Development and Application of a Fully Automated Chemiluminescence Enzyme Immunoassay for the Detection of Antibodies Against Porcine Circovirus 3 Cap
Abstract
:1. Introduction
2. Results
2.1. Preparation of PCV3 Cap
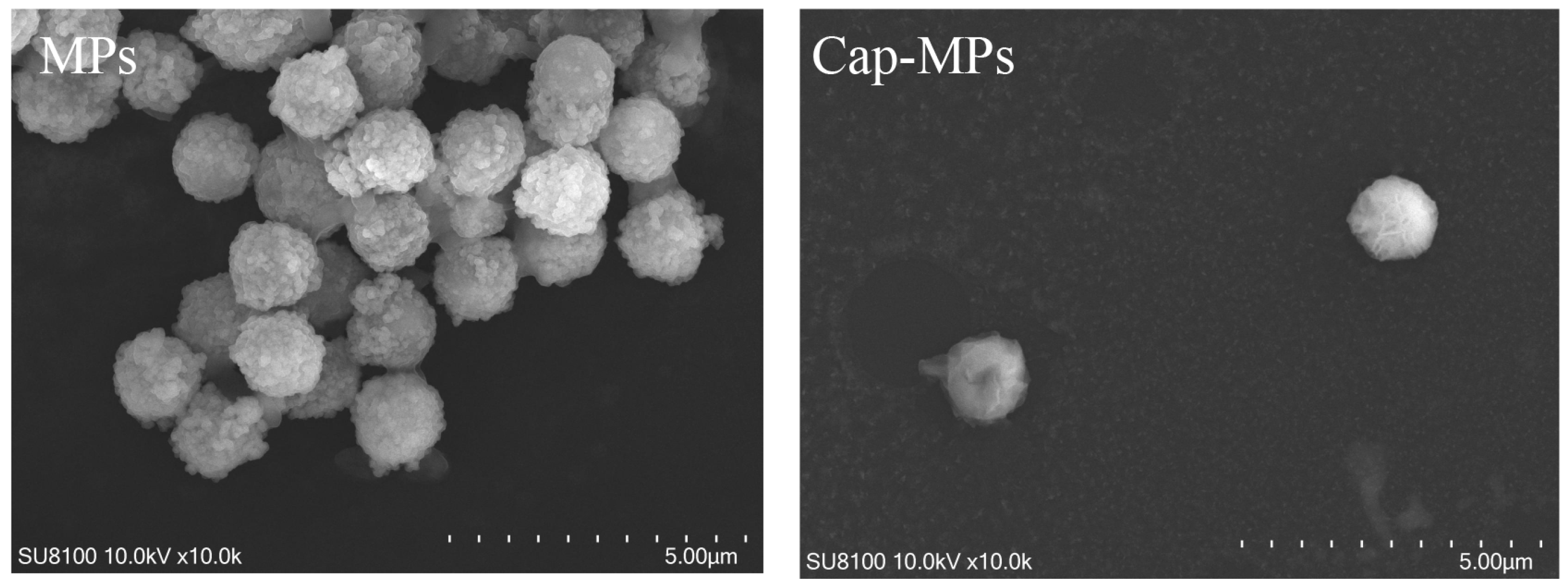
2.2. Characterization of Cap-MPs
2.3. Optimization of Working Conditions
2.3.1. Doses of Recombinant Cap Coupled to MPs
2.3.2. Dilutions of Cap-MPs and the AP-Conjugated Antibody
2.3.3. Reaction Time of Substrate Solution
2.3.4. Detection Procedure (One-Step and Two-Step)
2.4. Determination of Cut-Off Value
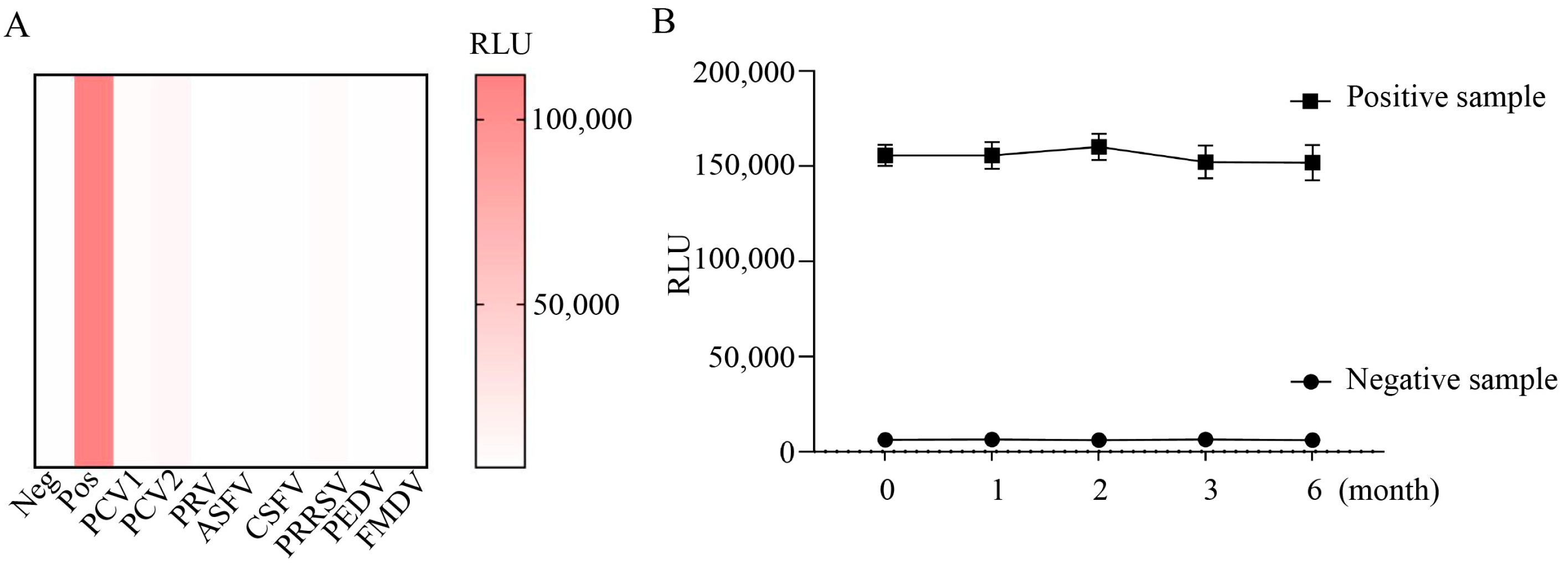
2.5. Assessment of Cross-Reactivity of CLEIA
2.6. Stability Test
2.7. Repeatability Test
2.8. Comparison of CLEIA with Commercial Kit
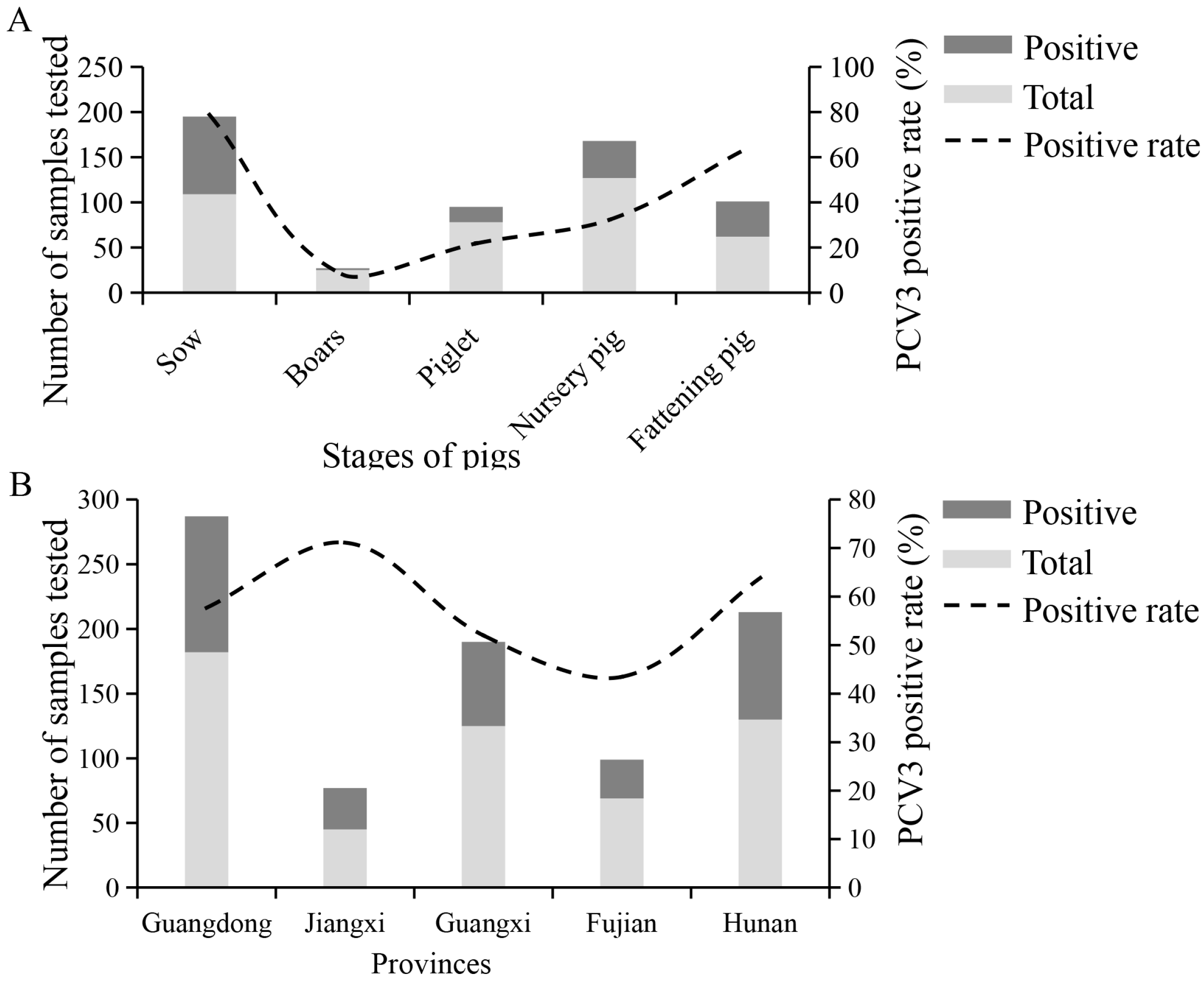
2.9. Serological Evidence for PCV3 Infection in South China
3. Discussion
4. Materials and Methods
4.1. Serum Samples
4.2. Expression and Purification of PCV3 Cap Using Baculovirus
4.3. Expression and Purification of Recombinant Cap Using E. coli
4.4. Conjugation of Cap Protein to MPs
4.5. CLEIA Procedure
4.6. Optimization of Parameters
4.6.1. Optimization of Doses of Coated Antigen
4.6.2. Optimization of Dilutions of Cap-MPs
4.6.3. Optimization of Dilutions of AP-Conjugated Antibody
4.6.4. Optimization of Reaction Time
4.6.5. Optimization of Detection Procedure
4.7. Determination of Cut-Off Value, Diagnostic Sensitivity, and Specificity
4.8. Determination of Specificity
4.9. Determination of Stability
4.10. Determination of Reproducibility
4.11. Analysis of Clinical Serum Samples and Comparison of CLEIA with Commercial Kit
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palinski, R.; Pineyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, e01879-16. [Google Scholar] [CrossRef]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016, 13, 184. [Google Scholar] [CrossRef]
- Rodrigues, I.; Cruz, A.; Souza, A.E.; Knackfuss, F.B.; Costa, C.; Silveira, R.L.; Castro, T.X. Retrospective study of porcine circovirus 3 (PCV3) in swine tissue from Brazil (1967–2018). Braz. J. Microbiol. 2020, 51, 1391–1397. [Google Scholar] [CrossRef]
- Mai, J.; Wang, D.; Zou, Y.; Zhang, S.; Meng, C.; Wang, A.; Wang, N. High Co-infection Status of Novel Porcine Parvovirus 7 With Porcine Circovirus 3 in Sows That Experienced Reproductive Failure. Front. Vet. Sci. 2021, 8, 695553. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Ruan, H.; Qiao, S.; Deng, R.; Zhang, G. Co-infection status of porcine circoviruses (PCV2 and PCV3) and porcine epidemic diarrhea virus (PEDV) in pigs with watery diarrhea in Henan province, central China. Microb. Pathog. 2020, 142, 104047. [Google Scholar] [CrossRef]
- Chen, N.; Huang, Y.; Ye, M.; Li, S.; Xiao, Y.; Cui, B.; Zhu, J. Co-infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circoviruses (PCV2 and PCV3) in eight regions of China from 2016 to 2018. Infect. Genet. Evol. 2019, 68, 127–135. [Google Scholar] [CrossRef]
- Kruger, L.; Langin, M.; Reichart, B.; Fiebig, U.; Kristiansen, Y.; Prinz, C.; Kessler, B.; Egerer, S.; Wolf, E.; Abicht, J.M.; et al. Transmission of Porcine Circovirus 3 (PCV3) by Xenotransplantation of Pig Hearts into Baboons. Viruses 2019, 11, 650. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, D.; Wang, J.; Zhu, S.; She, R.; Ren, X.; Tian, J.; Quan, R.; Hou, L.; Li, Z.; et al. Induction of Porcine Dermatitis and Nephropathy Syndrome in Piglets by Infection with Porcine Circovirus Type 3. J. Virol. 2019, 93, e02045-18. [Google Scholar] [CrossRef]
- Deng, J.; Li, X.; Zheng, D.; Wang, Y.; Chen, L.; Song, H.; Wang, T.; Huang, Y.; Pang, W.; Tian, K. Establishment and application of an indirect ELISA for porcine circovirus 3. Arch. Virol. 2018, 163, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, L.; Xu, S. Pathogenicity and immune modulation of porcine circovirus 3. Front. Vet. Sci. 2023, 10, 1280177. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, F.; Correa-Fiz, F.; Franzo, G.; Sibila, M.; Nunez, J.I.; Segales, J. Current Knowledge on Porcine circovirus 3 (PCV3): A Novel Virus With a Yet Unknown Impact on the Swine Industry. Front. Vet. Sci. 2018, 5, 315. [Google Scholar] [CrossRef]
- Cao, X.; Huang, M.; Wang, Y.; Chen, Y.; Yang, H.; Quan, F. Immunogenicity Analysis of PCV3 Recombinant Capsid Protein Virus-like Particles and Their Application in Antibodies Detection. Int. J. Mol. Sci. 2023, 24, 10377. [Google Scholar] [CrossRef]
- Azim, M.A.U.; Hasan, M.; Ansari, I.H.; Nasreen, F. Chemiluminescence Immunoassay: Basic Mechanism and Applications. Bangladesh J. Nucl. Med. 2018, 18, 171–178. [Google Scholar] [CrossRef]
- Chen, G.; Jin, M.; Du, P.; Zhang, C.; Cui, X.; Zhang, Y.; Wang, J.; Jin, F.; She, Y.; Shao, H.; et al. A review of enhancers for chemiluminescence enzyme immunoassay. Food Agric. Immunol. 2017, 28, 315–327. [Google Scholar] [CrossRef]
- Xin, T.B.; Wang, X.; Jin, H.; Liang, S.X.; Lin, J.M.; Li, Z.J. Development of magnetic particle-based chemiluminescence enzyme immunoassay for the detection of 17beta-estradiol in environmental water. Appl. Biochem. Biotechnol. 2009, 158, 582–594. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.J.; Yuan, L.P.; Shen, Y.D.; Liu, Y.X.; Liu, B.; Zhang, S.W.; Xie, Z.X.; Lei, H.T.; Sun, Y.M.; Xu, Z.L. A full-automated magnetic particle-based chemiluminescence immunoassay for rapid detection of cortisol in milk. Anal. Chim. Acta 2018, 1035, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; He, J.; Chen, W.; Ho, H.P.; Kong, S.K.; Wang, C.; Long, J.; Loo, J.; Gu, D. Development of peptide-based chemiluminescence enzyme immunoassay (CLEIA) for diagnosis of dengue virus infection in human. Anal. Biochem. 2018, 556, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ouyang, H.; Yang, S.; Wang, L.; Fu, Z. Multiplexed detection of two proteins by a reaction kinetics-resolved chemiluminescence immunoassay strategy. Analyst 2015, 140, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Arruda, B.; Pineyro, P.; Derscheid, R.; Hause, B.; Byers, E.; Dion, K.; Long, D.; Sievers, C.; Tangen, J.; Williams, T.; et al. PCV3-associated disease in the United States swine herd. Emerg. Microbes Infect. 2019, 8, 684–698. [Google Scholar] [CrossRef]
- Hosseini, S.; Vázquez-Villegas, P.; Rito-Palomares, M.; Martinez-Chapa, S.O. Advantages, disadvantages and modifications of conventional ELISA. In Enzyme-Linked Immunosorbent Assay (ELISA); Springer: Singapore, 2018; pp. 67–115. [Google Scholar]
- Yang, Y.; Lv, C.; Fan, J.; Zhao, Y.; Jiang, L.; Sun, X.; Zhang, Q.; Jin, M. Development of a chemiluminescence immunoassay to accurately detect African swine fever virus antibodies in serum. J. Virol. Methods 2021, 298, 114269. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Cao, L.; Luo, J.; Zhou, G.; Zuo, Q.; Liu, X.; Hu, Y.; Tian, H.; Zheng, H. A chemiluminescent magnetic microparticle immunoassay for the detection of antibody against African swine fever virus. Appl. Microbiol. Biotechnol. 2023, 107, 3779–3788. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Z.; Zhao, F.R.; Gao, S.D.; Shao, J.J.; Zhang, Y.G.; Chang, H.Y. Development of a chemiluminescence immunoassay using recombinant non-structural epitope-based proteins to accurately differentiate foot-and-mouth disease virus-infected and vaccinated bovines. Transbound. Emerg. Dis. 2018, 65, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Shi, D.; Zhang, Z.; Bian, L.; Li, Z.; Liu, T.; He, C.; Xu, S.; Wu, Y.; Lin, G. A chemiluminescence immunoassay for precise automatic quality control of glycoprotein in human rabies vaccine. Vaccine 2021, 39, 7470–7476. [Google Scholar] [CrossRef]
- Zhu, J.; Dou, L.; Shao, S.; Kou, J.; Yu, X.; Wen, K.; Wang, Z.; Yu, W. An Automated and Highly Sensitive Chemiluminescence Immunoassay for Diagnosing Mushroom Poisoning. Front. Chem. 2021, 9, 813219. [Google Scholar] [CrossRef]
- Ma, Z.; Lv, J.; Zhang, Z.; Zhao, Y.; Pan, L.; Zhang, Y. A chemiluminescence immunoassay for rapid detection of classical swine fever virus E2 antibodies in pig serum samples. Transbound. Emerg. Dis. 2020, 67, 1797–1803. [Google Scholar] [CrossRef]
- Cinquanta, L.; Fontana, D.E.; Bizzaro, N. Chemiluminescent immunoassay technology: What does it change in autoantibody detection? Auto. Immun. Highlights 2017, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.C.; Lin, W.L.; Wu, C.M.; Chi, J.N.; Chien, M.S.; Huang, C. Characterization of porcine circovirus type 2 (PCV2) capsid particle assembly and its application to virus-like particle vaccine development. Appl. Microbiol. Biotechnol. 2012, 95, 1501–1507. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Terron, M.C.; Khandokar, Y.; Aragao, D.; Hardy, J.M.; Radjainia, M.; Jimenez-Zaragoza, M.; de Pablo, P.J.; Coulibaly, F.; Luque, D.; et al. Structural insights into the assembly and regulation of distinct viral capsid complexes. Nat. Commun. 2016, 7, 13014. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.B.; Li, Y.F.; Guo, J.Q.; Wang, Z.T.; Chen, Q.X.; Shen, H.G.; Zhou, J.Y. Development and validation of a recombinant capsid protein-based ELISA for detection of antibody to porcine circovirus type 2. Res. Vet. Sci. 2008, 84, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Mai, J.; Lei, B.; Zhang, Y.; Yang, Y.; Wang, N. Structure, Antigenic Properties, and Highly Efficient Assembly of PCV4 Capsid Protein. Front. Vet. Sci. 2021, 8, 695466. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; He, Q.; Zhang, Z.; Chen, H.; Gimenez-Lirola, L.; Yuan, F.; Bei, W. Detection of Porcine Circovirus Type 3 in Serum, Semen, Oral Fluid, and Preputial Fluid Samples of Boars. Vet. Sci. 2023, 10, 689. [Google Scholar] [CrossRef]
- Temeeyasen, G.; Lierman, S.; Arruda, B.L.; Main, R.; Vannucci, F.; Gimenez-Lirola, L.G.; Pineyro, P.E. Pathogenicity and immune response against porcine circovirus type 3 infection in caesarean-derived, colostrum-deprived pigs. J. Gen. Virol. 2021, 102, jgv001502. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, D.; Jiang, Y.; Li, Z.; Zou, Y.; Li, M.; Yu, H.; Huang, K.; Yang, Y.; Wang, N. Development and application of a baculovirus-expressed capsid protein-based indirect ELISA for detection of porcine circovirus 3 IgG antibodies. BMC Vet. Res. 2019, 15, 79. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Huang, Y.; Guo, Y.; Wang, L.; Zhang, Y.; Zhou, L.; Ge, X.; Han, J.; Guo, X.; Yang, H. Prevalence and Evolution Analysis of Porcine Circovirus 3 in China from 2018 to 2022. Animals 2022, 12, 1588. [Google Scholar] [CrossRef]
- Tang, C.; Verwilligen, A.; Sadoff, J.; Brandenburg, B.; Sneekes-Vriese, E.; van den Kerkhof, T.; Dillen, L.; Rutten, L.; Juraszek, J.; Callewaert, K.; et al. Absolute quantitation of binding antibodies from clinical samples. NPJ Vaccines 2024, 9, 8. [Google Scholar] [CrossRef]
- Tannous, B.A.; Verhaegen, M.; Christopoulos, T.K.; Kourakli, A. Combined flash- and glow-type chemiluminescent reactions for high-throughput genotyping of biallelic polymorphisms. Anal. Biochem. 2003, 320, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, D.; Liu, Y.; Zhang, L.; Peng, G.; Xu, Z.; Jia, H.; Song, C. Development of an effective one-step double-antigen sandwich ELISA based on p72 to detect antibodies against African swine fever virus. Front. Vet. Sci. 2023, 10, 1160583. [Google Scholar] [CrossRef] [PubMed]



| Sample No. | Intra-Assay | Inter-Assay | ||
|---|---|---|---|---|
| M ± SD | CV (%) | M ± SD | CV (%) | |
| 1 | 6107 ± 158.11 | 2.59 | 6110 ± 200.06 | 3.27 |
| 2 | 6975 ± 127.22 | 1.82 | 6868 ± 285.22 | 4.15 |
| 3 | 56,298 ± 2339.61 | 4.14 | 57,042 ± 3074.56 | 5.39 |
| 4 | 58,624 ± 1829.07 | 3.12 | 59,414 ± 4782.83 | 8.05 |
| 5 | 170,571 ± 6259.96 | 3.67 | 184,555 ± 13,103.41 | 7.10 |
| 6 | 182,180 ± 4645.59 | 2.55 | 193,328 ± 12,004.99 | 6.21 |
| Method | Commercial ELISA | ||||
|---|---|---|---|---|---|
| No. Positive | No. Negative | Total | Coincidence Rate | ||
| CLEIA | No. Positive | 95 | 8 | 103 | 92.23% (95/103) |
| No. Negative | 1 | 83 | 84 | 98.81% (83/84) | |
| Total | 96 | 91 | 187 | 95.19% (95 + 83)/187) | |
| Year | No. Samples Tested | No. Positive | Positive Rate (%) |
|---|---|---|---|
| 2021 | 112 | 59 | 52.68 |
| 2022 | 150 | 80 | 53.33 |
| 2023 | 289 | 176 | 60.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, D.; Zeng, D.; Wang, X.; Liu, Y.; Peng, G.; Xu, Z.; Song, C. Development and Application of a Fully Automated Chemiluminescence Enzyme Immunoassay for the Detection of Antibodies Against Porcine Circovirus 3 Cap. Viruses 2024, 16, 1925. https://doi.org/10.3390/v16121925
Wang L, Li D, Zeng D, Wang X, Liu Y, Peng G, Xu Z, Song C. Development and Application of a Fully Automated Chemiluminescence Enzyme Immunoassay for the Detection of Antibodies Against Porcine Circovirus 3 Cap. Viruses. 2024; 16(12):1925. https://doi.org/10.3390/v16121925
Chicago/Turabian StyleWang, Lei, Duan Li, Daoping Zeng, Xiaomin Wang, Yanlin Liu, Guoliang Peng, Zheng Xu, and Changxu Song. 2024. "Development and Application of a Fully Automated Chemiluminescence Enzyme Immunoassay for the Detection of Antibodies Against Porcine Circovirus 3 Cap" Viruses 16, no. 12: 1925. https://doi.org/10.3390/v16121925
APA StyleWang, L., Li, D., Zeng, D., Wang, X., Liu, Y., Peng, G., Xu, Z., & Song, C. (2024). Development and Application of a Fully Automated Chemiluminescence Enzyme Immunoassay for the Detection of Antibodies Against Porcine Circovirus 3 Cap. Viruses, 16(12), 1925. https://doi.org/10.3390/v16121925






