Safety, Immunogenicity, and Mechanism of a Rotavirus mRNA-LNP Vaccine in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval of the Study Protocol
2.2. Cells and Viruses
2.3. Generation of mRNA and mRNA-LNP
2.4. Transfection and Viral Protein Expression
2.5. Mouse Experiments
2.6. Detection of IgG Antibody and Neutralizing Antibody
2.7. Flow Cytometry
2.8. Enzyme-Linked Immunosorbent Spot (ELISpot)
2.9. Detection of Cytokines in Serum
2.10. Transcriptome Sequencing of Peripheral Blood Mononuclear Cells (PBMCs)
2.11. Statistical Analyses
3. Results
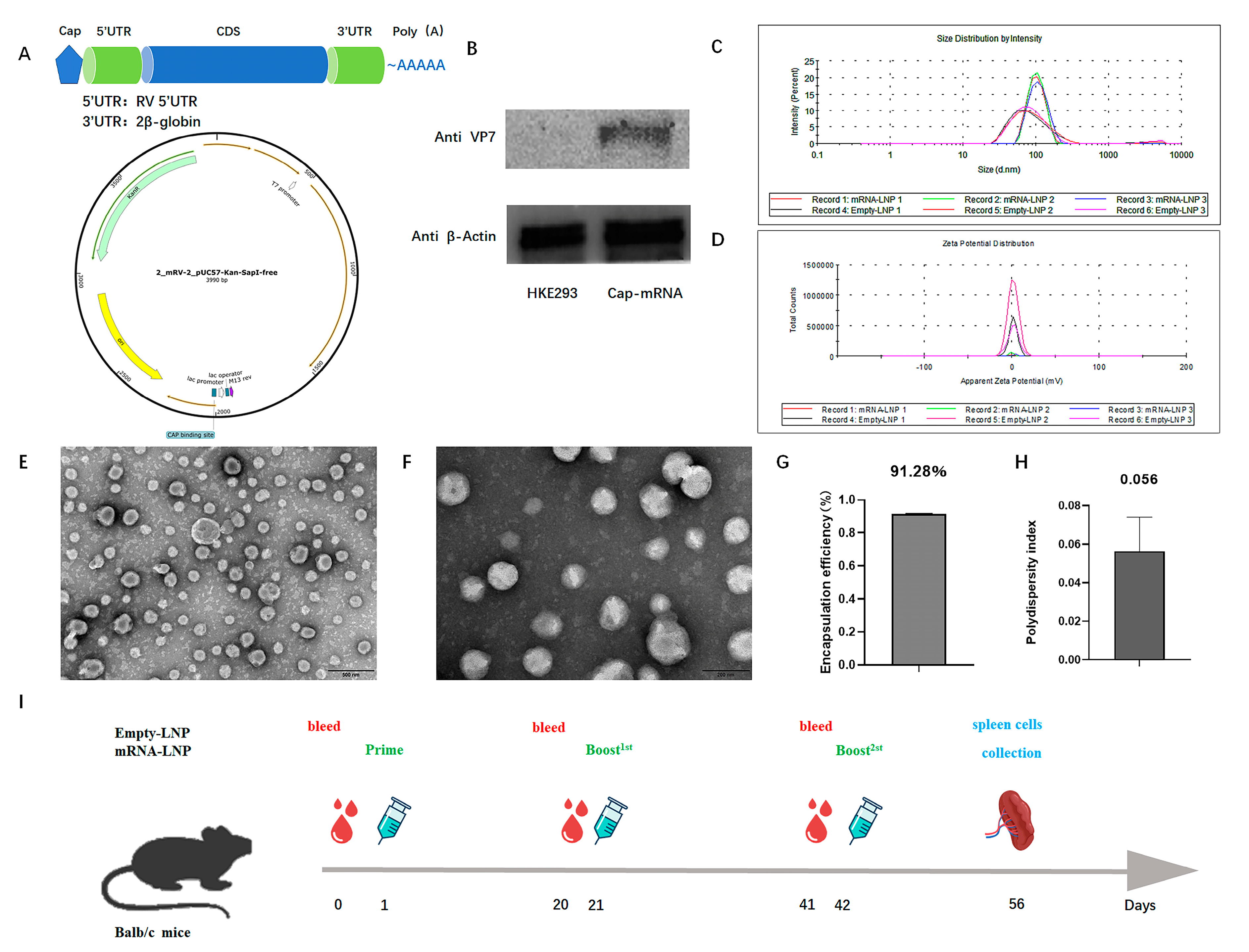
3.1. Construction, Characterization, and Protein Expression of RV VP7 mRNA-LNP
3.2. RV mRNA Vaccine Elicited Effective Humoral and Cellular Immune Responses
3.3. Transcriptome Sequencing of Peripheral Blood Mononuclear Cells (PBMCs)
3.4. Vaccine Safety
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sadiq, A.; Bostan, N.; Yinda, K.C.; Naseem, S.; Sattar, S. Rotavirus: Genetics, pathogenesis and vaccine advances. Rev. Med. Virol. 2018, 28, e2003. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Naghavi, M.; Allen, C.; Barber, R.M.; Bhutta, Z.A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; Coates, M.M.; et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef] [PubMed]
- O’Ryan, M.; Linhares, A. Update on Rotarix: An oral human rotavirus vaccine. Expert Rev. Vaccines 2009, 8, 1627–1641. [Google Scholar] [CrossRef]
- Chandran, A.; Santosham, M. RotaTeq: A three-dose oral pentavalent reassortant rotavirus vaccine. Expert Rev. Vaccines 2009, 7, 1475–1480. [Google Scholar] [CrossRef]
- Changotra, H.; Vij, A. Rotavirus virus-like particles (RV-VLPs) vaccines: An update. Rev. Med. Virol. 2017, 27, e1954. [Google Scholar] [CrossRef]
- Skansberg, A.; Sauer, M.; Tan, M.; Santosham, M.; Jennings, M.C. Product review of the rotavirus vaccines ROTASIIL, ROTAVAC, and Rotavin-M1. Hum. Vaccines Immunother. 2020, 17, 1223–1234. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Yang, Y.; Liang, Z.; Tian, Y.; Liu, B.; Gao, Z.; Jia, L.; Chen, L.; Wang, Q. Effectiveness of Lanzhou lamb rotavirus vaccine in preventing gastroenteritis among children younger than 5 years of age. Vaccine 2019, 37, 3611–3616. [Google Scholar] [CrossRef]
- Xia, S.; Du, J.; Su, J.; Liu, Y.; Huang, L.; Yu, Q.; Xie, Z.; Gao, J.; Xu, B.; Gao, X.; et al. Efficacy, immunogenicity and safety of a trivalent live human-lamb reassortant rotavirus vaccine (LLR3) in healthy Chinese infants: A randomized, double-blind, placebo-controlled trial. Vaccine 2020, 38, 7393–7400. [Google Scholar] [CrossRef]
- Burnett, E.; Parashar, U.; Tate, J. Rotavirus Vaccines: Effectiveness, Safety, and Future Directions. Pediatr. Drugs 2018, 20, 223–233. [Google Scholar] [CrossRef]
- Weaver, E.A.; Chilengi, R.; Simuyandi, M.; Beach, L.; Mwila, K.; Becker-Dreps, S.; Emperador, D.M.; Velasquez, D.E.; Bosomprah, S.; Jiang, B. Association of Maternal Immunity with Rotavirus Vaccine Immunogenicity in Zambian Infants. PLoS ONE 2016, 11, e0150100. [Google Scholar]
- Murphy, T.V.; Gargiullo, P.M.; Massoudi, M.S.; Nelson, D.B.; Jumaan, A.O.; Okoro, C.A.; Zanardi, L.R.; Setia, S.; Fair, E.; LeBaron, C.W.; et al. Intussusception among infants given an oral rotavirus vaccine. N. Engl. J. Med. 2001, 344, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Youssef, M.; Hitti, C.; Puppin Chaves Fulber, J.; Kamen, A.A. Enabling mRNA Therapeutics: Current Landscape and Challenges in Manufacturing. Biomolecules 2023, 13, 1497. [Google Scholar] [CrossRef] [PubMed]
- Barbier, A.J.; Jiang, A.Y.; Zhang, P.; Wooster, R.; Anderson, D.G. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 2022, 40, 840–854. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Tang, X.; Chen, Y.; Chen, K.; Fan, N.; Xiao, W.; Zheng, Q.; Li, G.; Teng, Y.; Wu, M.; et al. mRNA-based therapeutics: Powerful and versatile tools to combat diseases. Signal Transduct. Target Ther. 2022, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Guevara, M.L.; Persano, F.; Persano, S. Advances in Lipid Nanoparticles for mRNA-Based Cancer Immunotherapy. Front. Chem. 2020, 8, 589959. [Google Scholar] [CrossRef]
- Freyn, A.W.; Ramos da Silva, J.; Rosado, V.C.; Bliss, C.M.; Pine, M.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; de Souza Ferreira, L.C.; Weissman, D.; et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020, 28, 1569–1584. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Pelc, R.S.; Muramatsu, H.; Andersen, H.; DeMaso, C.R.; Dowd, K.A.; Sutherland, L.L.; Scearce, R.M.; Parks, R.; et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. [Google Scholar] [CrossRef]
- Zhang, P.; Narayanan, E.; Liu, Q.; Tsybovsky, Y.; Boswell, K.; Ding, S.; Hu, Z.; Follmann, D.; Lin, Y.; Miao, H.; et al. A multiclade env–gag VLP mRNA vaccine elicits tier-2 HIV-1-neutralizing antibodies and reduces the risk of heterologous SHIV infection in macaques. Nat. Med. 2021, 27, 2234–2245. [Google Scholar] [CrossRef] [PubMed]
- Aldon, Y.; McKay, P.F.; Moreno Herrero, J.; Vogel, A.B.; Lévai, R.; Maisonnasse, P.; Dereuddre-Bosquet, N.; Haas, H.; Fábián, K.; Le Grand, R.; et al. Immunogenicity of stabilized HIV-1 Env trimers delivered by self-amplifying mRNA. Mol. Ther.-Nucleic Acids 2021, 25, 483–493. [Google Scholar] [CrossRef]
- Mu, Z.; Wiehe, K.; Saunders, K.O.; Henderson, R.; Cain, D.W.; Parks, R.; Martik, D.; Mansouri, K.; Edwards, R.J.; Newman, A.; et al. mRNA-encoded HIV-1 Env trimer ferritin nanoparticles induce monoclonal antibodies that neutralize heterologous HIV-1 isolates in mice. Cell Rep. 2022, 38, 110514. [Google Scholar] [CrossRef] [PubMed]
- Gomez, C.E.; Perdiguero, B.; Usero, L.; Marcos-Villar, L.; Miralles, L.; Leal, L.; Sorzano, C.O.S.; Sanchez-Corzo, C.; Plana, M.; Garcia, F.; et al. Enhancement of the HIV-1-Specific Immune Response Induced by an mRNA Vaccine through Boosting with a Poxvirus MVA Vector Expressing the Same Antigen. Vaccines 2021, 9, 959. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Xu, S.; Lu, Y.; Luo, Z.; Yan, Y.; Wang, C.; Ji, J. Development of mRNA vaccines against respiratory syncytial virus (RSV). Cytokine Growth Factor Rev. 2022, 68, 37–53. [Google Scholar] [CrossRef]
- Awasthi, S.; Knox, J.J.; Desmond, A.; Alameh, M.G.; Gaudette, B.T.; Lubinski, J.M.; Naughton, A.; Hook, L.M.; Egan, K.P.; Tam, Y.K.; et al. Trivalent nucleoside-modified mRNA vaccine yields durable memory B cell protection against genital herpes in preclinical models. J. Clin. Investig. 2021, 131, e152310. [Google Scholar] [CrossRef]
- Monslow, M.A.; Elbashir, S.; Sullivan, N.L.; Thiriot, D.S.; Ahl, P.; Smith, J.; Miller, E.; Cook, J.; Cosmi, S.; Thoryk, E.; et al. Immunogenicity generated by mRNA vaccine encoding VZV gE antigen is comparable to adjuvanted subunit vaccine and better than live attenuated vaccine in nonhuman primates. Vaccine 2020, 38, 5793–5802. [Google Scholar] [CrossRef]
- Nelson, C.S.; Jenks, J.A.; Pardi, N.; Goodwin, M.; Roark, H.; Edwards, W.; McLellan, J.S.; Pollara, J.; Weissman, D.; Permar, S.R. Human Cytomegalovirus Glycoprotein B Nucleoside-Modified mRNA Vaccine Elicits Antibody Responses with Greater Durability and Breadth than MF59-Adjuvanted gB Protein Immunization. J. Virol. 2020, 94, e00186-20. [Google Scholar] [CrossRef]
- Alberer, M.; Gnad-Vogt, U.; Hong, H.S.; Mehr, K.T.; Backert, L.; Finak, G.; Gottardo, R.; Bica, M.A.; Garofano, A.; Koch, S.D.; et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: An open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet 2017, 390, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Wollner, C.J.; Richner, M.; Hassert, M.A.; Pinto, A.K.; Brien, J.D.; Richner, J.M. A Dengue Virus Serotype 1 mRNA-LNP Vaccine Elicits Protective Immune Responses. J. Virol. 2021, 95, e02482-20. [Google Scholar] [CrossRef]
- Roier, S.; Mangala Prasad, V.; McNeal, M.M.; Lee, K.K.; Petsch, B.; Rauch, S. mRNA-based VP8* nanoparticle vaccines against rotavirus are highly immunogenic in rodents. npj Vaccines 2023, 8, 190. [Google Scholar] [CrossRef]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus infection. Nat. Rev. Dis. Prim. 2017, 3, 17083. [Google Scholar] [CrossRef]
- Esona, M.D.; Gautam, R. Rotavirus. Clin. Lab. Med. 2015, 35, 363–391. [Google Scholar] [CrossRef]
- Spindler, K.R.; Long, C.P.; McDonald, S.M. Rotavirus genome replication: Some assembly required. PLoS Pathog. 2017, 13, e1006242. [Google Scholar]
- Morozova, O.V.; Sashina, T.A.; Fomina, S.G.; Novikova, N.A. Comparative characteristics of the VP7 and VP4 antigenic epitopes of the rotaviruses circulating in Russia (Nizhny Novgorod) and the Rotarix and RotaTeq vaccines. Arch. Virol. 2015, 160, 1693–1703. [Google Scholar] [CrossRef]
- Rodriguez, J.M.; Luque, D. Structural Insights into Rotavirus Entry. Adv. Exp. Med. Biol. 2019, 1215, 45–68. [Google Scholar]
- Zhao, B.; Pan, X.; Teng, Y.; Xia, W.; Wang, J.; Wen, Y.; Chen, Y. Rotavirus VP7 epitope chimeric proteins elicit cross-immunoreactivity in guinea pigs. Virol. Sin. 2015, 30, 363–370. [Google Scholar] [CrossRef]
- Zhuang, X.; Qi, Y.; Wang, M.; Yu, N.; Nan, F.; Zhang, H.; Tian, M.; Li, C.; Lu, H.; Jin, N. mRNA Vaccines Encoding the HA Protein of Influenza A H1N1 Virus Delivered by Cationic Lipid Nanoparticles Induce Protective Immune Responses in Mice. Vaccines 2020, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Türeci, O.z.; Sahin, U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef] [PubMed]
- Hekele, A.; Bertholet, S.; Archer, J.; Gibson, D.G.; Palladino, G.; Brito, L.A.; Otten, G.R.; Brazzoli, M.; Buccato, S.; Bonci, A.; et al. Rapidly produced SAM® vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2019, 2, e52. [Google Scholar] [CrossRef] [PubMed]
- Bahl, K.; Senn, J.J.; Yuzhakov, O.; Bulychev, A.; Brito, L.A.; Hassett, K.J.; Laska, M.E.; Smith, M.; Almarsson, Ö.; Thompson, J.; et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017, 25, 1316–1327. [Google Scholar] [CrossRef]
- Meyer, M.; Huang, E.; Yuzhakov, O.; Ramanathan, P.; Ciaramella, G.; Bukreyev, A. Modified mRNA-Based Vaccines Elicit Robust Immune Responses and Protect Guinea Pigs from Ebola Virus Disease. J. Infect. Dis. 2018, 217, 451–455. [Google Scholar] [CrossRef]
- Roth, C.; Cantaert, T.; Colas, C.; Prot, M.; Casadémont, I.; Levillayer, L.; Thalmensi, J.; Langlade-Demoyen, P.; Gerke, C.; Bahl, K.; et al. A Modified mRNA Vaccine Targeting Immunodominant NS Epitopes Protects Against Dengue Virus Infection in HLA Class I Transgenic Mice. Front. Immunol. 2019, 10, 1424. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D.; World Health Organization–Coordinated Global Rotavirus Surveillance Network; Agocs, M.; Serhan, F.; de Oliveira, L.; Mwenda, J.M.; Mihigo, R.; et al. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clin. Infect. Dis. 2016, 62 (Suppl. 2), S96–S105. [Google Scholar]
- Beres, L.K.; Tate, J.E.; Njobvu, L.; Chibwe, B.; Rudd, C.; Guffey, M.B.; Stringer, J.S.A.; Parashar, U.D.; Chilengi, R. A Preliminary Assessment of Rotavirus Vaccine Effectiveness in Zambia. Clin. Infect. Dis. 2016, 62, S175–S182. [Google Scholar] [CrossRef] [PubMed]
- Sahakyan, G.; Grigoryan, S.; Wasley, A.; Mosina, L.; Sargsyan, S.; Asoyan, A.; Gevorgyan, Z.; Kocharyan, K.; Avagyan, T.; Lopman, B.; et al. Impact and Effectiveness of Monovalent Rotavirus Vaccine in Armenian Children. Clin. Infect. Dis. 2016, 62, S147–S154. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Palacios, G.M.; Pérez-Schael, I.; Velázquez, F.R.; Abate, H.; Breuer, T.; Clemens, S.C.; Cheuvart, B.; Espinoza, F.; Gillard, P.; Innis, B.L.; et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 2006, 354, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Jonesteller, C.L.; Burnett, E.; Yen, C.; Tate, J.E.; Parashar, U.D. Effectiveness of Rotavirus Vaccination: A Systematic Review of the First Decade of Global Postlicensure Data, 2006–2016. Clin. Infect. Dis. 2017, 65, 840–850. [Google Scholar] [CrossRef]
- Carvalho, M.F.; Gill, D. Rotavirus vaccine efficacy: Current status and areas for improvement. Hum. Vaccines Immunother. 2019, 15, 1237–1250. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Li, Y.; Chen, R.; Hu, X.; Leng, Q.; Song, X.; Lin, X.; Ye, J.; Wang, J.; Li, J.; et al. Safety, Immunogenicity, and Mechanism of a Rotavirus mRNA-LNP Vaccine in Mice. Viruses 2024, 16, 211. https://doi.org/10.3390/v16020211
Lu C, Li Y, Chen R, Hu X, Leng Q, Song X, Lin X, Ye J, Wang J, Li J, et al. Safety, Immunogenicity, and Mechanism of a Rotavirus mRNA-LNP Vaccine in Mice. Viruses. 2024; 16(2):211. https://doi.org/10.3390/v16020211
Chicago/Turabian StyleLu, Chenxing, Yan Li, Rong Chen, Xiaoqing Hu, Qingmei Leng, Xiaopeng Song, Xiaochen Lin, Jun Ye, Jinlan Wang, Jinmei Li, and et al. 2024. "Safety, Immunogenicity, and Mechanism of a Rotavirus mRNA-LNP Vaccine in Mice" Viruses 16, no. 2: 211. https://doi.org/10.3390/v16020211





