Characterization of a Thermostable Endolysin of the Aeribacillus Phage AeriP45 as a Potential Staphylococcus Biofilm-Removing Agent
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Phage AP45 Propagation and Host Range Experiments
2.3. In Silico Structural Analysis of Putative Endolysins
2.4. Production and Purification of the Recombinant Endolysin LysAP45
2.5. Zymography Using Peptidoglycans of S. aureus, S. haemolyticus, S. epidermidis, S. warneri, and S. saprophyticus
2.6. Antimicrobial Activity Assay
2.7. S. aureus, S. haemolyticus, and S. epedermidis Biofilm Assay
2.8. Thermal Stability of LysAP45 Assay
2.9. Statistical Analysis
3. Results
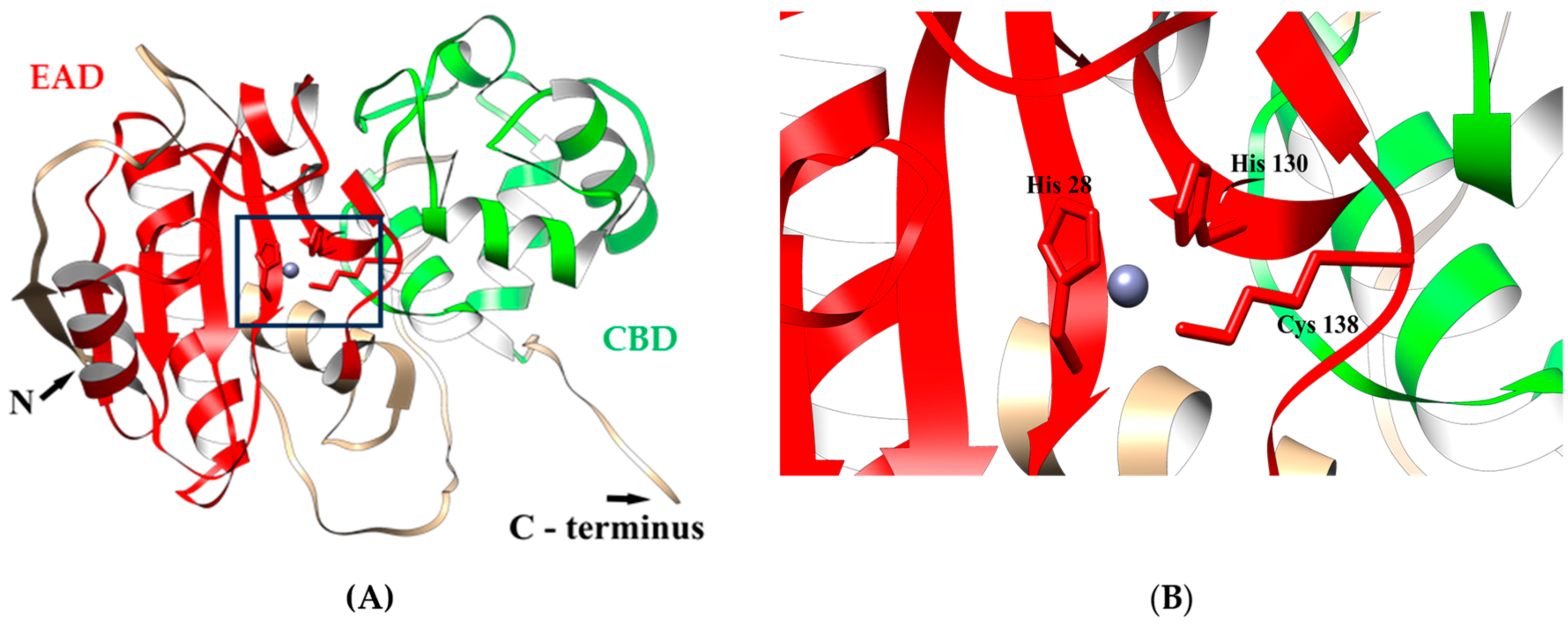
3.1. In Silico Characterization of LysAP45
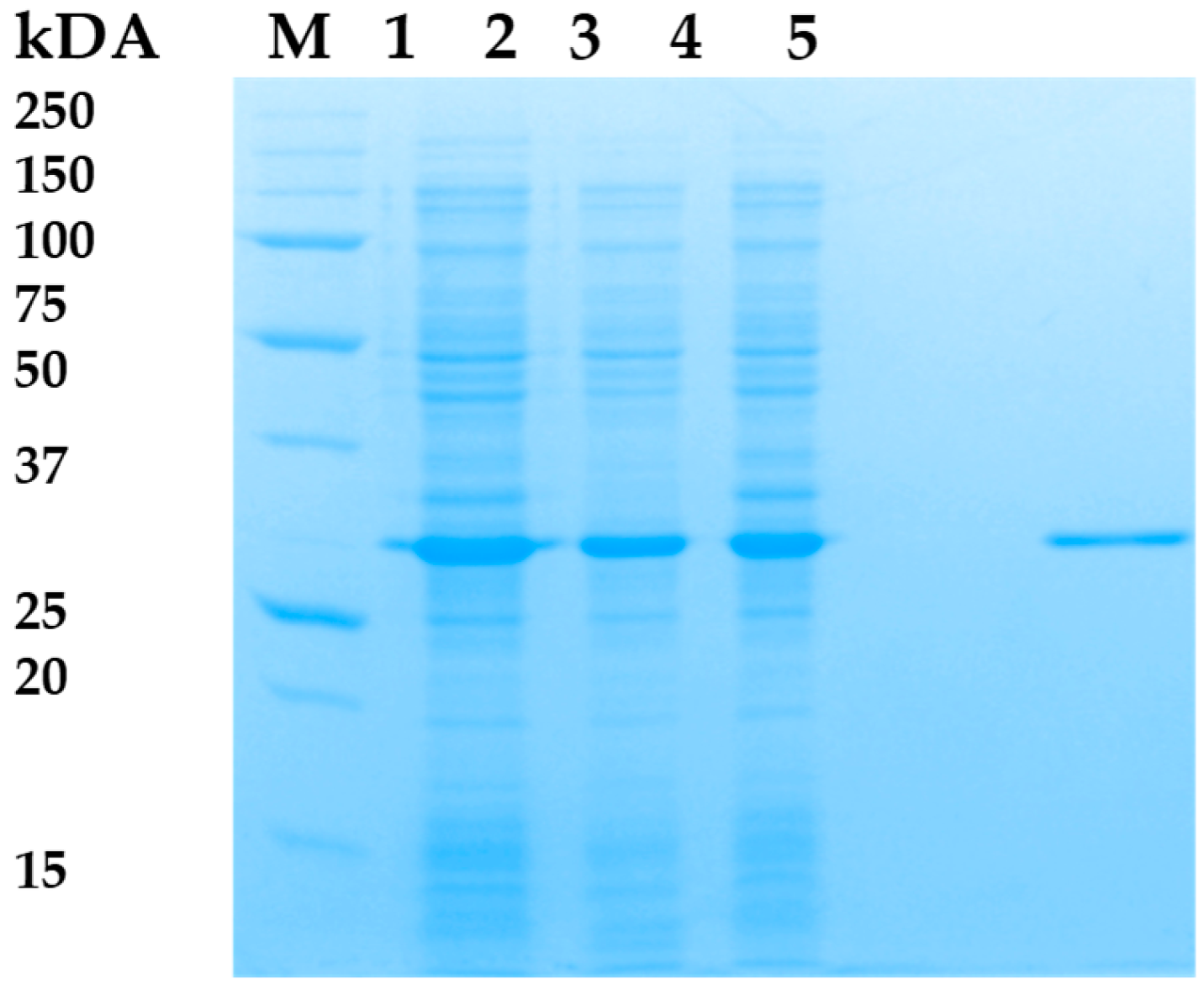
3.2. Expression and Purification of LysAP45
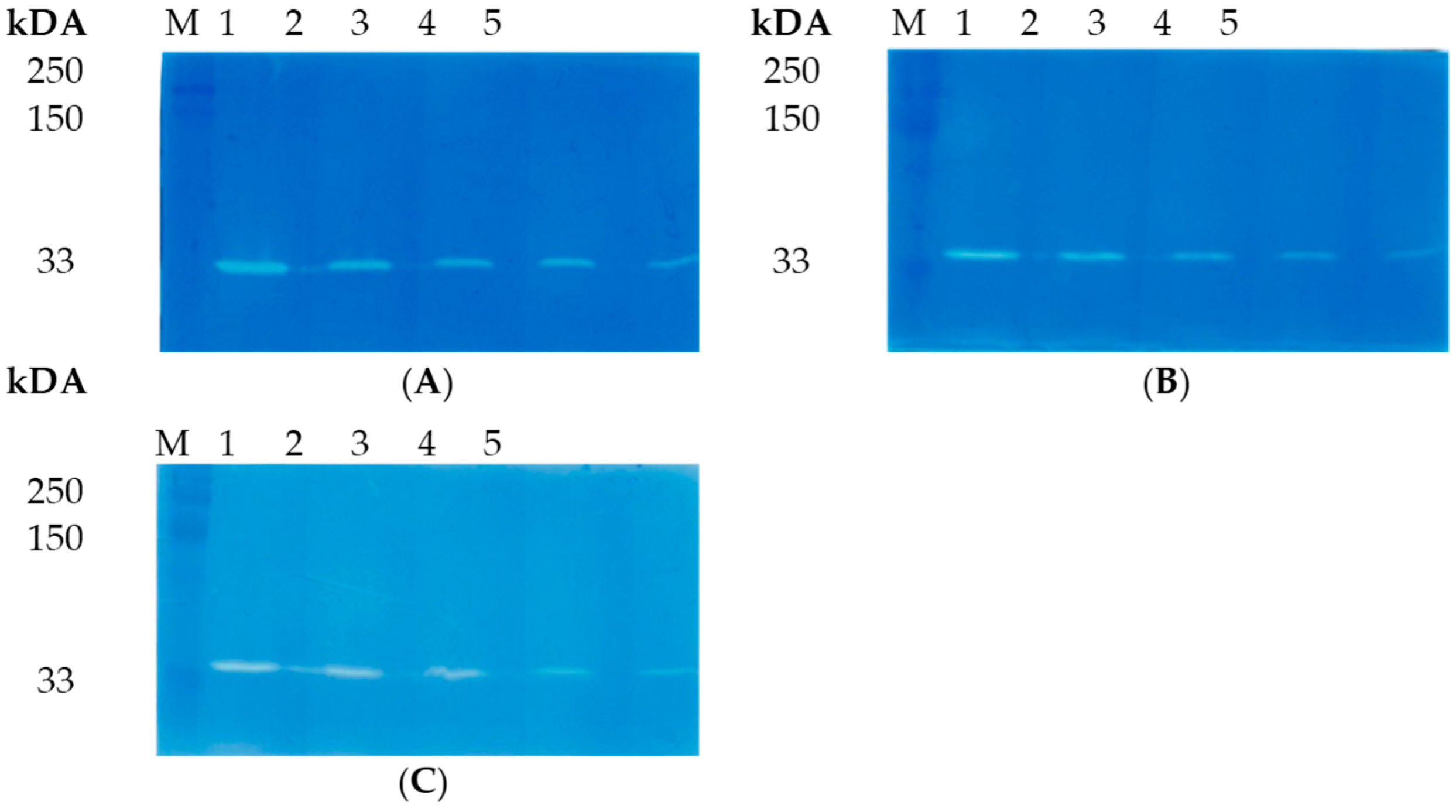
3.3. Enzymatic Activity of LysAP45
3.4. Phage AP45 Host Range
3.5. Anti-Staphylococcal Activity of LysAP45
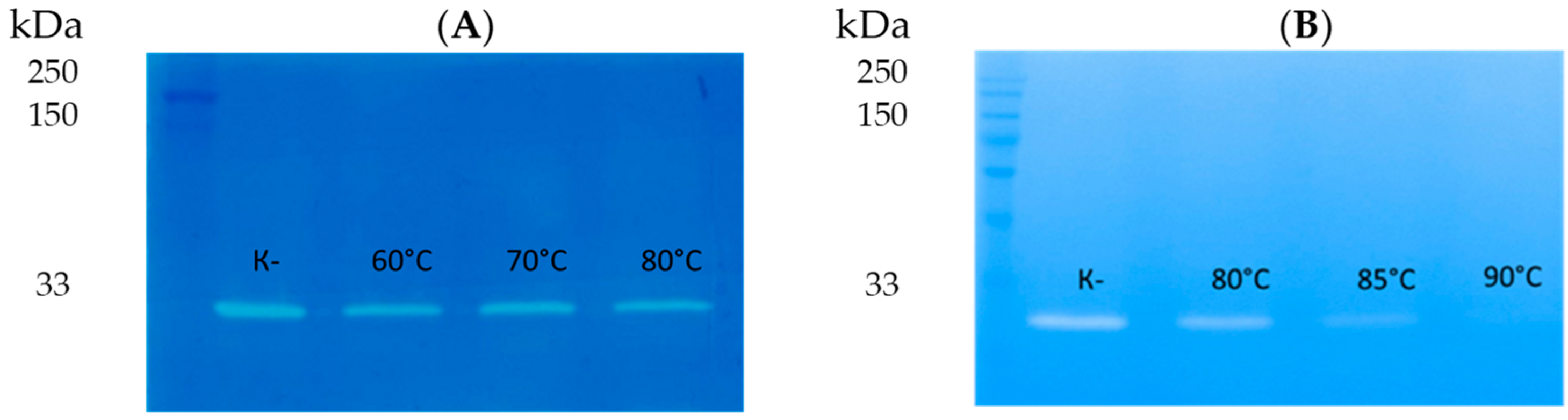
3.6. Thermal Stability of LysAP45
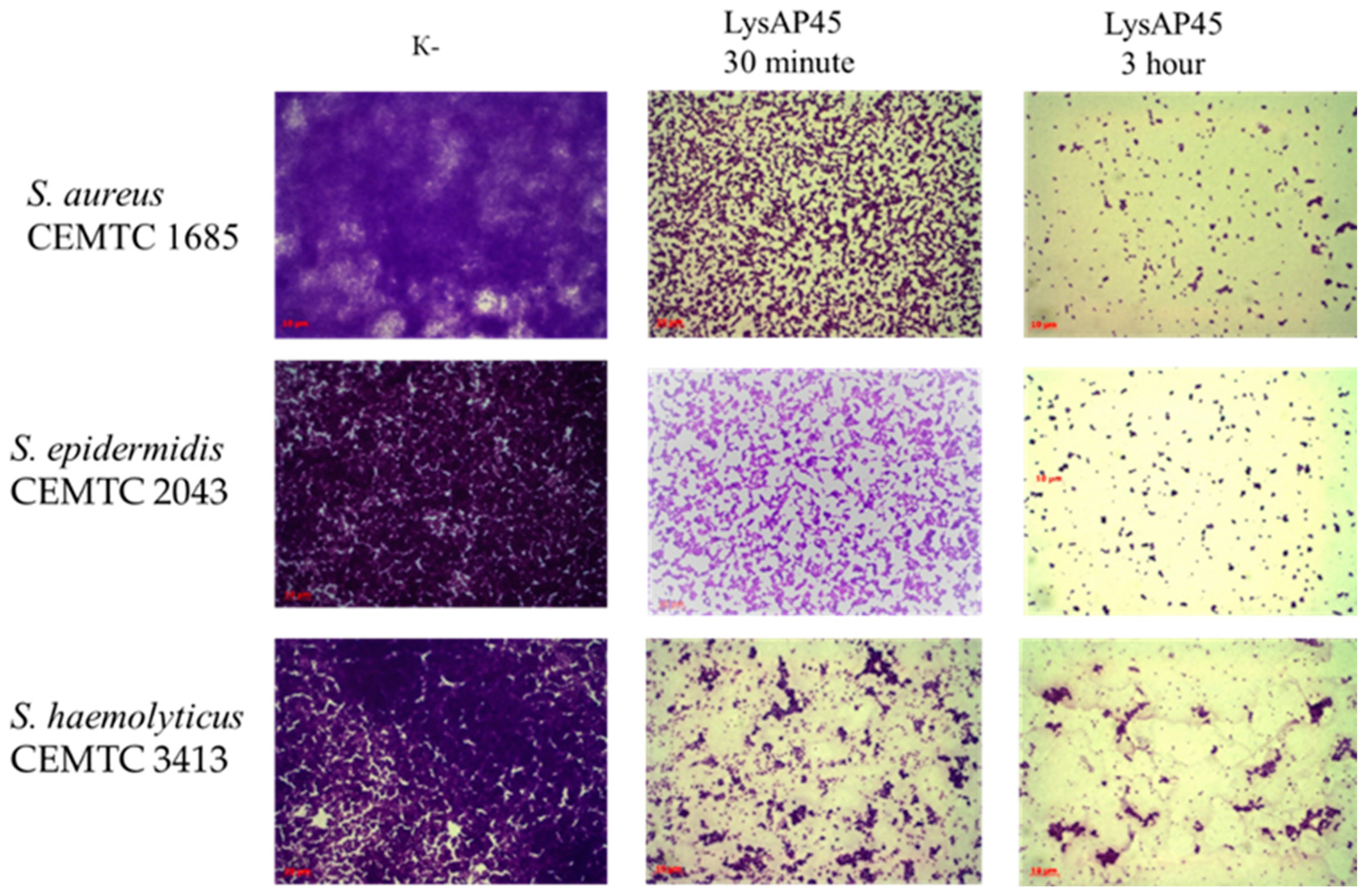
3.7. Biofilm Disruption Activity of LysAP45
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barber, M. Methicillin-resistant staphylococci. J. Clin. Pathol. 1961, 14, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Szweda, P.; Schielmann, M.; Kotlowski, R.; Gorczyca, G.; Zalewska, M.; Milewski, S. Peptidoglycan hydrolases-potential weapons against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2012, 96, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed]
- Severn, M.M.; Horswill, A.R. Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nature Rev. Microbiol. 2023, 21, 97–111. [Google Scholar] [CrossRef]
- Schilcher, K.; Horswill, A.R. Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev MMBR 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- Oliveira, W.F.; Silva, P.M.S.; Silva, R.C.S.; Silva, G.M.M.; Machado, G.; Coelho, L.C.B.B.; Correia, M.T.S. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J. Hosp. Infect. 2018, 98, 111–117. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-Negative Staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis Pathogenesis. Methods Mol. Biol. 2014, 1106, 17–31. [Google Scholar]
- Paharik, A.E.; Horswill, A.R. The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response. Virulence Mech. Bact. Pathog. 2016, 50, 529–566. [Google Scholar]
- Bardasheva, A.; Tikunov, A.; Kozlova, Y.; Zhirakovskaia, E.; Fedorets, V.; Fomenko, N.; Kalymbetova, T.; Chretien, S.; Pavlov, V.; Tikunova, N.; et al. Antibiotic Resistance and Pathogenomics of Staphylococci Circulating in Novosibirsk, Russia. Microorganisms 2021, 9, 2487. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-Encoded Endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Murray, E.; Draper, L.A.; Ross, R.P.; Hill, C. The Advantages and Challenges of Using Endolysins in a Clinical Setting. Viruses 2021, 13, 680. [Google Scholar] [CrossRef] [PubMed]
- Gondil, V.S.; Harjai, K.; Chhibber, S. Endolysins as Emerging Alternative Therapeutic Agents to Counter Drug-Resistant Infections. Int. J. Antimicrob. Agents 2020, 55, 105844. [Google Scholar] [CrossRef] [PubMed]
- Dams, D.; Briers, Y. Enzybiotics: Enzyme-Based Antibacterials as Therapeutics. In Therapeutic Enzymes: Function and Clinical Implications; Labrou, N., Ed.; Springer: Singapore, 2019; Volume 1148, pp. 233–253. ISBN 9789811377082. [Google Scholar]
- Jeong, T.-H.; Hong, H.-W.; Kim, M.S.; Song, M.; Myung, H. Characterization of Three Different Endolysins Effective against Gram-Negative Bacteria. Viruses 2023, 15, 679. [Google Scholar] [CrossRef] [PubMed]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan. Front. Microbiol. 2019, 10, 331. [Google Scholar] [CrossRef]
- Premetis, G.E.; Stathi, A.; Papageorgiou, A.C.; Labrou, N.E. Characterization of a glycoside hydrolase endolysin from Acinetobacter baumannii phage AbTZA1 with high antibacterial potency and novel structural features. FEBS J. 2023, 290, 2146–2164. [Google Scholar] [CrossRef]
- Antonova, N.P.; Vasina, D.V.; Lendel, A.M.; Usachev, E.V.; Makarov, V.V.; Gintsburg, A.L.; Tkachuk, A.P.; Gushchin, V.A. Broad Bactericidal Activity of the Myoviridae Bacteriophage Lysins LysAm24, LysECD7, and LysSi3 against Gram-Negative ESKAPE Pathogens. Viruses 2019, 11, 284. [Google Scholar] [CrossRef]
- Low, L.Y.; Yang, C.; Perego, M.; Osterman, A.; Liddington, R. Role of Net Charge on Catalytic Domain and Influence of Cell Wall Binding Domain on Bactericidal Activity, Specificity, and Host Range of Phage Lysins. J. Biol. Chem. 2011, 286, 34391–34403. [Google Scholar] [CrossRef]
- Morozova, V.; Bokovaya, O.; Kozlova, Y.; Kurilshikov, A.; Babkin, I.; Tupikin, A.; Bondar, A.; Ryabchikova, E.; Brayanskaya, A.; Peltek, S.; et al. A novel thermophilic Aeribacillus bacteriophage AP45 isolated from the Valley of Geysers, Kamchatka: Genome analysis suggests the existence of a new genus within the Siphoviridae family. Extrem. Life Under Extrem. Cond. 2019, 23, 599–612. [Google Scholar] [CrossRef]
- Kutter, E. Phage host range and efficiency of plating. Methods Mol. Biol. 2009, 501, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. Colab-Fold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Hon, J.; Marusiak, M.; Martinek, T.; Kunka, A.; Zendulka, J.; Bednar, D.; Damborsky, J. SoluProt: Prediction of soluble protein expression in Escherichia coli. Bioinformatics 2021, 37, 23–28. [Google Scholar] [CrossRef]
- Hebditch, M.; Carballo-Amador, M.A.; Charonis, S.; Curtis, R.; Warwicker, J. Protein–Sol: A web tool for predicting protein solubility from sequence. Bioinformatics 2017, 33, 3098–3100. [Google Scholar] [CrossRef]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved Protein Function Prediction by Combining Structure, Sequence and Protein–Protein Interaction Information. Nucleic Acids Res. 2017, 45, W291–W299. [Google Scholar] [CrossRef]
- Fukushima, T.; Sekiguchi, J. Zymographic Techniques for the Analysis of Bacterial Cell Wall in Bacillus. Methods Mol. Biol. 2016, 1440, 87–98. [Google Scholar]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Liu, H.; Kheirvari, M.; Tumban, E. Potential Applications of Thermophilic Bacteriophages in One Health. Int. J. Mol. Sci. 2023, 24, 8222. [Google Scholar] [CrossRef]
- Kim, H.; Seo, J. A Novel Strategy to Identify Endolysins with Lytic Activity against Methicillin-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2023, 24, 5772. [Google Scholar] [CrossRef] [PubMed]
- Miernikiewicz, P.; Barylski, J.; Wilczak, A.; Dragoš, A.; Rybicka, I.; Bałdysz, S.; Szymczak, A.; Dogsa, I.; Rokush, K.; Harhala, M.A.; et al. New Phage-Derived Antibacterial Enzyme PolaR Targeting Rothia spp. Cells 2023, 12, 1997. [Google Scholar] [CrossRef] [PubMed]
- Dudev, T.; Lim, C. Metal-binding affinity and selectivity of nonstandard natural amino acid residues from DFT/CDM calculations. J. Phys. Chem. B 2009, 113, 11754–11764. [Google Scholar] [CrossRef] [PubMed]
- Zebrowska, J.; Witkowska, M.; Struck, A.; Laszuk, P.E.; Raczuk, E.; Ponikowska, M.; Skowron, P.M.; Zylicz-Stachula, A. Antimicrobial Potential of the Genera Geobacillus and Parageobacillus, as Well as Endolysins Biosynthesized by Their Bacteriophages. Antibiotics 2022, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, T.N.; Lin, Y.; Maung, A.T.; Shen, C.; Zhao, J.; El-Telbany, M.; Zayda, M.; Masuda, Y.; Honjoh, K.; Miyamoto, T. Characterization and antibacterial activity of highly thermo- and pH-stable endolysin LysCPQ7 and its application as a biocontrol agent against Clostridium perfringens in milk and cheese. Food Control 2024, 156, 110157. [Google Scholar] [CrossRef]
- Skorynina, A.V.; Piligrimova, E.G.; Kazantseva, O.A.; Kulyabin, V.A.; Baicher, S.D.; Ryabova, N.A.; Shadrin, A.M. Bacillus-infecting bacteriophage Izhevsk harbors thermostable endolysin with broad range specificity. PLoS ONE 2020, 15, e0242657. [Google Scholar] [CrossRef]
- Ha, E.; Son, B.; Ryu, S. Clostridium perfringens Virulent Bacteriophage CPS2 and Its Thermostable Endolysin LysCPS2. Viruses 2018, 10, 251. [Google Scholar] [CrossRef]
- Kong, M.; Na, H.; Ha, N.C.; Ryu, S. LysPBC2, a Novel Endolysin Harboring a Bacillus cereus Spore Binding Domain. Appl. Environ. Microbiol. 2019, 85, e02462-18. [Google Scholar] [CrossRef]
- Choi, D.; Kong, M. LysGR1, a novel thermostable endolysin from Geobacillus stearothermophilus bacteriophage GR1. Front. Microbiol. 2023, 14, 1178748. [Google Scholar] [CrossRef]
- Plotka, M.; Kaczorowska, A.K.; Stefanska, A.; Morzywolek, A.; Fridjonsson, O.H.; Dunin-Horkawicz, S.; Kozlowski, L.; Hreggvidsson, G.O.; Kristjansson, J.K.; Dabrowski, S.; et al. Novel highly thermostable endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 with amino acid sequence similarity to eukaryotic peptidoglycan recognition proteins. Appl. Environ. Microbiol. 2014, 80, 886–895. [Google Scholar] [CrossRef]
- Żebrowska, J.; Żołnierkiewicz, O.; Ponikowska, M.; Puchalski, M.; Krawczun, N.; Makowska, J.; Skowron, P. Cloning and Characterization of a Thermostable Endolysin of Bacteriophage TP-84 as a Potential Disinfectant and Biofilm-Removing Biological Agent. Int. J. Mol. Sci. 2022, 23, 7612. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ji, X.; Li, Q.; Zhang, G.; Peng, J.; Hai, J.; Zhang, Y.; Ci, B.; Li, H.; Xiong, Y.; et al. TSPphg Lysin from the Extremophilic Thermus Bacteriophage TSP4 as a Potential Antimicrobial Agent against Both Gram-Negative and Gram-Positive Pathogenic Bacteria. Viruses 2020, 12, 192. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, L.; Martínez, B.; Zhou, Y.; Rodríguez, A.; Donovan, D.M.; García, P. Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88. BMC Microbiol. 2011, 11, 138. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhu, C.; Chen, J.; Ye, X.; Huang, Y.P. Antibacterial Activity of Stenotrophomonas maltophilia Endolysin P28 against both Gram-positive and Gram-negative Bacteria. Front. Microbiol. 2015, 6, 1299. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golosova, N.N.; Khlusevich, Y.A.; Morozova, V.V.; Matveev, A.L.; Kozlova, Y.N.; Tikunov, A.Y.; Panina, E.A.; Tikunova, N.V. Characterization of a Thermostable Endolysin of the Aeribacillus Phage AeriP45 as a Potential Staphylococcus Biofilm-Removing Agent. Viruses 2024, 16, 93. https://doi.org/10.3390/v16010093
Golosova NN, Khlusevich YA, Morozova VV, Matveev AL, Kozlova YN, Tikunov AY, Panina EA, Tikunova NV. Characterization of a Thermostable Endolysin of the Aeribacillus Phage AeriP45 as a Potential Staphylococcus Biofilm-Removing Agent. Viruses. 2024; 16(1):93. https://doi.org/10.3390/v16010093
Chicago/Turabian StyleGolosova, Natalia N., Yana A. Khlusevich, Vera V. Morozova, Andrey L. Matveev, Yulia N. Kozlova, Artem Y. Tikunov, Elizaveta A. Panina, and Nina V. Tikunova. 2024. "Characterization of a Thermostable Endolysin of the Aeribacillus Phage AeriP45 as a Potential Staphylococcus Biofilm-Removing Agent" Viruses 16, no. 1: 93. https://doi.org/10.3390/v16010093
APA StyleGolosova, N. N., Khlusevich, Y. A., Morozova, V. V., Matveev, A. L., Kozlova, Y. N., Tikunov, A. Y., Panina, E. A., & Tikunova, N. V. (2024). Characterization of a Thermostable Endolysin of the Aeribacillus Phage AeriP45 as a Potential Staphylococcus Biofilm-Removing Agent. Viruses, 16(1), 93. https://doi.org/10.3390/v16010093





