Research Progress into the Biological Functions of IFITM3
Abstract
:1. Introduction
2. Discovery of the IFITM Genes
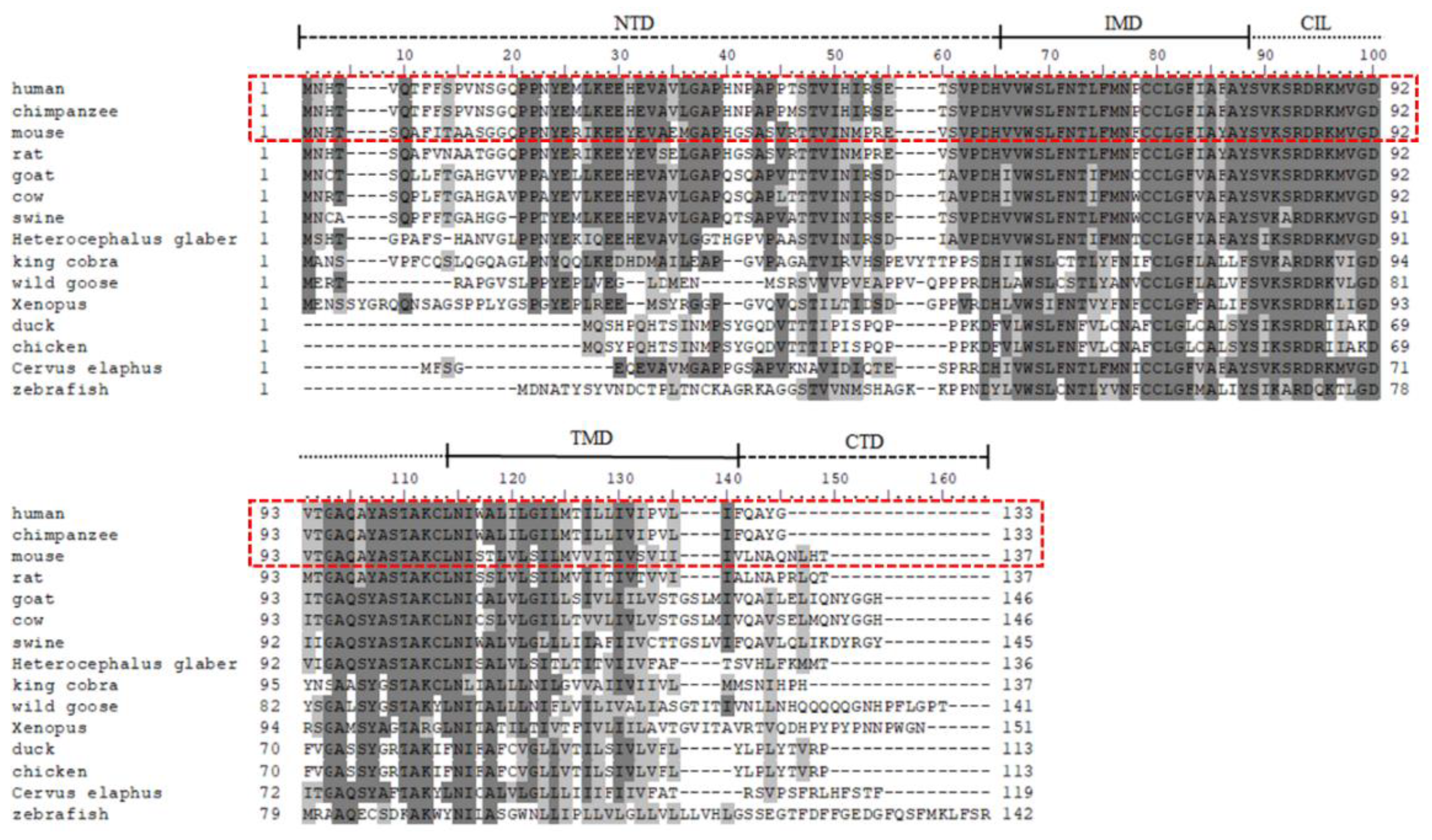
3. Molecular Evolution of IFITM3 Protein
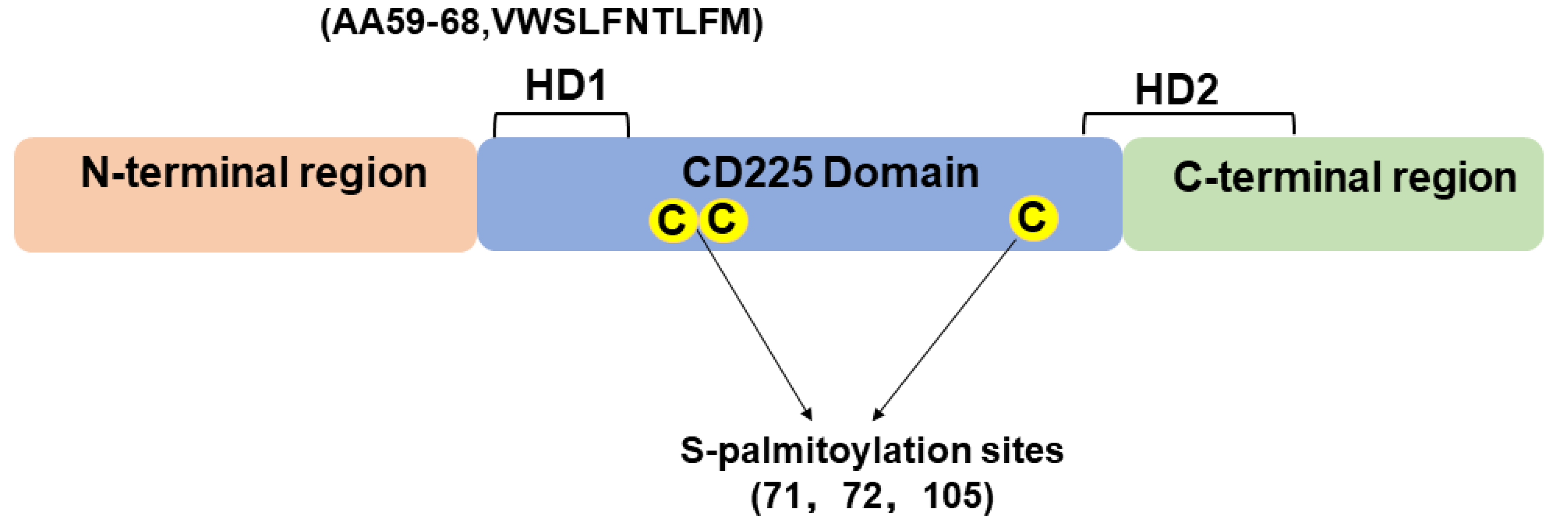
4. Structure of IFITM3 Protein
5. Biological Functions of IFITM3
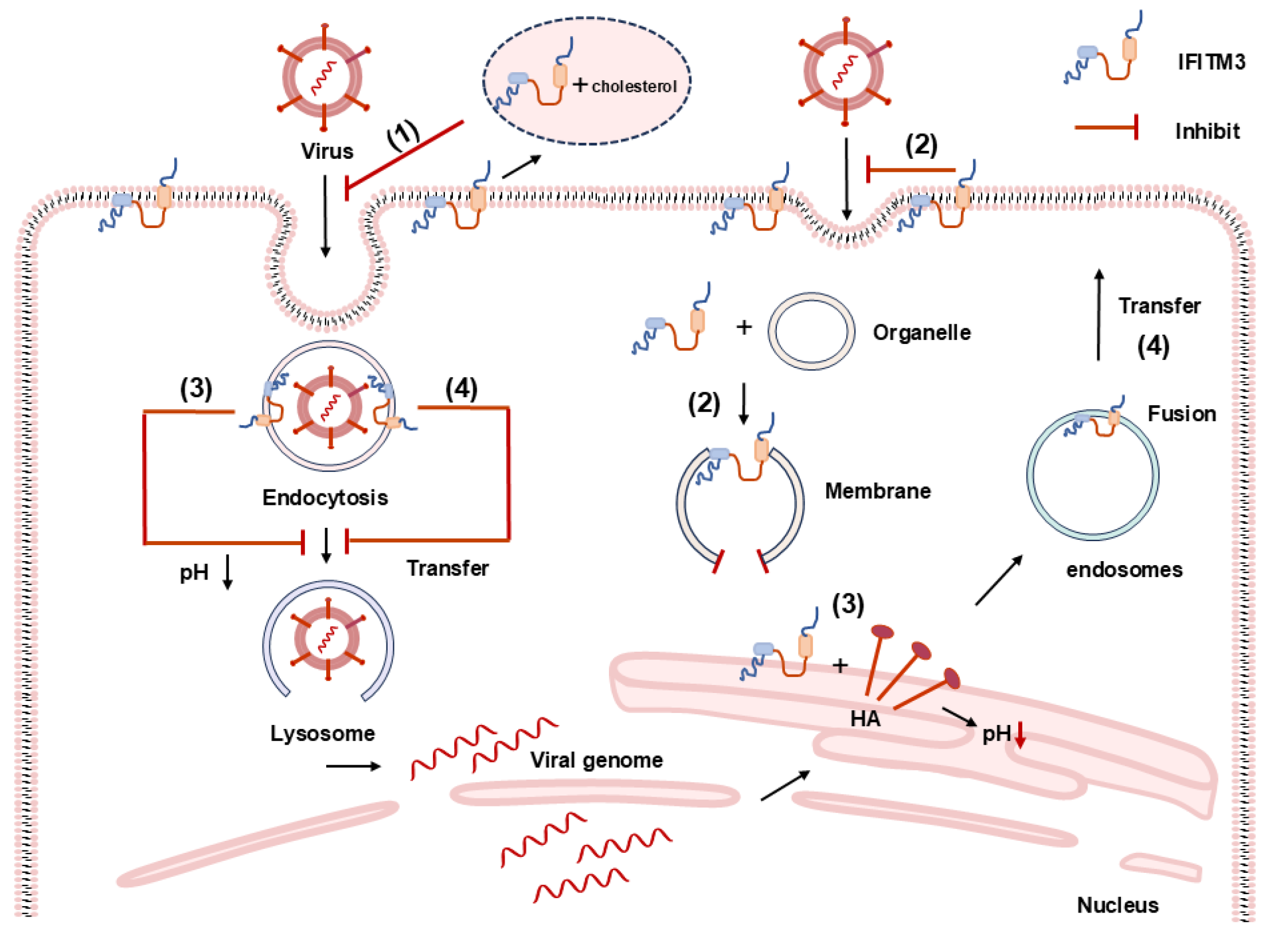
5.1. Antiviral Effects of IFITM3
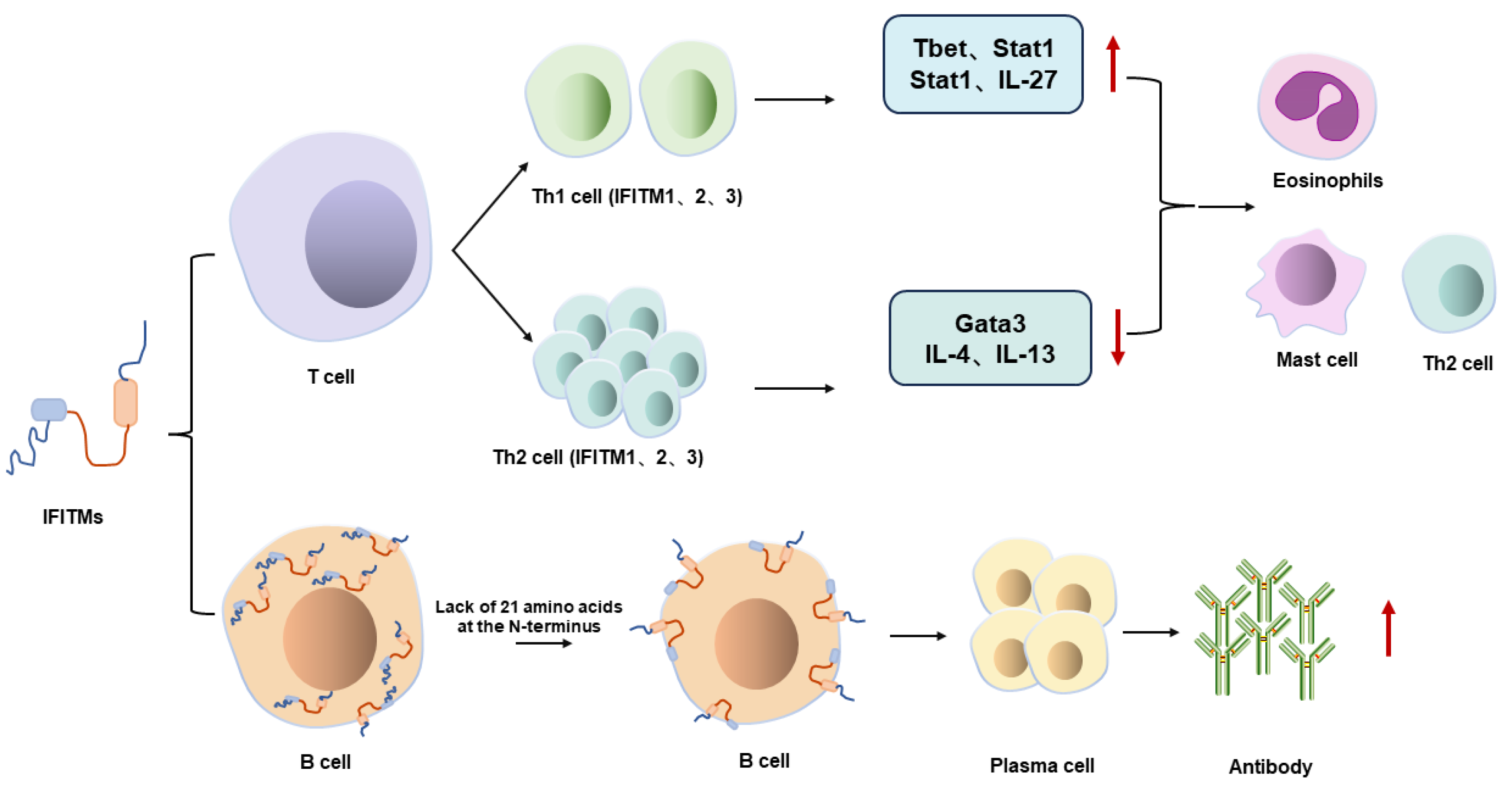
5.2. Immunomodulatory Effects of IFITMs
5.3. The Role of IFITM3 in Tumorigenesis
5.4. The Other Biological Functions of IFITM3
6. Future Directions in IFITM Research
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, X.; Li, J.; Winkler, C.A.; An, P.; Guo, J.T. IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections. Front. Microbiol. 2018, 9, 3228. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, I.C.; Bailey, C.C.; Weyer, J.L.; Radoshitzky, S.R.; Becker, M.M.; Chiang, J.J.; Brass, A.L.; Ahmed, A.A.; Chi, X.; Dong, L.; et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011, 7, e1001258. [Google Scholar] [CrossRef] [PubMed]
- Wrensch, F.; Winkler, M.; Pöhlmann, S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: Evidence for cholesterol-independent mechanisms. Viruses 2014, 6, 3683–3698. [Google Scholar] [CrossRef]
- Lu, J.; Pan, Q.; Rong, L.; He, W.; Liu, S.-L.; Liang, C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011, 85, 2126–2137. [Google Scholar] [CrossRef] [PubMed]
- Brass, A.L.; Huang, I.C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef]
- Perreira, J.M.; Chin, C.R.; Feeley, E.M.; Brass, A.L. IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 2013, 425, 4937–4955. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Dong, H.; Zhu, H.; Nelson, D.; Liu, C.; Lambiase, L.; Li, X. Identification of the IFITM3 gene as an inhibitor of hepatitis C viral translation in a stable STAT1 cell line. J. Viral Hepat. 2011, 18, e523–e529. [Google Scholar] [CrossRef] [PubMed]
- Wrensch, F.; Karsten, C.B.; Gnirß, K.; Hoffmann, M.; Lu, K.; Takada, A.; Winkler, M.; Simmons, G.; Pöhlmann, S. Interferon-Induced Transmembrane Protein-Mediated Inhibition of Host Cell Entry of Ebolaviruses. J. Infect. Dis. 2015, 212, S210–S218. [Google Scholar] [CrossRef]
- Majdoul, S.; Compton, A.A. Lessons in self-defence: Inhibition of virus entry by intrinsic immunity. Nat. Rev. Immunol. 2022, 22, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Yánez, D.C.; Sahni, H.; Ross, S.; Solanki, A.; Lau, C.I.; Papaioannou, E.; Barbarulo, A.; Powell, R.; Lange, U.C.; Adams, D.J.; et al. IFITM proteins drive type 2 T helper cell differentiation and exacerbate allergic airway inflammation. Eur. J. Immunol. 2019, 49, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.-Y.; Frost, G.R.; Wu, X.; Crump, C.; Pan, S.J.; Wong, E.; Barros, M.; Li, T.; Nie, P.; Zhai, Y.; et al. The innate immunity protein IFITM3 modulates γ-secretase in Alzheimer’s disease. Nature 2020, 586, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Pyun, J.M.; Park, Y.H.; Hodges, A.; Jang, J.W.; Bice, P.J.; Kim, S.; Saykin, A.J.; Nho, K. Immunity gene IFITM3 variant: Relation to cognition and Alzheimer’s disease pathology. Alzheimers Dement. 2022, 14, e12317. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.K.; Randolph, A.G.; Bhangale, T.; Dogra, P.; Ohlson, M.; Oshansky, C.M.; Zamora, A.E.; Shannon, J.P.; Finkelstein, D.; Dressen, A.; et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 2017, 23, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, Y.P.; Deng, H.L.; Wang, M.Q.; Chen, Y.; Zhang, Y.F.; Wang, J.; Dang, S.S. DNA methylation and SNP in IFITM3 are correlated with hand, foot and mouth disease caused by enterovirus 71. Int. J. Infect. Dis. 2021, 4, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.A.-O.; Luo, Q.L.; Guan, Y.G.; Fan, D.Y.; Luan, G.M.; Jing, A.A.-O. HCMV infection and IFITM3 rs12252 are associated with Rasmussen’s encephalitis disease progression. Ann. Clin. Transl. Neurol. 2021, 8, 558–570. [Google Scholar] [CrossRef]
- Lee, J.; Robinson, M.E.; Ma, N.; Artadji, D.; Ahmed, M.A.; Xiao, G.; Sadras, T.; Deb, G.; Winchester, J.; Cosgun, K.N.; et al. IFITM3 functions as a PIP3 scaffold to amplify PI3K signalling in B cells. Nature 2020, 588, 491–497. [Google Scholar] [CrossRef]
- Everitt, A.R.; Clare, S.; Pertel, T.; John, S.P.; Wash, R.S.; Smith, S.E.; Chin, C.R.; Feeley, E.M.; Sims, J.S.; Adams, D.J.; et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 2012, 484, 519–523. [Google Scholar] [CrossRef]
- Williams, D.E.J.; Wu, W.-L.; Grotefend, C.R.; Radic, V.; Chung, C.; Chung, Y.-H.; Farzan, M.; Huang, I.C. IFITM3 polymorphism rs12252-C restricts influenza A viruses. PLoS ONE 2014, 9, e110096. [Google Scholar] [CrossRef]
- Pan, Y.; Yang, P.; Dong, T.; Zhang, Y.; Shi, W.; Peng, X.; Cui, S.; Zhang, D.; Lu, G.; Liu, Y.; et al. IFITM3 Rs12252-C Variant Increases Potential Risk for Severe Influenza Virus Infection in Chinese Population. Front. Cell Infect. Microbiol. 2017, 7, 294. [Google Scholar] [CrossRef]
- Xu, F.; Wang, G.; Zhao, F.; Huang, Y.; Fan, Z.; Mei, S.; Xie, Y.; Wei, L.; Hu, Y.; Wang, C.; et al. IFITM3 Inhibits SARS-CoV-2 Infection and Is Associated with COVID-19 Susceptibility. Viruses 2022, 14, 2553. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, I.; Afifipour, A.; Sakhaee, F.; Zamani, M.S.; Mirzaei Gheinari, F.; Anvari, E.; Fateh, A. Impact of interferon-induced transmembrane protein 3 gene rs12252 polymorphism on COVID-19 mortality. Cytokine 2022, 157, 155957. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, E.A.; Armellino, D.; Lam, G.; Cordon-Cardo, C.; Murray, H.W.; Evans, R.L. IFN-gamma and IFN-alpha induce the expression and synthesis of Leu 13 antigen by cultured human endothelial cells. J. Immunol. 1989, 143, 3961–3966. [Google Scholar] [CrossRef] [PubMed]
- Forero, A.; Ozarkar, S.; Li, H.; Lee, C.H.; Hemann, E.A.; Nadjsombati, M.S.; Hendricks, M.R.; So, L.; Green, R.; Roy, C.N.; et al. Differential Activation of the Transcription Factor IRF1 Underlies the Distinct Immune Responses Elicited by Type I and Type III Interferons. Immunity 2019, 51, 451–464.e6. [Google Scholar] [CrossRef] [PubMed]
- Alber, D.; Staeheli, P. Partial inhibition of vesicular stomatitis virus by the interferon-induced human 9-27 protein. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 1996, 16, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Griffin, L.M.; Little, A.S.; Huang, I.C.; Farzan, M.; Pyeon, D. The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus. PLoS ONE 2014, 9, e96579. [Google Scholar] [CrossRef]
- Rabbani, M.A.; Ribaudo, M.; Guo, J.T.; Barik, S. Identification of Interferon-Stimulated Gene Proteins That Inhibit Human Parainfluenza Virus Type 3. J. Virol. 2016, 90, 11145–11156. [Google Scholar] [CrossRef]
- McMichael, T.M.; Zhang, Y.; Kenney, A.D.; Zhang, L.; Zani, A.; Lu, M.; Chemudupati, M.; Li, J.; Yount, J.S. IFITM3 Restricts Human Metapneumovirus Infection. J. Infect. Dis. 2018, 218, 1582–1591. [Google Scholar] [CrossRef]
- Smith, S.E.; Busse, D.A.-O.; Binter, S.; Weston, S.; Diaz Soria, C.; Laksono, B.M.; Clare, S.; Van Nieuwkoop, S.; Van den Hoogen, B.G.; Clement, M.; et al. Interferon-Induced Transmembrane Protein 1 Restricts Replication of Viruses That Enter Cells via the Plasma Membrane. J. Virol. 2019, 93, e02003-18. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, L.; Zan, Y.; Du, N.; Yang, Y.; Tien, P. Human respiratory syncytial virus infection is inhibited by IFN-induced transmembrane proteins. J. Gen. Virol. 2015, 96, 170–182. [Google Scholar] [CrossRef]
- Everitt, A.R.; Clare, S.; McDonald, J.U.; Kane, L.; Harcourt, K.; Ahras, M.; Lall, A.; Hale, C.; Rodgers, A.; Young, D.B.; et al. Defining the range of pathogens susceptible to Ifitm3 restriction using a knockout mouse model. PLoS ONE 2013, 8, e80723. [Google Scholar] [CrossRef] [PubMed]
- Narayana, S.K.; Helbig, K.J.; McCartney, E.M.; Eyre, N.S.; Bull, R.A.; Eltahla, A.; Lloyd, A.R.; Beard, M.R. The Interferon-induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry. J. Biol. Chem. 2015, 290, 25946–25959. [Google Scholar] [CrossRef] [PubMed]
- Monel, B.; Compton, A.A.; Bruel, T.; Amraoui, S.; Burlaud-Gaillard, J.; Roy, N.; Guivel-Benhassine, F.; Porrot, F.; Génin, P.; Meertens, L.; et al. Zika virus induces massive cytoplasmic vacuolization and paraptosis-like death in infected cells. EMBO J. 2017, 36, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Weidner, J.M.; Qing, M.; Pan, X.-B.; Guo, H.; Xu, C.; Zhang, X.; Birk, A.; Chang, J.; Shi, P.-Y.; et al. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol. 2010, 84, 8332–8341. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.A.-O.; Wang, L.; Li, S.; Dorf, M.A.-O. ZMPSTE24 defends against influenza and other pathogenic viruses. J. Exp. Med. 2017, 214, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Mudhasani, R.; Tran, J.P.; Retterer, C.; Radoshitzky, S.R.; Kota, K.P.; Altamura, L.A.; Smith, J.M.; Packard, B.Z.; Kuhn, J.H.; Costantino, J.; et al. IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J. Virol. 2013, 87, 8451–8464. [Google Scholar] [CrossRef]
- Poddar, S.; Hyde, J.L.; Gorman, M.J.; Farzan, M.; Diamond, M.S. The Interferon-Stimulated Gene IFITM3 Restricts Infection and Pathogenesis of Arthritogenic and Encephalitic Alphaviruses. J. Virol. 2016, 90, 8780–8794. [Google Scholar] [CrossRef] [PubMed]
- Weston, S.; Czieso, S.; White, I.J.; Smith, S.E.; Wash, R.S.; Diaz-Soria, C.; Kellam, P.; Marsh, M. Alphavirus Restriction by IFITM Proteins. Traffic 2016, 17, 997–1013. [Google Scholar] [CrossRef]
- Wilkins, J.; Zheng, Y.M.; Yu, J.; Liang, C.; Liu, S.A.-O. Nonhuman Primate IFITM Proteins Are Potent Inhibitors of HIV and SIV. PLoS ONE 2016, 11, e0156739. [Google Scholar] [CrossRef]
- Foster, T.L.; Wilson, H.; Iyer, S.S.; Coss, K.; Doores, K.; Smith, S.; Kellam, P.; Finzi, A.; Borrow, P.; Hahn, B.H.; et al. Resistance of Transmitted Founder HIV-1 to IFITM-Mediated Restriction. Cell Host Microbe 2016, 20, 429–442. [Google Scholar] [CrossRef]
- Zhao, X.; Sehgal, M.; Hou, Z.; Cheng, J.; Shu, S.; Wu, S.; Guo, F.; Le Marchand, S.J.; Lin, H.; Chang, J.; et al. Identification of Residues Controlling Restriction versus Enhancing Activities of IFITM Proteins on Entry of Human Coronaviruses. J. Virol. 2018, 92, e01535-17. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Kenney, A.D.; Kudryashova, E.; Zani, A.; Zhang, L.; Lai, K.K.; Hall-Stoodley, L.; Robinson, R.T.; Kudryashov, D.A.-O.; Compton, A.A.-O.; et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021, 40, e106501. [Google Scholar] [CrossRef] [PubMed]
- Hickford, D.; Frankenberg, S.; Shaw, G.; Renfree, M.B. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genom. 2012, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- Benfield, C.T.O.; MacKenzie, F.; Ritzefeld, M.; Mazzon, M.; Weston, S.; Tate, E.W.; Teo, B.H.; Smith, S.E.; Kellam, P.; Holmes, E.C.; et al. Bat IFITM3 restriction depends on S-palmitoylation and a polymorphic site within the CD225 domain. Life Sci. Alliance 2020, 3, e201900542. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.A.-O.; Mendivil Ramos, O.A.-O.; Kramer, M.A.-O.; Goodwin, S.A.-O.; Oppenheim, S.A.-O.; Becker, D.A.-O.; Schatz, M.A.-O.; Simmons, N.A.-O.; Siepel, A.A.-O.; McCombie, W.A.-O. Long-Read Sequencing Reveals Rapid Evolution of Immunity- and Cancer-Related Genes in Bats. Genome Biol. Evol. 2023, 15, evad148. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Smith, N.; Yu, L.; Paton, I.R.; Gutowska, M.W.; Forrest, H.L.; Danner, A.F.; Seiler, J.P.; Digard, P.; Webster, R.G.; et al. A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genom. 2015, 16, 574. [Google Scholar] [CrossRef]
- Bassano, I.; Ong, S.H.; Sanz-Hernandez, M.; Vinkler, M.; Kebede, A.; Hanotte, O.; Onuigbo, E.; Fife, M.; Kellam, P.A.-O. Comparative analysis of the chicken IFITM locus by targeted genome sequencing reveals evolution of the locus and positive selection in IFITM1 and IFITM3. BMC Genom. 2019, 20, 272. [Google Scholar] [CrossRef] [PubMed]
- Compton, A.A.-O.; Roy, N.; Porrot, F.; Billet, A.; Casartelli, N.; Yount, J.S.; Liang, C.; Schwartz, O. Natural mutations in IFITM3 modulate post-translational regulation and toggle antiviral specificity. EMBO Rep. 2016, 17, 1657–1671. [Google Scholar] [CrossRef]
- Siegrist, F.; Ebeling, M.; Certa, U. The small interferon-induced transmembrane genes and proteins. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2011, 31, 183–197. [Google Scholar] [CrossRef]
- Bailey, C.C.; Kondur, H.R.; Huang, I.C.; Farzan, M. Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J. Biol. Chem. 2013, 288, 32184–32193. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, J.; Li, M.; Yang, H.; Zhang, C. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS ONE 2012, 7, e49265. [Google Scholar] [CrossRef] [PubMed]
- Chesarino, N.M.; Compton, A.A.-O.; McMichael, T.M.; Kenney, A.D.; Zhang, L.; Soewarna, V.; Davis, M.; Schwartz, O.A.-O.; Yount, J.A.-O. IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep. 2017, 18, 1740–1751. [Google Scholar] [CrossRef] [PubMed]
- Yount, J.S.; Moltedo, B.; Yang, Y.Y.; Charron, G.; Moran, T.M.; López, C.B.; Hang, H.C. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 2010, 6, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Chesarino, N.M.; McMichael, T.M.; Hach, J.C.; Yount, J.S. Phosphorylation of the antiviral protein interferon-inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J. Biol. Chem. 2014, 289, 11986–11992. [Google Scholar] [CrossRef] [PubMed]
- Yount, J.S.; Karssemeijer, R.A.; Hang, H.C. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 2012, 287, 19631–19641. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Han, Q.; Nie, J.; Cao, X.; Chen, Z.; Yin, S.; Gao, Y.; Lin, F.; Zhou, X.; Xu, K.; et al. Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J. Biol. Chem. 2013, 288, 35093–35103. [Google Scholar] [CrossRef]
- Jia, R.; Xu, F.; Qian, J.; Yao, Y.; Miao, C.; Zheng, Y.M.; Liu, S.L.; Guo, F.; Geng, Y.; Qiao, W.; et al. Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell. Microbiol. 2014, 16, 1080–1093. [Google Scholar] [CrossRef] [PubMed]
- John, S.P.; Chin, C.R.; Perreira, J.M.; Feeley, E.M.; Aker, A.M.; Savidis, G.; Smith, S.E.; Elia, A.E.H.; Everitt, A.R.; Vora, M.; et al. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J. Virol. 2013, 87, 7837–7852. [Google Scholar] [CrossRef]
- Jia, R.; Pan, Q.; Ding, S.; Rong, L.; Liu, S.-L.; Geng, Y.; Qiao, W.; Liang, C. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 2012, 86, 13697–13707. [Google Scholar] [CrossRef]
- Hach, J.C.; McMichael, T.; Chesarino, N.M.; Yount, J.S. Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J. Virol. 2013, 87, 9923–9927. [Google Scholar] [CrossRef]
- Ling, S.; Zhang, C.; Wang, W.; Cai, X.; Yu, L.; Wu, F.; Zhang, L.; Tian, C. Combined approaches of EPR and NMR illustrate only one transmembrane helix in the human IFITM3. Sci. Rep. 2016, 5, 24029. [Google Scholar] [CrossRef] [PubMed]
- Friedlová, N.; Zavadil Kokáš, F.; Hupp, T.R.; Vojtěšek, B.; Nekulová, M. IFITM protein regulation and functions: Far beyond the fight against viruses. Front. Immunol. 2022, 13, 1042368. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.C.; Huang, I.C.; Kam, C.; Farzan, M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012, 8, e1002909. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Lei, N.; Lu, J.; Gao, R.B.; Li, Z.; Liu, L.Q.; Sun, Y.; Guo, J.F.; Wang, D.Y.; Shu, Y.L. Interferon-induced Transmembrane Protein 3 Prevents Acute Influenza Pathogenesis in Mice. Biomed. Environ. Sci. BES 2020, 33, 295–305. [Google Scholar] [PubMed]
- Chmielewska, A.A.-O.; Gómez-Herranz, M.; Gach, P.; Nekulova, M.; Bagnucka, M.A.-O.; Lipińska, A.D.; Rychłowski, M.; Hoffmann, W.; Król, E.; Vojtesek, B.; et al. The Role of IFITM Proteins in Tick-Borne Encephalitis Virus Infection. J. Virol. 2022, 96, e0113021. [Google Scholar] [CrossRef]
- Unali, G.; Crivicich, G.; Pagani, I.A.-O.; Abou-Alezz, M.A.-O.; Folchini, F.; Valeri, E.; Matafora, V.A.-O.X.; Reisz, J.A.; Giordano, A.M.S.; Cuccovillo, I.A.-O.; et al. Interferon-inducible phospholipids govern IFITM3-dependent endosomal antiviral immunity. EMBO J. 2023, 42, e112234. [Google Scholar] [CrossRef] [PubMed]
- Amini-Bavil-Olyaee, S.; Choi, Y.J.; Lee, J.H.; Shi, M.; Huang, I.C.; Farzan, M.; Jung, J.U. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 2013, 13, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.; Datta, S.A.K.; Beaven, A.H.; Jolley, A.A.; Sodt, A.J.; Compton, A.A. Cholesterol Binds the Amphipathic Helix of IFITM3 and Regulates Antiviral Activity. J. Mol. Biol. 2022, 434, 167759. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.A.-O.; Coomer, C.A.-O.; Majdoul, S.A.-O.; Ding, S.A.-O.X.; Padilla-Parra, S.A.-O.; Compton, A.A.-O. Homology-guided identification of a conserved motif linking the antiviral functions of IFITM3 to its oligomeric state. eLife 2020, 9, e58537. [Google Scholar] [CrossRef]
- Desai, T.M.; Marin, M.; Chin, C.R.; Savidis, G.; Brass, A.L.; Melikyan, G.B. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014, 10, e1004048. [Google Scholar] [CrossRef]
- Li, K.; Markosyan, R.M.; Zheng, Y.M.; Golfetto, O.; Bungart, B.; Li, M.; Ding, S.; He, Y.; Liang, C.; Lee, J.C.; et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013, 9, e1003124. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.A.-O.; Steinkühler, J.A.-O.; Marin, M.; Li, X.; Lu, W.A.-O.; Dimova, R.; Melikyan, G.A.-O. Interferon-Induced Transmembrane Protein 3 Blocks Fusion of Diverse Enveloped Viruses by Altering Mechanical Properties of Cell Membranes. ACS Nano 2021, 15, 8155–8170. [Google Scholar] [CrossRef] [PubMed]
- Anafu, A.A.; Bowen, C.H.; Chin, C.R.; Brass, A.L.; Holm, G.H. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 2013, 288, 17261–17271. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.S.; He, R.; Hoffmann, H.H.; Das, T.; Thinon, E.; Rice, C.M.; Peng, T.; Chandran, K.; Hang, H.C. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 2019, 15, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.A.-O.; Liao, X.; Huang, B.; Wang, L.; Liao, G.; Luo, C.; Wen, S.; Fang, S.; Luo, H.; Shu, Y. The truncated IFITM3 facilitates the humoral immune response in inactivated influenza vaccine-vaccinated mice via interaction with CD81. Emerg. Microbes Infect. 2023, 12, 2246599. [Google Scholar] [CrossRef]
- Rahman, K.; Wilt, I.; Jolley, A.A.; Chowdhury, B.; Datta, S.A.K.; Compton, A.A. SNARE mimicry by the CD225 domain of IFITM3 enables regulation of homotypic late endosome fusion. bioRxiv 2024. [Google Scholar] [CrossRef]
- Chen, L.; Li, X.; Deng, Y.; Bi, Y.; Yan, Z.; Yang, Y.; Zhang, X.; Li, H.; Xie, J.; Feng, R.A.-O. IFITM2 Presents Antiviral Response through Enhancing Type I IFN Signaling Pathway. Viruses 2023, 15, 866. [Google Scholar] [CrossRef]
- Gómez-Herranz, M.; Taylor, J.; Sloan, R.D. IFITM proteins: Understanding their diverse roles in viral infection, cancer, and immunity. J. Biol. Chem. 2023, 299, 102741. [Google Scholar] [CrossRef] [PubMed]
- Bedford, J.G.; O’Keeffe, M.; Reading, P.C.; Wakim, L.M. Rapid interferon independent expression of IFITM3 following T cell activation protects cells from influenza virus infection. PLoS ONE 2019, 14, e0210132. [Google Scholar] [CrossRef]
- Wakim, L.M.; Gupta, N.; Mintern, J.D.; Villadangos, J.A. Enhanced survival of lung tissue-resident memory CD8⁺ T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol. 2013, 14, 238–245. [Google Scholar] [CrossRef]
- Chen, Y.X.; Welte, K.; Gebhard, D.H.; Evans, R.L. Induction of T cell aggregation by antibody to a 16kd human leukocyte surface antigen. J. Immunol. 1984, 133, 2496–2501. [Google Scholar] [CrossRef] [PubMed]
- Stacey, M.A.; Clare, S.; Clement, M.; Marsden, M.; Abdul-Karim, J.; Kane, L.; Harcourt, K.; Brandt, C.; Fielding, C.A.; Smith, S.E.; et al. The antiviral restriction factor IFN-induced transmembrane protein 3 prevents cytokine-driven CMV pathogenesis. J. Clin. Investig. 2017, 127, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.A.; Forbester, J.L.; Marsden, M.; Sabberwal, P.; Sommerville, M.S.; Wellington, D.A.; Dimonte, S.; Clare, S.; Harcourt, K.; Yin, Z.A.; et al. IFITM3 restricts virus-induced inflammatory cytokine production by limiting Nogo-B mediated TLR responses. Nat. Commun. 2022, 13, 5294. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Young, J.; Weis, J.J.; Weis, J.H. Expression of the mouse fragilis gene products in immune cells and association with receptor signaling complexes. Genes Immun. 2006, 7, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Frey, M.; Appenheimer, M.M.; Evans, S.S. Tyrosine kinase-dependent regulation of L-selectin expression through the Leu-13 signal transduction molecule: Evidence for a protein kinase C-independent mechanism of L-selectin shedding. J. Immunol. 1997, 158, 5424–5434. [Google Scholar] [CrossRef]
- Yang, N.; Liu, Z.; Pang, S.; Wu, J.; Liang, J.; Sun, L. Predicative value of IFITM2 in renal clear cell carcinoma: IFITM2 is associated with lymphatic metastasis and poor clinical outcome. Biochem. Biophys. Res. Commun. 2021, 534, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, A.; Wan, Y.; Liu, X.; Qiu, C.; Xi, X.; Ren, Y.; Wang, J.; Dong, Y.; Bao, M.; et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. USA 2014, 111, 769–774. [Google Scholar] [CrossRef]
- Amet, T.; Son, Y.M.; Jiang, L.; Cheon, I.S.; Huang, S.; Gupta, S.K.; Dent, A.L.; Montaner, L.J.; Yu, Q.; Sun, J. BCL6 represses antiviral resistance in follicular T helper cells. J. Leukoc. Biol. 2017, 102, 527–536. [Google Scholar] [CrossRef]
- Levine, S.; Xian, C.Y.; Agocha, B.; Allopenna, J.; Welte, K.; Armstrong, D.; Yang, S.Y.; Evans, R.L. Differential modulation of the CD-2 and CD-3 T cell activation pathways by a monoclonal antibody to Leu-13. Cell. Immunol. 1991, 132, 366–376. [Google Scholar] [CrossRef]
- Bradbury, L.E.; Kansas, G.S.; Levy, S.; Evans, R.L.; Tedder, T.F. The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules. J. Immunol. 1992, 149, 2841–2850. [Google Scholar] [CrossRef] [PubMed]
- Furmanski, A.L.; Barbarulo, A.; Solanki, A.; Lau, C.I.; Sahni, H.; Saldana, J.I.; D’Acquisto, F.; Crompton, T. The transcriptional activator Gli2 modulates T-cell receptor signalling through attenuation of AP-1 and NFκB activity. J. Cell Sci. 2015, 128, 2085–2095. [Google Scholar] [CrossRef] [PubMed]
- Wakim, L.M.; Woodward-Davis, A.; Liu, R.; Hu, Y.; Villadangos, J.; Smyth, G.; Bevan, M.J. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J. Immunol. 2012, 189, 3462–3471. [Google Scholar] [CrossRef] [PubMed]
- Chesarino, N.M.; McMichael, T.M.; Yount, J.S. Regulation of the trafficking and antiviral activity of IFITM3 by post-translational modifications. Future Microbiol. 2014, 9, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Chesarino, N.M.; McMichael, T.M.; Yount, J.S. E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3. PLoS Pathog. 2015, 11, e1005095. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, E.; Yánez, D.C.; Ross, S.; Lau, C.I.; Solanki, A.; Chawda, M.M.; Virasami, A.; Ranz, I.; Ono, M.; O’Shaughnessy, R.F.L.; et al. Sonic Hedgehog signaling limits atopic dermatitis via Gli2-driven immune regulation. J. Clin. Investig. 2019, 129, 3153–3170. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Dassopoulos, T.; Cope, L.; Maitra, A.; Brant, S.R.; Harris, M.L.; Bayless, T.M.; Parmigiani, G.; Chakravarti, S. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: Insights into distinctive pathogenesis. Inflamm. Bowel Dis. 2007, 13, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Hisamatsu, T.; Watanabe, M.; Ogata, H.; Ezaki, T.; Hozawa, S.; Ishii, H.; Kanai, T.; Hibi, T. Interferon-inducible gene family 1-8U expression in colitis-associated colon cancer and severely inflamed mucosa in ulcerative colitis. Cancer Res. 1999, 59, 5927–5931. [Google Scholar]
- Mo, J.S.; Na, K.S.; Yu, J.I.; Chae, S.C. Identification of the polymorphisms in IFITM1 gene and their association in a Korean population with ulcerative colitis. Immunol. Lett. 2013, 156, 118–122. [Google Scholar] [CrossRef]
- Seo, G.S.; Lee, J.K.; Yu, J.I.; Yun, K.J.; Chae, S.C.; Choi, S.C. Identification of the polymorphisms in IFITM3 gene and their association in a Korean population with ulcerative colitis. Exp. Mol. Med. 2010, 42, 99–104. [Google Scholar] [CrossRef]
- Alteber, Z.; Sharbi-Yunger, A.; Pevsner-Fischer, M.; Blat, D.; Roitman, L.; Tzehoval, E.; Elinav, E.; Eisenbach, L. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol. Cell Biol. 2018, 96, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; Monier, M.N.; Fradagrada, A.; Mitchell, K.; Baychelier, F.; Eid, P.; Johannes, L.; Lamaze, C. Stat-mediated signaling induced by type I and type II interferons (IFNs) is differentially controlled through lipid microdomain association and clathrin-dependent endocytosis of IFN receptors. Mol. Biol. Cell 2006, 17, 2896–2909. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Wang, D.; Li, D.; Zhao, Y.; Peng, Y.; Wellington, D.; Dai, Y.; Sun, H.; Sun, J.; Liu, G.; et al. High Level Antibody Response to Pandemic Influenza H1N1/09 Virus Is Associated With Interferon-Induced Transmembrane Protein-3 rs12252-CC in Young Adults. Front. Cell Infect. Microbiol. 2018, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Lei, N.; Li, Y.; Sun, Q.; Lu, J.; Zhou, J.; Li, Z.; Liu, L.; Guo, J.; Qin, K.; Wang, H.; et al. IFITM3 affects the level of antibody response after influenza vaccination. Emerg. Microbes Infect. 2020, 9, 976–987. [Google Scholar] [CrossRef]
- Shi, G.; Schwartz, O.A.-O.; Compton, A.A.-O. More than meets the I: The diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology 2017, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, W.; Han, Y.; Cheng, H.; Liu, Q.; Ke, S.; Zhu, F.A.-O.; Lu, Y.; Dai, X.; Wang, C.; et al. FOXP3(+) regulatory T cell perturbation mediated by the IFNγ-STAT1-IFITM3 feedback loop is essential for anti-tumor immunity. Nat. Commun. 2024, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Deblandre, G.A.; Marinx, O.P.; Evans, S.S.; Majjaj, S.; Leo, O.; Caput, D.; Huez, G.A.; Wathelet, M.G. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J. Biol. Chem. 1995, 270, 23860–23866. [Google Scholar] [CrossRef]
- Yang, G.; Xu, Y.; Chen, X.; Hu, G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene 2007, 26, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Gan, C.P.; Sam, K.K.; Yee, P.S.; Zainal, N.S.; Lee, B.K.B.; Abdul Rahman, Z.A.; Patel, V.; Tan, A.C.; Zain, R.B.; Cheong, S.A.-O. IFITM3 knockdown reduces the expression of CCND1 and CDK4 and suppresses the growth of oral squamous cell carcinoma cells. Cell. Oncol. 2019, 42, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.A.-O.; Wang, S.; Gao, M.; Chang, J.; Sun, J.; Qin, L.; Li, A.A.-O.; Lv, F.; Lou, J.; Zhang, Y.A.-O.; et al. Interferon-Induced Transmembrane Protein 3 Expression Upregulation Is Involved in Progression of Hepatocellular Carcinoma. BioMed Res. Int. 2021, 2021, 5612138. [Google Scholar] [CrossRef]
- Rajapaksa, U.S.; Jin, C.; Dong, T. Malignancy and IFITM3: Friend or Foe? Front Oncol. 2020, 10, 593245. [Google Scholar] [CrossRef]
- He, J.D.; Luo, H.-L.; Li, J.; Feng, W.-T.; Chen, L.-B. Influences of the interferon induced transmembrane protein 1 on the proliferation, invasion, and metastasis of the colorectal cancer SW480 cell lines. Chin. Med. J. 2012, 125, 517–522. [Google Scholar] [PubMed]
- Sari, I.N.; Yang, Y.G.; Phi, L.T.; Kim, H.; Baek, M.J.; Jeong, D.; Kwon, H.Y. Interferon-induced transmembrane protein 1 (IFITM1) is required for the progression of colorectal cancer. Oncotarget 2016, 7, 86039–86050. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.G.; Koh, Y.W.; Sari, I.N.; Jun, N.; Lee, S.; Phi, L.T.H.; Kim, K.S.; Wijaya, Y.T.; Lee, S.H.; Baek, M.J.; et al. Interferon-induced transmembrane protein 1-mediated EGFR/SOX2 signaling axis is essential for progression of non-small cell lung cancer. Int. J. Cancer 2018, 144, 2020–2032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Z.; Kong, D.; Zhao, X.; Chen, X.; Chai, W. Knockdown of interferon-induced transmembrane protein 1 inhibited proliferation, induced cell cycle arrest and apoptosis, and suppressed MAPK signaling pathway in pancreatic cancer cells. Biosci. Biotechnol. Biochem. 2020, 84, 1603–1613. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, L.; Fan, Y.; Hong, Y.; Yang, X.; Li, Y.; Lu, J.; Lv, J.; Pan, X.; Qu, F.; et al. IFITM3 promotes bone metastasis of prostate cancer cells by mediating activation of the TGF-β signaling pathway. Cell Death Dis. 2019, 10, 517. [Google Scholar] [CrossRef]
- Wang, H.A.-O.; Tang, F.; Bian, E.; Zhang, Y.; Ji, X.; Yang, Z.; Zhao, B. IFITM3/STAT3 axis promotes glioma cells invasion and is modulated by TGF-β. Mol. Biol. Rep. 2020, 47, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.; Siegrist, F.; Foser, S.; Certa, U. Interferon-alpha induces reversible DNA demethylation of the interferon-induced transmembrane protein-3 core promoter in human melanoma cells. J. Interferon Cytokine Res. 2011, 31, 601–608. [Google Scholar] [CrossRef]
- Horváth, S.; Mirnics, K. Immune system disturbances in schizophrenia. Biol. Psychiatry 2013, 75, 316–323. [Google Scholar] [CrossRef]
- Daniel-Carmi, V.; Makovitzki-Avraham, E.; Reuven, E.-M.; Goldstein, I.; Zilkha, N.; Rotter, V.; Tzehoval, E.; Eisenbach, L. The human 1-8D gene (IFITM2) is a novel p53 independent pro-apoptotic gene. Int. J. Cancer 2009, 125, 2810–2819. [Google Scholar] [CrossRef]
- Fumoto, S.; Shimokuni, T.; Tanimoto, K.; Hiyama, K.; Otani, K.; Ohtaki, M.; Hihara, J.; Yoshida, K.; Hiyama, E.; Noguchi, T.; et al. Selection of a novel drug-response predictor in esophageal cancer: A novel screening method using microarray and identification of IFITM1 as a potent marker gene of CDDP response. Int. J. Oncol. 2008, 32, 413–423. [Google Scholar] [CrossRef]
- Khodarev, N.N.; Beckett, M.; Labay, E.; Darga, T.; Roizman, B.; Weichselbaum, R.R. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1714–1719. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, L.; Xi, Y.; Sun, R.; Wang, H.; Ren, Y.; Zhao, L.; Wang, X.; Li, X.A.-O. Combination of IFITM1 knockdown and radiotherapy inhibits the growth of oral cancer. Cancer Sci. 2018, 109, 3115–3128. [Google Scholar] [CrossRef] [PubMed]
- Lui, A.J.; Geanes, E.S.; Ogony, J.; Behbod, F.; Marquess, J.; Valdez, K.; Jewell, W.; Tawfik, O.; Lewis-Wambi, J. IFITM1 suppression blocks proliferation and invasion of aromatase inhibitor-resistant breast cancer in vivo by JAK/STAT-mediated induction of p21. Cancer Lett. 2017, 399, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Lui, A.; Ogony, J.; Jan, R.; Sims, P.J.; Lewis-Wambi, J. Targeting interferon response genes sensitizes aromatase inhibitor resistant breast cancer cells to estrogen-induced cell death. Breast Cancer Res. 2015, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Gao, H.; Chen, P.; Jia, J.; Wu, S. Knockdown of interferon-induced transmembrane protein 3 expression suppresses breast cancer cell growth and colony formation and affects the cell cycle. Oncol. Rep. 2013, 30, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T. Perinatal innate immune activation and neuropsychological development. Nihon Shinkei Seishin Yakurigaku Zasshi Jpn. J. Psychopharmacol. 2013, 33, 149–154. [Google Scholar]
- Saetre, P.; Emilsson, L.; Axelsson, E.; Kreuger, J.; Lindholm, E.; Jazin, E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry 2007, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Lang, R.; Li, H.; Luo, X.; Liu, C.; Zhang, Y.; Guo, S.; Xu, J.; Bao, C.; Dong, W.; Yu, Y. Expression and mechanisms of interferon-stimulated genes in viral infection of the central nervous system (CNS) and neurological diseases. Front. Immunol. 2022, 13, 1008072. [Google Scholar] [CrossRef]

| Inhibited | Resistant [2,27] |
|---|---|
| orthomyxoviruses (such as IAV [6]), paramyxoviruses (parainfluenza virus [28], metapneumovirus [29], and respiratory syncytial virus [30,31,32]), rhabdoviruses (vesicular stomatitis virus (VSV), flaviviruses (WNV [6], DENV [6], hepatitis C virus (HCV) [33], Zika virus (ZIKV) [34] and yellow fever virus [35]), filoviruses (Ebola virus (EBOV) [3,9] and Marburg virus [3]), poxviruses (vaccinia virus and cowpox virus (CPXV) [36], bunyaviruses (Rift Valley fever virus and La Crosse virus) [37], alphaviruses (chikungunya virus [38], Sindbis virus [39], Semliki Forest virus [39]), lentiviruses (human and simian immunodeficiency viruses) [5,40,41], and coronaviruses (human coronavirus 229E (hCoV-229E) [42], severe acute respiratory syndrome coronavirus (SARS-CoV) [3], Middle East respiratory syndrome coronavirus (MERS-CoV) [4] and SARS-CoV-2 [43]) | amphotropic murine leukemia virus, Sendai virus, papillomavirus, cytomegalovirus, adenovirus, and the arenaviruses Lassa virus (LASV), Machupo virus, and lymphocyticchoriomeningitis virus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Q.; Wang, L.; Liao, X.; Huang, B.; Luo, C.; Liao, G.; Yuan, L.; Liu, X.; Luo, H.; Shu, Y. Research Progress into the Biological Functions of IFITM3. Viruses 2024, 16, 1543. https://doi.org/10.3390/v16101543
Xie Q, Wang L, Liao X, Huang B, Luo C, Liao G, Yuan L, Liu X, Luo H, Shu Y. Research Progress into the Biological Functions of IFITM3. Viruses. 2024; 16(10):1543. https://doi.org/10.3390/v16101543
Chicago/Turabian StyleXie, Qian, Liangliang Wang, Xinzhong Liao, Bi Huang, Chuming Luo, Guancheng Liao, Lifang Yuan, Xuejie Liu, Huanle Luo, and Yuelong Shu. 2024. "Research Progress into the Biological Functions of IFITM3" Viruses 16, no. 10: 1543. https://doi.org/10.3390/v16101543






