Isolation and Pathogenicity Analysis of a G5P[23] Porcine Rotavirus Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples, Cells, and Antibodies
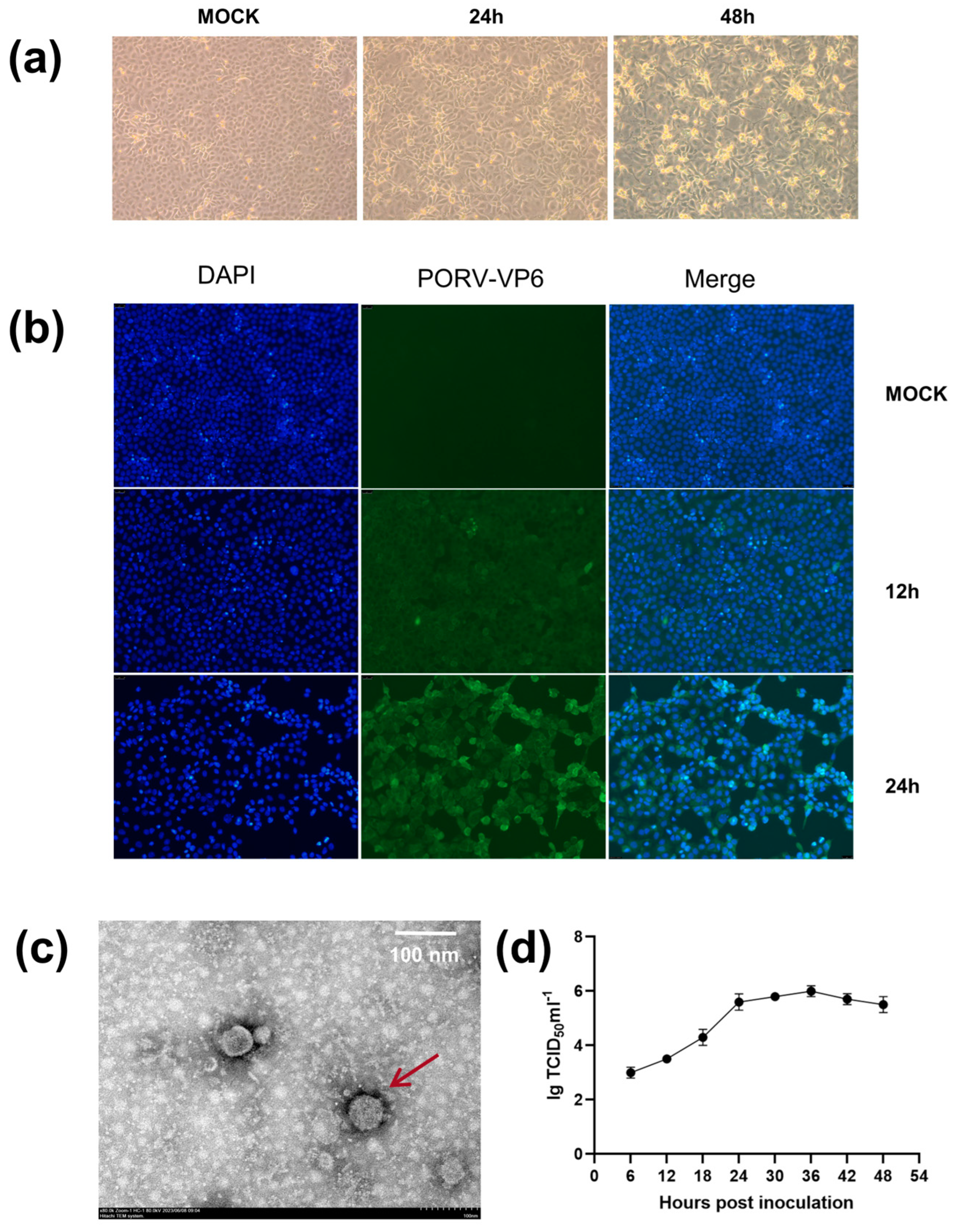
2.2. Virus Isolation Assay
2.3. Titration and Growth Curve Determination
2.4. Immunofluorescence Assay (IFA)
2.5. Transmission Electron Microscopy (TEM)
2.6. Sequence Analysis
2.7. Recombination Analysis
2.8. Animal Experiments
2.9. Statistical Analysis
3. Results
3.1. Virus Isolation
3.2. Sequence Analysis
3.3. Recombination Analysis
3.4. Clinical Signs and Histological Changes
3.5. Viral Load in Stool Samples and Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Anderson, E.J.; Weber, S.G. Rotavirus infection in adults. Lancet Infect. Dis. 2004, 4, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Rheingans, R.D.; Antil, L.; Dreibelbis, R.; Podewils, L.J.; Bresee, J.S.; Parashar, U.D. Economic Costs of Rotavirus Gastroenteritis and Cost-Effectiveness of Vaccination in Developing Countries. J. Infect. Dis. 2009, 200, S16–S27. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.; Bridger, J.; Kendall, K.; Gomara, M.I.; El-Attar, L.; Gray, J. The zoonotic potential of rotavirus. J. Infect. 2004, 48, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Bányai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010, 140, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Mebus, C.A.; Nr, U.; Rhodes, M.B.; Twiehaus, M.J. Calf Diarrhea (Scours): Reproduced with a Virus from a Field Outbreak; University of Nebraska: Lincoln, NE, USA, 1969. [Google Scholar]
- Bishop, R.F.; Davidson, G.P.; Holmes, I.H.; Ruck, B.J. Virus particles in epithelial cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet 1973, 2, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- Chasey, D.; Bridger, J.C.; McCrae, M.A. A new type of atypical rotavirus in pigs. Arch. Virol. 1986, 89, 235–243. [Google Scholar] [CrossRef]
- McNulty, M.S.; Allan, G.M.; McFerran, J.B. Prevalence of antibody to conventional and atypical rotaviruses in chickens. Vet. Rec. 1984, 114, 219. [Google Scholar] [CrossRef]
- Bohl, E.H.; Theil, K.W.; Saif, L.J. Isolation and serotyping of porcine rotaviruses and antigenic comparison with other rotaviruses. J. Clin. Microbiol. 1984, 19, 105–111. [Google Scholar] [CrossRef]
- Welter, M.W.; Welter, C.J.; Chambers, D.M.; Svensson, L. Adaptation and serial passage of porcine group C rotavirus in ST-cells, an established diploid swine testicular cell line. Arch. Virol. 1991, 120, 297–304. [Google Scholar] [CrossRef]
- Esona, M.D.; Foytich, K.; Wang, Y.H.; Shin, G.; Wei, G.; Gentsch, J.R.; Glass, R.I.; Jiang, B.M. Molecular characterization of human rotavirus vaccine strain CDC-9 during sequential passages in Vero cells. Hum. Vaccines 2010, 6, 247–253. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Lv, C.C.; Xu, X.; Li, X.D.; Yao, Y.L.; Gao, X.J.; Sun, Z.; Wang, Y.Z.; Sun, Y.J.; Xiao, Y.; et al. The dynamics of a Chinese porcine G9P 23 rotavirus production in MA-104 cells and intestines of 3-day-old piglets. J. Vet. Med. Sci. 2018, 80, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Mertens, N.; Theuss, T.; Köchling, M.; Dohmann, K.; Lillie-Jaschniski, K. Pathogens Detected in 205 German Farms with Porcine Neonatal Diarrhea in 2017. Vet. Sci. 2022, 9, 44. [Google Scholar] [CrossRef]
- Jacobson, M. On the Infectious Causes of Neonatal Piglet Diarrhoea-A Review. Vet. Sci. 2022, 9, 422. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Amimo, J.O.; Saif, L.J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef] [PubMed]
- Díaz, Y.; Chemello, M.E.; Peña, F.; Aristimuño, O.C.; Zambrano, J.L.; Rojas, H.; Bartoli, F.; Salazar, L.; Chwetzoff, S.; Sapin, C.; et al. Expression of Nonstructural Rotavirus Protein NSP4 Mimics Ca2+ Homeostasis Changes Induced by Rotavirus Infection in Cultured Cells. J. Virol. 2008, 82, 11331–11343. [Google Scholar] [CrossRef]
- Lundgren, O.; Peregrin, A.T.; Persson, K.; Kordasti, S.; Uhnoo, I.; Svensson, L. Role of the enteric nervous system in the fluid and electrolyte secretion of rotavirus diarrhea. Science 2000, 287, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Otto, P.H.; Ciarlet, M.; Desselberger, U.; Van Ranst, M.; Johne, R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 2012, 157, 1177–1182. [Google Scholar] [CrossRef]
- Papp, H.; László, B.; Jakab, F.; Ganesh, B.; De Grazia, S.; Matthijnssens, J.; Ciarlet, M.; Martella, V.; Bányai, K. Review of group A rotavirus strains reported in swine and cattle. Vet. Microbiol. 2013, 165, 190–199. [Google Scholar] [CrossRef]
- Maes, P.; Matthijnssens, J.; Rahman, M.; Van Ranst, M. RotaC: A web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009, 9, 238. [Google Scholar] [CrossRef]
- Ren, X.L.; Saleem, W.; Haes, R.; Xie, J.X.; Theuns, S.; Nauwynck, H.J. Milk lactose protects against porcine group A rotavirus infection. Front. Microbiol. 2022, 13, 989242. [Google Scholar] [CrossRef]
- Zhu, J.H.; Rawal, G.; Aljets, E.; Yim-Im, W.; Yang, Y.L.; Huang, Y.W.; Krueger, K.; Gauger, P.; Main, R.; Zhang, J.Q. Development and Clinical Applications of a 5-Plex Real-Time RT-PCR for Swine Enteric Coronaviruses. Viruses 2022, 14, 1536. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, J.N.; Yao, G.; Guo, Q.Y.; Wang, J.Q.; Liu, G.L. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl. Microbiol. Biotechnol. 2019, 103, 4943–4952. [Google Scholar] [CrossRef] [PubMed]
- Marthaler, D.; Homwong, N.; Rossow, K.; Culhane, M.; Goyal, S.; Collins, J.; Matthijnssens, J.; Ciarlet, M. Rapid detection and high occurrence of porcine rotavirus A, B, and C by RT-qPCR in diagnostic samples. J. Virol. Methods 2014, 209, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Park, G.N.; Kim, D.I.; Choe, S.; Shin, J.; An, B.H.; Kim, K.S.; Hyun, B.H.; Lee, J.S.; An, D.J. Genetic Diversity of Porcine Group A Rotavirus Strains from Pigs in South Korea. Viruses 2022, 14, 2522. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Pickett, B.E.; Sadat, E.L.; Zhang, Y.; Noronha, J.M.; Squires, R.B.; Hunt, V.; Liu, M.Y.; Kumar, S.; Zaremba, S.; Gu, Z.P.; et al. ViPR: An open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012, 40, D593–D598. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Liu, X.S.; Zhang, Q.L.; Zhang, L.P.; Zhou, P.; Yang, J.; Fang, Y.Z.; Dong, Z.L.; Zhao, D.H.; Li, W.Y.; Feng, J.X.; et al. A newly isolated Chinese virulent genotype GIIb porcine epidemic diarrhea virus strain: Biological characteristics, pathogenicity and immune protective effects as an inactivated vaccine candidate. Virus Res. 2019, 259, 18–27. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Rahman, M.; Attoui, H.; Bányai, K.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gómara, M.; Kirkwood, C.D.; Martella, V.; et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008, 153, 1621–1629. [Google Scholar] [CrossRef]
- Yodmeeklin, A.; Khamrin, P.; Chuchaona, W.; Saikruang, W.; Kongkaew, A.; Vachirachewin, R.; Kumthip, K.; Okitsu, S.; Ushijima, H.; Maneekarn, N. Great genetic diversity of rotaviruses detected in piglets with diarrhea in Thailand. Arch. Virol. 2016, 161, 2843–2849. [Google Scholar] [CrossRef]
- Heiman, E.M.; McDonald, S.M.; Barro, M.; Taraporewala, Z.F.; Bar-Magen, T.; Patton, J.T. Group A Human Rotavirus Genomics: Evidence that Gene Constellations Are Influenced by Viral Protein Interactions. J. Virol. 2008, 82, 11106–11116. [Google Scholar] [CrossRef] [PubMed]
- Liprandi, F.; Gerder, M.; Bastidas, Z.; López, J.A.; Pujol, F.H.; Ludert, J.E.; Joelsson, D.B.; Ciarlet, M. A novel type of VP4 carried by a porcine rotavirus strain. Virology 2003, 315, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Than, V.T.; Lim, I.; Kim, W. Whole-Genome Analysis of a Rare Human Korean G3P 9 Rotavirus Strain Suggests a Complex Evolutionary Origin Potentially Involving Reassortment Events between Feline and Bovine Rotaviruses. PLoS ONE 2014, 9, e97127. [Google Scholar] [CrossRef] [PubMed]
- Sieg, M.; Rückner, A.; Köhler, C.; Burgener, I.; Vahlenkamp, T.W. A bovine G8P 1 group A rotavirus isolated from an asymptomatically infected dog. J. Gen. Virol. 2015, 96, 106–114. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D.; S, W.H.O.C.G.R. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Parashar, U.D.; Nelson, E.A.S.; Kang, G. Diagnosis, management, and prevention of rotavirus gastroenteritis in children. BMJ-Br. Med. J. 2013, 347, f7204. [Google Scholar] [CrossRef]
- Miao, Q.; Pan, Y.D.; Gong, L.; Guo, L.J.; Wu, L.; Jing, Z.Y.; Zhang, G.H.; Tian, J.; Feng, L. Full genome characterization of a human-porcine reassortment G12P 7 rotavirus and its pathogenicity in piglets. Transbound. Emerg. Dis. 2022, 69, 3506–3517. [Google Scholar] [CrossRef]
- da Silva, M.F.M.; Tort, L.F.L.; Goméz, M.M.; Assis, R.M.S.; Volotao, E.D.; de Mendonça, M.C.L.; Bello, G.; Leite, J.P.G. VP7 Gene of Human Rotavirus A Genotype G5: Phylogenetic Analysis Reveals the Existence of Three Different Lineages Worldwide. J. Med. Virol. 2011, 83, 357–366. [Google Scholar] [CrossRef]
- Gouvea, V.; de Castro, L.; Timenetsky, M.C.; Greenberg, H.; Santos, N. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J. Clin. Microbiol. 1994, 32, 1408–1409. [Google Scholar] [CrossRef]
- Zhang, H.W.; Zhang, Z.; Wang, Y.F.; Wang, X.; Xia, M.Q.; Wu, H. Isolation, molecular characterization and evaluation of the pathogenicity of a porcine rotavirus isolated from Jiangsu Province, China. Arch. Virol. 2015, 160, 1333–1338. [Google Scholar] [CrossRef]
- Kim, H.H.; Park, J.G.; Matthijnssens, J.; Kim, H.J.; Kwon, H.J.; Son, K.Y.; Ryu, E.H.; Kim, D.S.; Lee, W.S.; Kang, M.I.; et al. Pathogenicity of porcine G9P 23 and G9P 7 rotaviruses in piglets. Vet. Microbiol. 2013, 166, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Ciarlet, M.; Conner, M.E.; Finegold, M.J.; Estes, M.K. Group A rotavirus infection and age-dependent diarrheal disease in rats: A new animal model to study the pathophysiology of rotavirus infection. J. Virol. 2002, 76, 41–57. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Shen, H.; Zhao, S.; Chen, S.; Zhu, P.; Lin, W.; Chen, F. Isolation and Pathogenicity Analysis of a G5P[23] Porcine Rotavirus Strain. Viruses 2024, 16, 21. https://doi.org/10.3390/v16010021
Gao L, Shen H, Zhao S, Chen S, Zhu P, Lin W, Chen F. Isolation and Pathogenicity Analysis of a G5P[23] Porcine Rotavirus Strain. Viruses. 2024; 16(1):21. https://doi.org/10.3390/v16010021
Chicago/Turabian StyleGao, Liguo, Hanqin Shen, Sucan Zhao, Sheng Chen, Puduo Zhu, Wencheng Lin, and Feng Chen. 2024. "Isolation and Pathogenicity Analysis of a G5P[23] Porcine Rotavirus Strain" Viruses 16, no. 1: 21. https://doi.org/10.3390/v16010021





