Development and Implementation of a Quadruple RT-qPCR Method for the Identification of Porcine Reproductive and Respiratory Syndrome Virus Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses and Clinical Samples
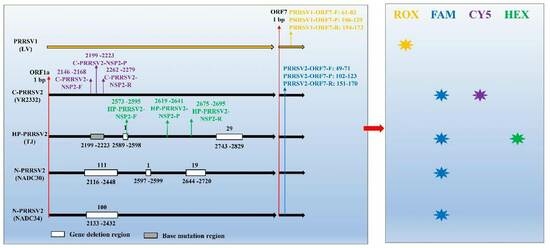
2.2. Primers and Probes
2.3. RNA Extraction and Reverse Transcription
2.4. Establishment of the Standard Curve
2.5. Optimization of Amplification Conditions
2.6. Specificity, Sensitivity, Repeatability, and Stability
2.7. The Standard Curve and the LOD of This Multiplex Amplification
2.8. Simulation of Clinical Sample
2.9. Comparison with Referent RT-qPCR
2.10. Clinical Application
3. Results
3.1. Designation of Primers and Probes
3.2. Identification of Recombinant Plasmids
3.3. Optimization of Reaction Conditions
3.3.1. Annealing Temperature
3.3.2. Optimal Primer and Probe Concentrations
3.4. Establishment of Standard Curve
3.5. Specificity Tests
3.6. Sensitivity Test
3.7. Repeatability Test
3.8. Standard Curve and LOD
3.9. LODs of Clinical Simulants
3.10. Comparison between the National Reference Method and RT-qPCR Methods
3.11. Clinical Application
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, N.; Cao, Z.; Yu, X.; Deng, X.; Zhao, T.; Wang, L.; Liu, Q.; Li, X.; Tian, K. Emergence of novel European genotype porcine reproductive and respiratory syndrome virus in mainland China. J. Gen. Virol. 2011, 92, 880–892. [Google Scholar] [CrossRef]
- Ming, S.; Yongying, M.; Bohua, L.; Huiying, L.; Xiaoyu, D.; Qiaorong, L.; Mingming, Q.; Xi, C.; Xinyan, Y.; Xizhao, C. Pathogenic Characterization of European Genotype Porcine Reproductive and Respiratory Syndrome Virus Recently Isolated in Mainland China. Open Virol. J. 2017, 11, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yu, X.; He, D.; Ku, X.; Hong, B.; Zeng, W.; Zhang, H.; He, Q. Investigation and analysis of etiology associated with porcine respiratory disease complex in China from 2017 to 2021. Front. Vet. Sci. 2022, 9, 960033. [Google Scholar] [CrossRef] [PubMed]
- Wensvoort, G.; de Kluyver, E.P.; Pol, J.M.; Wagenaar, F.; Moormann, R.J.; Hulst, M.M.; Bloemraad, R.; den Besten, A.; Zetstra, T.; Terpstra, C. Lelystad virus, the cause of porcine epidemic abortion and respiratory syndrome: A review of mystery swine disease research at Lelystad. Vet. Microbiol. 1992, 33, 185–193. [Google Scholar] [CrossRef]
- Gao, J.C.; Xiong, J.Y.; Ye, C.; Chang, X.B.; Guo, J.C.; Jiang, C.G.; Zhang, G.H.; Tian, Z.J.; Cai, X.H.; Tong, G.Z.; et al. Genotypic and geographical distribution of porcine reproductive and respiratory syndrome viruses in mainland China in 1996–2016. Vet. Microbiol. 2017, 208, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Music, N.; Gagnon, C.A. The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim. Health Res. Rev. 2010, 11, 135–163. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, C.; Xu, Y.; Yang, Y.; Li, J.; Dai, A.; Huang, C.; Luo, M.; Wei, C. Molecular Characteristics and Pathogenicity of a Novel Recombinant Porcine Reproductive and Respiratory Syndrome Virus Strain from NADC30-, NADC34-, and JXA1-Like Strains That Emerged in China. Microbiol. Spectr. 2022, 10, e266722. [Google Scholar] [CrossRef]
- Zhao, K.; Ye, C.; Chang, X.B.; Jiang, C.G.; Wang, S.J.; Cai, X.H.; Tong, G.Z.; Tian, Z.J.; Shi, M.; An, T.Q. Importation and Recombination Are Responsible for the Latest Emergence of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus in China. J. Virol. 2015, 89, 10712–10716. [Google Scholar] [CrossRef]
- Sun, Q.; Xu, H.; Li, C.; Gong, B.; Li, Z.; Tian, Z.J.; Zhang, H. Emergence of a novel PRRSV-1 strain in mainland China: A recombinant strain derived from the two commercial modified live viruses Amervac and DV. Front. Vet. Sci. 2022, 9, 974743. [Google Scholar] [CrossRef]
- Chen, N.; Xiao, Y.; Ye, M.; Li, X.; Li, S.; Xie, N.; Wei, Y.; Wang, J.; Zhu, J. High genetic diversity of Chinese porcine reproductive and respiratory syndrome viruses from 2016 to 2019. Res. Vet. Sci. 2020, 131, 38–42. [Google Scholar] [CrossRef]
- Kick, A.R.; Amaral, A.F.; Frias-De-Diego, A.; Cortes, L.M.; Fogle, J.E.; Crisci, E.; Almond, G.W.; Kaser, T. The Local and Systemic Humoral Immune Response Against Homologous and Heterologous Strains of the Type 2 Porcine Reproductive and Respiratory Syndrome Virus. Front. Immunol. 2021, 12, 637613. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bai, J.; Sun, Y.; Liu, X.; Gao, Y.; Wang, X.; Yang, Y.; Jiang, P. Comparison of pathogenicity of different subgenotype porcine reproductive and respiratory syndrome viruses isolated in China. Microb. Pathog. 2022, 168, 105607. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Kleiboeker, S.B.; Schommer, S.K.; Lee, S.M.; Watkins, S.; Chittick, W.; Polson, D. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. J. Vet. Diagn. Investig. 2005, 17, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Tian, Y.; Zhou, D.; Duan, Z.; Guo, R.; Liu, Z.; Yuan, F.; Liu, W. A Multiplex RT-PCR Assay to Detect and Discriminate Porcine Reproductive and Respiratory Syndrome Viruses in Clinical Specimens. Viruses 2017, 9, 205. [Google Scholar] [CrossRef]
- Egli, C.; Thur, B.; Liu, L.; Hofmann, M.A. Quantitative TaqMan RT-PCR for the detection and differentiation of European and North American strains of porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2001, 98, 63–75. [Google Scholar] [CrossRef]
- GB/T 1809-2023; Diagnostic Techniques for Porcine Reproductive and Respiratory Syndrome. Standards Press of China: Beijing, China, 2023.
- GB/T 35912-2018; Real-Time RT-PCR Method for Detection of Porcine Reproductive and Respiratory Syndrome Virus. Standards Press of China: Beijing, China, 2018.
- Rupasinghe, R.; Lee, K.; Liu, X.; Gauger, P.C.; Zhang, J.; Martinez-Lopez, B. Molecular Evolution of Porcine Reproductive and Respiratory Syndrome Virus Field Strains from Two Swine Production Systems in the Midwestern United States from 2001 to 2020. Microbiol. Spectr. 2022, 10, e263421. [Google Scholar] [CrossRef]
- Liu, J.; Xu, Y.; Lin, Z.; Fan, J.; Dai, A.; Deng, X.; Mao, W.; Huang, X.; Yang, X.; Wei, C. Epidemiology investigation of PRRSV discharged by faecal and genetic variation of ORF5. Transbound. Emerg. Dis. 2021, 68, 2334–2344. [Google Scholar] [CrossRef]
- Liu, J.; Lai, L.; Xu, Y.; Yang, Y.; Li, J.; Liu, C.; Hunag, C.; Wei, C. Evolutionary Analysis of Four Recombinant Viruses of the Porcine Reproductive and Respiratory Syndrome Virus From a Pig Farm in China. Front. Vet. Sci. 2022, 9, 933896. [Google Scholar] [CrossRef]
- Yu, F.; Yan, Y.; Shi, M.; Liu, H.Z.; Zhang, H.L.; Yang, Y.B.; Huang, X.Y.; Gauger, P.C.; Zhang, J.; Zhang, Y.H.; et al. Phylogenetics, Genomic Recombination, and NSP2 Polymorphic Patterns of Porcine Reproductive and Respiratory Syndrome Virus in China and the United States in 2014–2018. J. Virol. 2020, 94, 1110–1128. [Google Scholar] [CrossRef]
- Pan, J.; Zeng, M.; Zhao, M.; Huang, L. Research Progress on the detection methods of porcine reproductive and respiratory syndrome virus. Front. Microbiol. 2023, 14, 1097905. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, C.; Provost, C.; Gagnon, C.A. Whole-Genome Sequencing of Porcine Reproductive and Respiratory Syndrome Virus from Field Clinical Samples Improves the Genomic Surveillance of the Virus. J. Clin. Microbiol. 2020, 58, 1110–1128. [Google Scholar] [CrossRef]
- Li, H.; Wei, X.; Zhang, X.; Xu, H.; Zhao, X.; Zhou, S.; Huang, S.; Liu, X. Establishment of a multiplex RT-PCR assay for identification of atmospheric virus contamination in pig farms. Environ. Pollut. 2019, 253, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shi, K.; Liu, H.; Yin, Y.; Zhao, J.; Long, F.; Lu, W.; Si, H. Development of a multiplex qRT-PCR assay for detection of African swine fever virus, classical swine fever virus and porcine reproductive and respiratory syndrome virus. J. Vet. Sci. 2021, 22, e87. [Google Scholar] [CrossRef]
- Shi, K.; Chen, Y.; Yin, Y.; Long, F.; Feng, S.; Liu, H.; Qu, S.; Si, H. A Multiplex Crystal Digital PCR for Detection of African Swine Fever Virus, Classical Swine Fever Virus, and Porcine Reproductive and Respiratory Syndrome Virus. Front. Vet. Sci. 2022, 9, 926881. [Google Scholar] [CrossRef]
- Chen, N.H.; Chen, X.Z.; Hu, D.M.; Yu, X.L.; Wang, L.L.; Han, W.; Wu, J.J.; Cao, Z.; Wang, C.B.; Zhang, Q.; et al. Rapid differential detection of classical and highly pathogenic North American Porcine Reproductive and Respiratory Syndrome virus in China by a duplex real-time RT-PCR. J. Virol. Methods 2009, 161, 192–198. [Google Scholar] [CrossRef]
- Le Gall, A.; Legeay, O.; Bourhy, H.; Arnauld, C.; Albina, E.; Jestin, A. Molecular variation in the nucleoprotein gene (ORF7) of the porcine reproductive and respiratory syndrome virus (PRRSV). Virus Res. 1998, 54, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Zaulet, M.; Gurau, M.R.; Petrovan, V.; Buburuzan, L. Genetic diversity characterization of porcine reproductive and respiratory syndrome virus isolates in Romania, based on phylogenetic analysis. Int. J. Mol. Sci. 2012, 13, 12046–12061. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Li, D.; Hu, Y.; Gao, P.; Zhang, Y.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Yang, H. The Genetic Variation of Porcine Reproductive and Respiratory Syndrome Virus Replicase Protein nsp2 Modulates Viral Virulence and Persistence. J. Virol. 2023, 97, e168922. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Ye, M.; Xiao, Y.; Li, S.; Huang, Y.; Li, X.; Tian, K.; Zhu, J. Development of universal and quadruplex real-time RT-PCR assays for simultaneous detection and differentiation of porcine reproductive and respiratory syndrome viruses. Transbound. Emerg. Dis. 2019, 66, 2271–2278. [Google Scholar] [CrossRef]
- Kittawornrat, A.; Prickett, J.; Chittick, W.; Wang, C.; Engle, M.; Johnson, J.; Patnayak, D.; Schwartz, T.; Whitney, D.; Olsen, C.; et al. Porcine reproductive and respiratory syndrome virus (PRRSV) in serum and oral fluid samples from individual boars: Will oral fluid replace serum for PRRSV surveillance? Virus Res. 2010, 154, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, M.; Cui, X.; Gao, X.; Sun, W.; Ge, X.; Zhang, Y.; Guo, X.; Han, J.; Zhou, L.; et al. Attenuated Porcine Reproductive and Respiratory Syndrome Virus Regains Its Fatal Virulence by Serial Passaging in Pigs or Porcine Alveolar Macrophages To Increase Its Adaptation to Target Cells. Microbiol. Spectr. 2022, 10, e308422. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Z.; Ding, Y.; Ge, X.; Guo, X.; Yang, H. NADC30-like Strain of Porcine Reproductive and Respiratory Syndrome Virus, China. Emerg. Infect. Dis. 2015, 21, 2256–2257. [Google Scholar] [CrossRef]
- Zhou, L.; Kang, R.; Yu, J.; Xie, B.; Chen, C.; Li, X.; Xie, J.; Ye, Y.; Xiao, L.; Zhang, J.; et al. Genetic Characterization and Pathogenicity of a Novel Recombined Porcine Reproductive and Respiratory Syndrome Virus 2 among Nadc30-Like, Jxa1-Like, and Mlv-Like Strains. Viruses 2018, 10, 551. [Google Scholar] [CrossRef] [PubMed]










| Genes | Primers | Sequences (5′-3′) | Amplicons Size (bp) | Upstream Primer Positions | Downstream Primer Positions | Reference |
|---|---|---|---|---|---|---|
| PRRSV (ORF7) | Forward Reverse | ATGGCCAGCCAGTCAATCA TCGCCCTAATTGAATAGGTG | 398 (PRRSV1) | 14,653–14,761 | 15,031–15,050 | GB/T 1809-2023 [17] |
| 433 (PRRSV2) | 14,933–14,951 | 15,346–15,365 | ||||
| PRRSV (NSP2) | Forward Reverse | SGACACCTYCTTTGATTGG CTTGACARGGAGCTGCTTGA | 1041 (C-PRRSV2) | 2014–2032 | 3035–3051 | Designed by our laboratory |
| 951 (HP-PRRSV2) | 2184–2202 | 3115–3134 | ||||
| 648 (N-PRRSV2) | 2186–2240 | 2814–2833 |
| Genes | Primers/Probes | Sequences (5′-3′) | Positions | Amplicon Size |
|---|---|---|---|---|
| PRRSV1 (ORF7) | PRRSV1-F | CAGCCAGTCAATCARCTGTGCCA | 14,658–14,680 | 113 bp |
| PRRSV1-R | AGRGGRAAATGKGGCTTCTC | 14,751–14,770 | ||
| PRRSV1-P | ROX-TCCCAGCGCCAGCARCCTAGRGGA-BHQ2 | 14,703–14,726 | ||
| PRRSV2 (ORF7) | PRRSV2-F | CAGCCAGTCAATCARCTGTGCCA | 14,847–14,869 | 122 bp |
| PRRSV2-R | AGRGGRAAATGKGGCTTCTC | 14,949–14,968 | ||
| PRRSV2-P | FAM-CCAGTCCAGAGGCAAGGGACCG-BHQ1 | 14,900–14,921 | ||
| C-PRRSV2 (NSP2) | C-PRRSV2-F | TCACTGGCTAACTACTACTACCG | 2165–2187 | 134 bp |
| C-PRRSV2-R | CGTGGACCAGGCTCGGTTGG | 2279–2298 | ||
| C-PRRSV2-P | CY5-AAGACTAACCGCCGTGCTCTCCAAG-BHQ2 | 2218–2242 | ||
| HP-PRRSV2 (NSP2) | HP-PRRSV2-F | CTAACGGTTCGGAAGAAACT | 2762–2781 | 120 bp |
| HP-PRRSV2-R | TCATCAGCTTGGGGACACGTC | 2861–2881 | ||
| HP-PRRSV2-P | HEX-GTGTCATCGGCTCGGATGGTGT-BHQ1 | 2806–2827 |
| Composition | Volume (μL) |
|---|---|
| Animal Detection U + Probe Master Mix | 10.0 |
| PRRSV1-ORF7-F | 0.8 |
| PRRSV1-ORF7-R | 0.8 |
| PRRSV1-ORF7-P | 0.6 |
| PRRSV2-ORF7-F | 0.8 |
| PRRSV2-ORF7-R | 0.8 |
| PRRSV2-ORF7-P | 0.6 |
| C-PRRSV2-NSP2-F | 0.8 |
| C-PRRSV2-NSP2-R | 0.8 |
| C-PRRSV2-NSP2-P | 0.6 |
| HP-PRRSV2-NSP2-F | 0.8 |
| HP-PRRSV2-NSP2-R | 0.8 |
| HP-PRRSV2-NSP2-P | 0.6 |
| Template | 2.0 |
| DEPC H2O | up to 25 μL |
| Total volume | 25.0 |
| Target | Standard Sample (Copies/μL) | Intrabatch Repeatability Test | Interbatch Repeatability Test | ||
|---|---|---|---|---|---|
| Intrarepeatability Ct ± SD | Coefficients of Variation CV × 100% | Intrarepeatability Ct ± SD | Coefficients of Variation CV × 100% | ||
| PRRSV1-ORF7 | 8.77 × 105 | 27.35 ± 0.04 | 0.10 | 27.61 ± 0.48 | 1.23 |
| 8.77 × 104 | 30.80 ± 0.84 | 1.93 | 31.2 ± 0.04 | 0.09 | |
| 8.77 × 103 | 33.82 ± 0.01 | 0.21 | 33.92 ± 2.30 | 4.79 | |
| PRRSV2-ORF7 | 4.79 × 105 | 27.02 ± 0.42 | 1.10 | 27.54 ± 0.62 | 1.59 |
| 4.79 × 104 | 31.69 ± 0.56 | 1.25 | 32.03 ± 1.24 | 2.74 | |
| 4.79 × 103 | 34.53 ± 1.22 | 2.50 | 35.63 ± 0.98 | 1.94 | |
| C-PRRSV2-NSP2 | 4.71 × 105 | 24.50 ± 1.24 | 3.58 | 24.35 ± 0.46 | 1.34 |
| 4.71 × 104 | 27.27 ± 0.76 | 1.97 | 28.67 ± 1.64 | 4.04 | |
| 4.71 × 103 | 30.85 ± 0.92 | 2.11 | 31.75 ± 0.88 | 1.96 | |
| HP-PRRSV2-NSP2 | 5.54 × 105 | 29.77 ± 1.64 | 3.90 | 30.17 ± 0.84 | 1.97 |
| 5.54 × 104 | 32.34 ± 1.17 | 3.80 | 31.66 ± 2.10 | 4.69 | |
| 5.54 × 103 | 34.25 ± 0.84 | 1.73 | 35.5 ± 1.82 | 3.63 | |
| Kappa Test | Developed RT-qPCR | Total | ||
|---|---|---|---|---|
| + | − | |||
| National reference method | + | 24 | 2 | 26 |
| − | 1 | 73 | 74 | |
| total | 25 | 75 | 100 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruan, S.; Ren, W.; Yu, B.; Yu, X.; Wu, H.; Li, W.; Jiang, Y.; He, Q. Development and Implementation of a Quadruple RT-qPCR Method for the Identification of Porcine Reproductive and Respiratory Syndrome Virus Strains. Viruses 2023, 15, 1946. https://doi.org/10.3390/v15091946
Ruan S, Ren W, Yu B, Yu X, Wu H, Li W, Jiang Y, He Q. Development and Implementation of a Quadruple RT-qPCR Method for the Identification of Porcine Reproductive and Respiratory Syndrome Virus Strains. Viruses. 2023; 15(9):1946. https://doi.org/10.3390/v15091946
Chicago/Turabian StyleRuan, Shengnan, Wenhui Ren, Bin Yu, Xuexiang Yu, Hao Wu, Wentao Li, Yunbo Jiang, and Qigai He. 2023. "Development and Implementation of a Quadruple RT-qPCR Method for the Identification of Porcine Reproductive and Respiratory Syndrome Virus Strains" Viruses 15, no. 9: 1946. https://doi.org/10.3390/v15091946
APA StyleRuan, S., Ren, W., Yu, B., Yu, X., Wu, H., Li, W., Jiang, Y., & He, Q. (2023). Development and Implementation of a Quadruple RT-qPCR Method for the Identification of Porcine Reproductive and Respiratory Syndrome Virus Strains. Viruses, 15(9), 1946. https://doi.org/10.3390/v15091946