 |
| ORF length maximum (bp) | 3777 |
| ORF length minimum (bp) | 111 |
| ORF average length (bp) | 656 |
| ORF density (number/kb) | 1.397 |
| ORF/genome (%) | 91.72 |
| Predicted tRNA number | 0 |
| Predicted CRISPR protospacer number | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Zhang, T.; Lai, Q.; Zhang, M.; Yu, M.; Zeng, R.; Jin, M. Characterization and Comparative Genomic Analysis of a Deep-Sea Bacillus Phage Reveal a Novel Genus. Viruses 2023, 15, 1919. https://doi.org/10.3390/v15091919
Chen Y, Zhang T, Lai Q, Zhang M, Yu M, Zeng R, Jin M. Characterization and Comparative Genomic Analysis of a Deep-Sea Bacillus Phage Reveal a Novel Genus. Viruses. 2023; 15(9):1919. https://doi.org/10.3390/v15091919
Chicago/Turabian StyleChen, Yuan, Tianyou Zhang, Qiliang Lai, Menghui Zhang, Meishun Yu, Runying Zeng, and Min Jin. 2023. "Characterization and Comparative Genomic Analysis of a Deep-Sea Bacillus Phage Reveal a Novel Genus" Viruses 15, no. 9: 1919. https://doi.org/10.3390/v15091919
APA StyleChen, Y., Zhang, T., Lai, Q., Zhang, M., Yu, M., Zeng, R., & Jin, M. (2023). Characterization and Comparative Genomic Analysis of a Deep-Sea Bacillus Phage Reveal a Novel Genus. Viruses, 15(9), 1919. https://doi.org/10.3390/v15091919





