Hypoxia and Activation of Neutrophil Degranulation-Related Genes in the Peripheral Blood of COVID-19 Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of Peripheral Gene Dysregulation in COVID-19
2.2. Analysis of the Link between Hypoxia and Gene Dysregulation
2.3. Analysis of Gene Dysregulation in Other Severe Respiratory Infection
2.4. Analysis of Hypoxia Sensing Pathways and Neutrophil Degranulation
3. Results
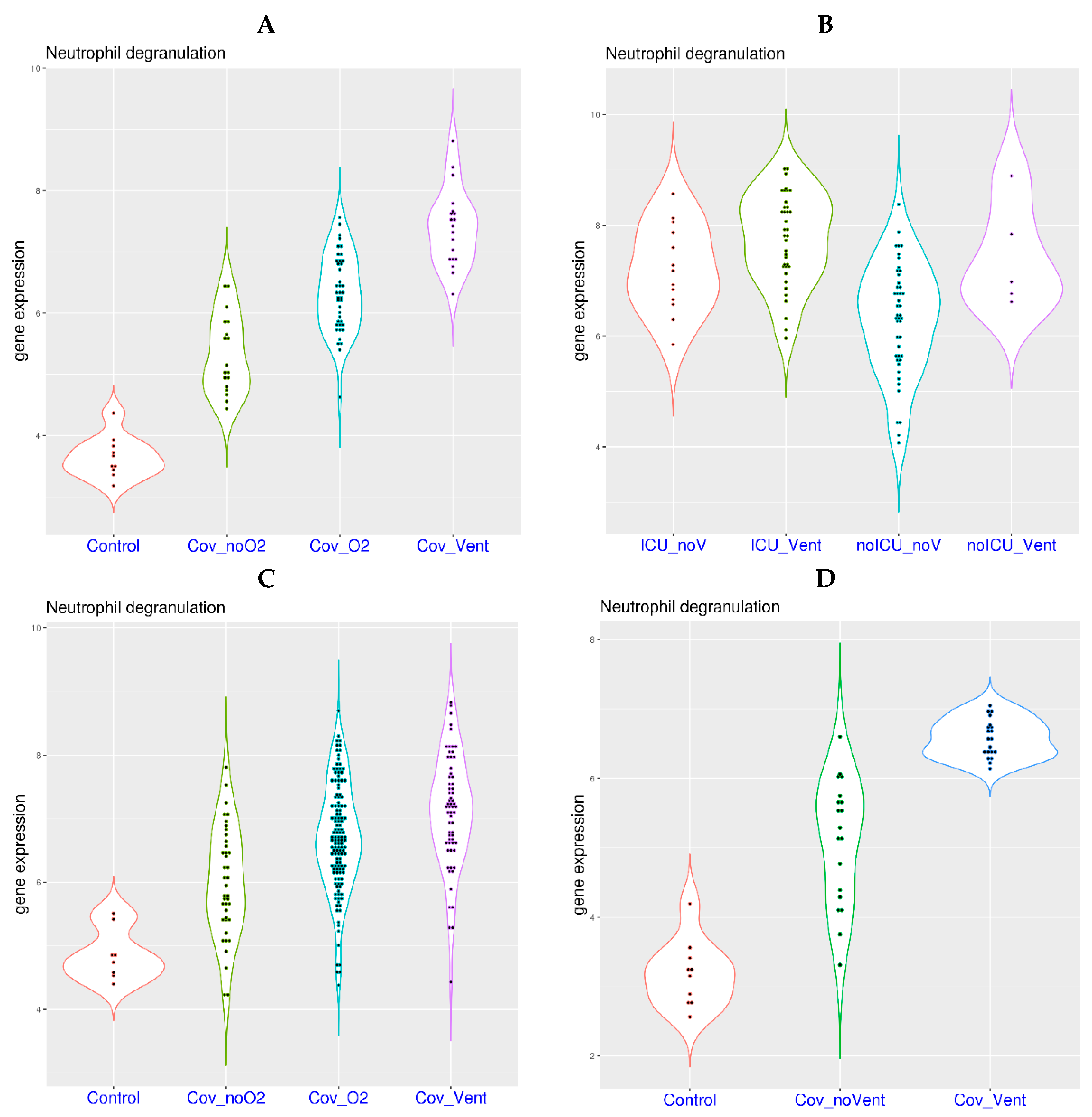
3.1. Activation of Neutrophil Degranulation-Related Genes in the Blood of COVID-19 Patients
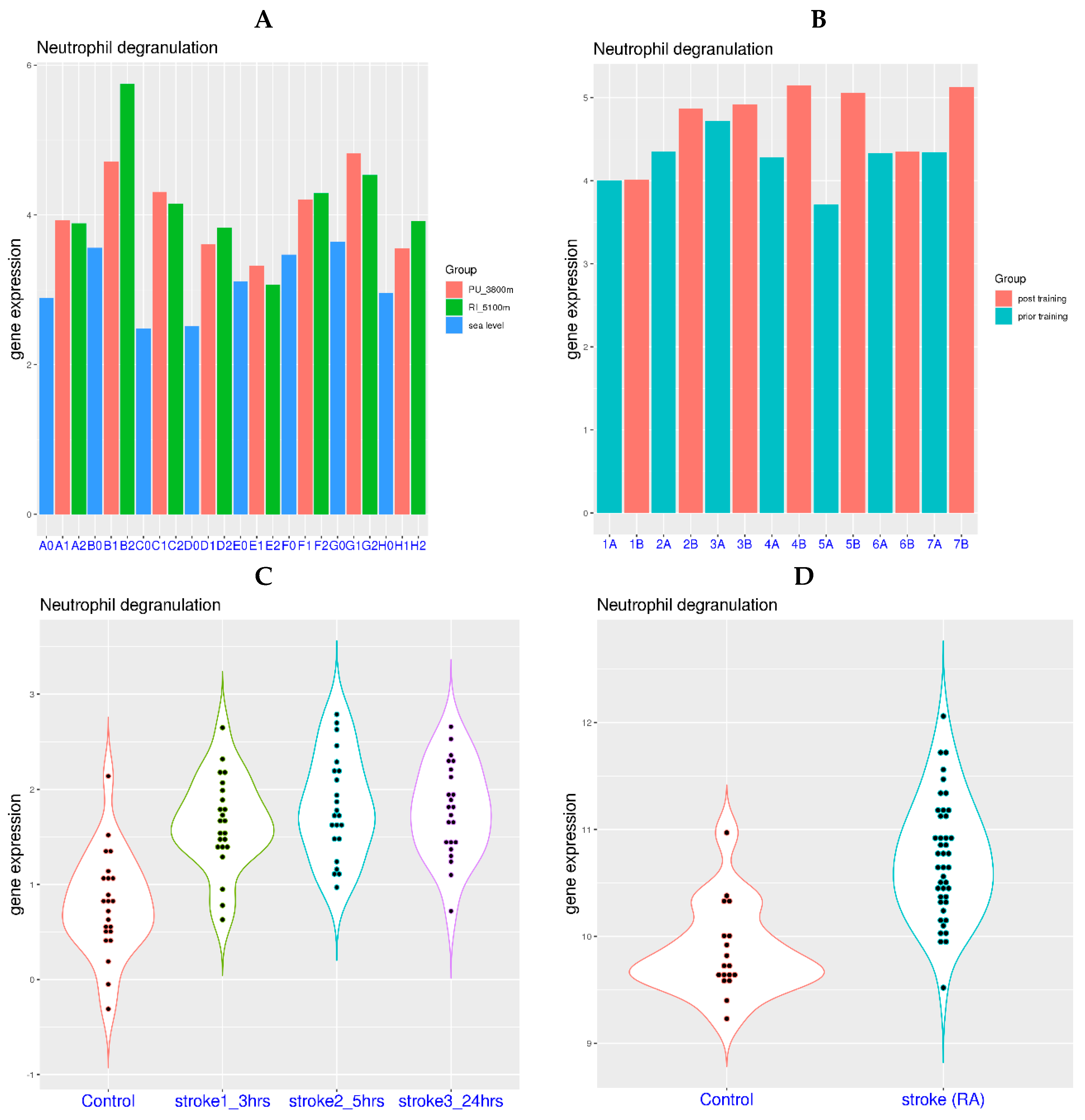
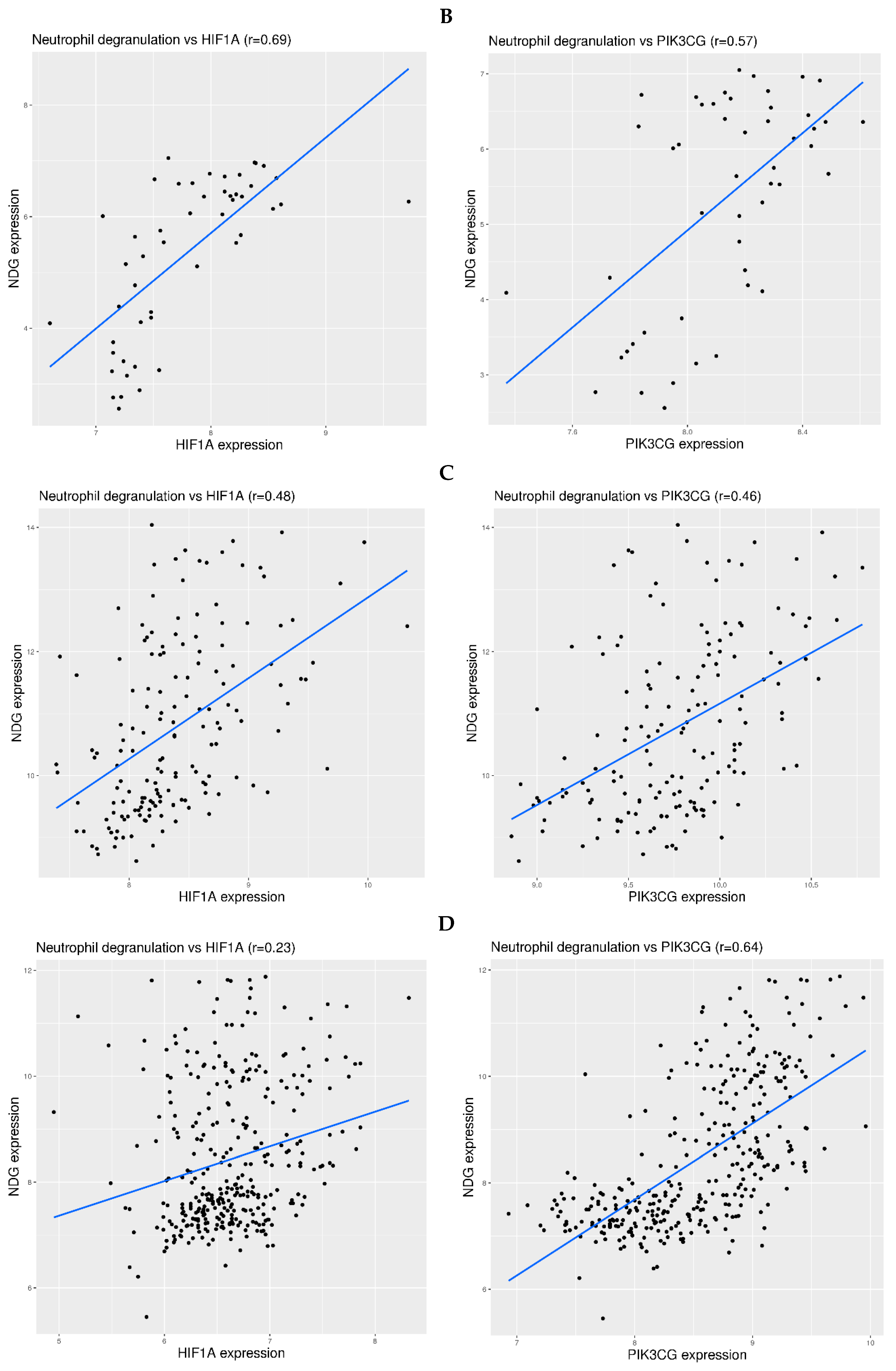
3.2. Neutrophil Degranulation-Related Genes Can Be Activated by Systemic Hypoxia
3.3. Oxygen Requirement and Gene Expression Related to Neutrophil Degranulation in COVID-19
3.4. Activation of Neutrophil Degranulation-Related Genes in Other Respiratory Diseases
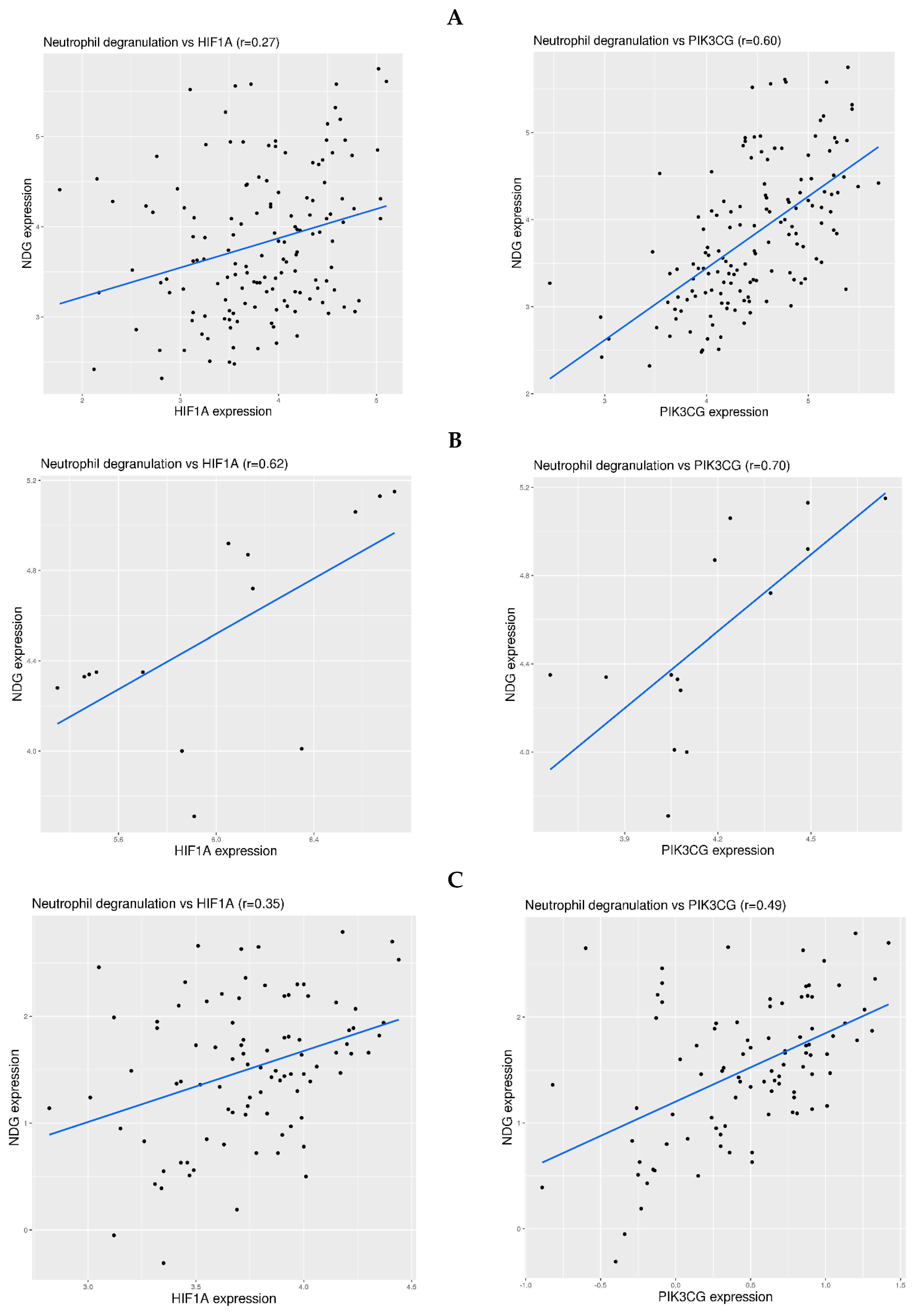
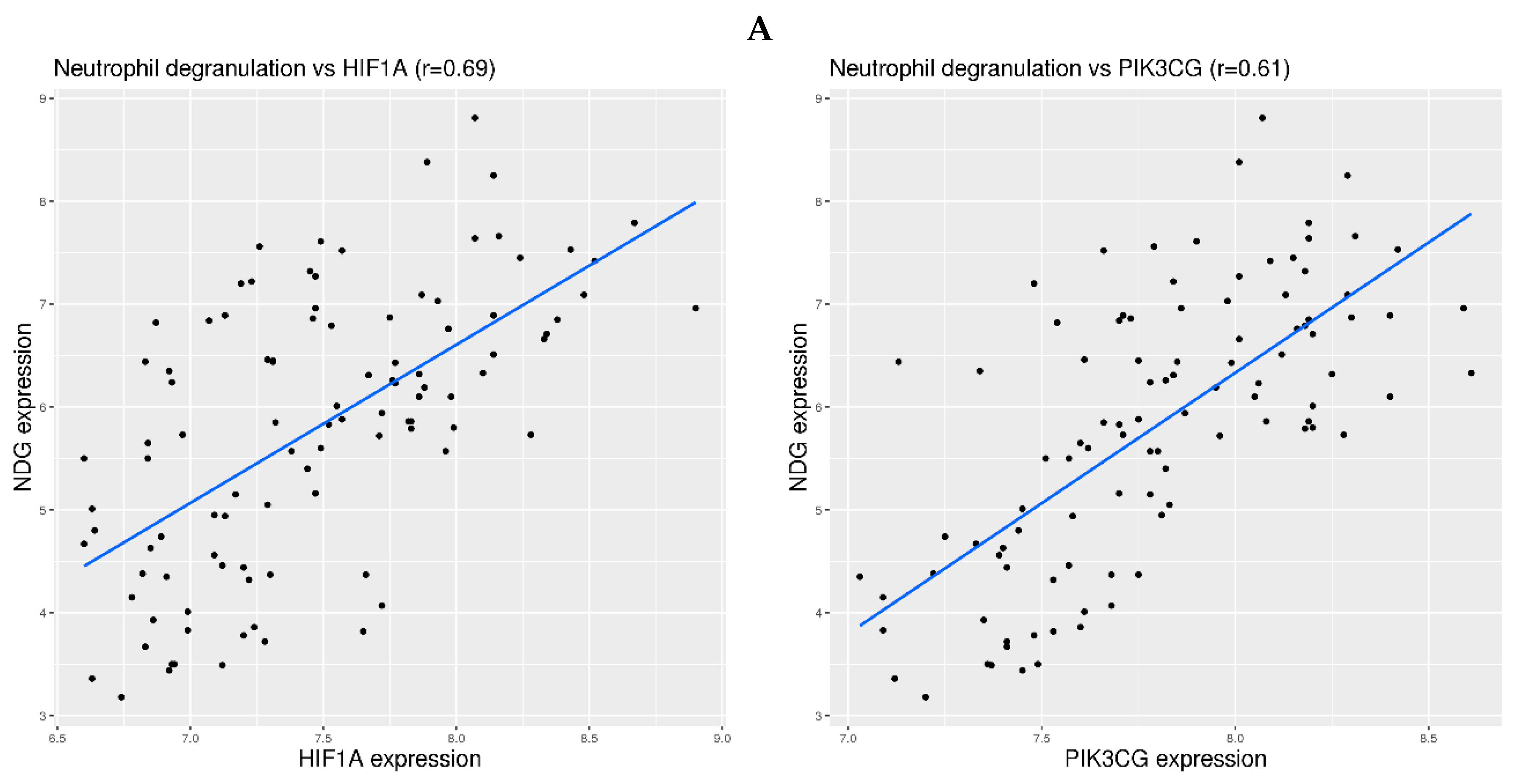
3.5. Examination of Hypoxia Sensing Pathways in the Activation of Neutrophil Degranulation-Related Genes in Blood
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dye, C. The benefits of large scale COVID-19 vaccination. BMJ 2022, 377, o867. [Google Scholar] [CrossRef] [PubMed]
- Rochwerg, B.; Agoritsas, T.; Lamontagne, F.C.C.; Leo, Y.-S.; Macdonald, H.; Agarwal, A.; Zeng, L.; Lytvyn, L.; Appiah, J.A.; Amin, W.A.; et al. living WHO guideline on drugs for COVID-19. BMJ 2022, 370, m3379. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Rivas, M.; Mora-Luján, J.M.; Formiga, F.; González, M.C.; Andreu, M.d.M.G.; Moreno-Torres, V.; García, G.M.G.; Pedrajas, J.N.A.; Boixeda, R.; Pérez-Lluna, L.; et al. Clusters of inflammation in COVID-19: Descriptive analysis and prognosis on more than 15,000 patients from the Spanish SEMI-COVID-19 Registry. Intern. Emerg. Med. 2022, 17, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Lei, H. A single transcript for the prognosis of disease severity in COVID-19 patients. Sci. Rep. 2021, 11, 12174. [Google Scholar] [CrossRef] [PubMed]
- Lei, H. A two-gene marker for the two-tiered innate immune response in COVID-19 patients. PLoS ONE 2023, 18, e0280392. [Google Scholar] [CrossRef]
- Santus, P.; Radovanovic, D.; Saderi, L.; Marino, P.; Cogliati, C.; De Filippis, G.; Rizzi, M.; Franceschi, E.; Pini, S.; Giuliani, F.; et al. Severity of respiratory failure at admission and in-hospital mortality in patients with COVID-19: A prospective observational multicentre study. BMJ Open 2020, 10, e043651. [Google Scholar] [CrossRef]
- Brouqui, P.; Amrane, S.; Million, M.; Cortaredona, S.; Parola, P.; Lagier, J.-C.; Raoult, D. Asymptomatic hypoxia in COVID-19 is associated with poor outcome. Int. J. Infect. Dis. 2021, 102, 233–238. [Google Scholar] [CrossRef]
- Thair, S.A.; He, Y.D.; Hasin-Brumshtein, Y.; Sakaram, S.; Pandya, R.; Toh, J.; Rawling, D.; Remmel, M.; Coyle, S.; Dalekos, G.N.; et al. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. iScience 2021, 24, 101947. [Google Scholar] [CrossRef]
- McClain, M.T.; Constantine, F.J.; Henao, R.; Liu, Y.; Tsalik, E.L.; Burke, T.W.; Steinbrink, J.M.; Petzold, E.; Nicholson, B.P.; Rolfe, R.; et al. Dysregulated transcriptional responses to SARS-CoV-2 in the periphery. Nat. Commun. 2021, 12, 1079. [Google Scholar] [CrossRef]
- Bernardes, J.P.; Mishra, N.; Tran, F.; Bahmer, T.; Best, L.; Blase, J.I.; Bordoni, D.; Franzenburg, J.; Geisen, U.; Josephs-Spaulding, J.; et al. Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity 2020, 53, 1296–1314.e9. [Google Scholar] [CrossRef] [PubMed]
- Lévy, Y.; Wiedemann, A.; Hejblum, B.P.; Durand, M.; Lefebvre, C.; Surénaud, M.; Lacabaratz, C.; Perreau, M.; Foucat, E.; Déchenaud, M.; et al. CD177, a specific marker of neutrophil activation, is associated with coronavirus disease 2019 severity and death. iScience 2021, 24, 102711. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Ishikawa, M.; Matsumoto, H.; Sugihara, F.; Okuzaki, D.; Hirata, H.; Ogura, H. Transcriptional differences between coronavirus disease 2019 and bacterial sepsis. Virol. J. 2022, 19, 198. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Flores, V.; Romero, R.; Xu, Y.; Theis, K.R.; Arenas-Hernandez, M.; Miller, D.; Peyvandipour, A.; Bhatti, G.; Galaz, J.; Gershater, M.; et al. Maternal-fetal immune responses in pregnant women infected with SARS-CoV-2. Nat. Commun. 2022, 13, 320. [Google Scholar] [CrossRef] [PubMed]
- Wargodsky, R.; Cruz, P.D.; LaFleur, J.; Yamane, D.; Kim, J.S.; Benjenk, I.; Heinz, E.; Irondi, O.O.; Farrar, K.; Toma, I.; et al. RNA Sequencing in COVID-19 patients identifies neutrophil activation biomarkers as a promising diagnostic platform for infections. PLoS ONE 2022, 17, e0261679. [Google Scholar] [CrossRef]
- Banerjee, U.; Chunchanur, S.; Ambica, R.; Balaji, K.N.; Singh, A.; Chakravortty, D.; Chandra, N. Systems-level profiling of early peripheral host-response landscape variations across COVID-19 severity states in an Indian cohort. Genes Immun. 2023, 24, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Prebensen, C.; Lefol, Y.; Myhre, P.L.; Lüders, T.; Jonassen, C.; Blomfeldt, A.; Omland, T.; Nilsen, H. Longitudinal whole blood transcriptomic analysis characterizes neutrophil activation and interferon signaling in moderate and severe COVID-19. Sci. Rep. 2023, 13, 10368. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Schughart, K.; Pelaia, T.M.; Chew, T.; Kim, K.; Karvunidis, T.; Knippenberg, B.; Teoh, S.; Phu, A.L.; Short, K.R.; et al. Blood transcriptome responses in patients correlate with severity of COVID-19 disease. Front. Immunol. 2022, 13, 1043219. [Google Scholar] [CrossRef]
- Gelemanovic, A.; Catipovic Ardalic, T.; Pribisalic, A.; Hayward, C.; Kolcic, I.; Polasek, O. Genome-Wide Meta-Analysis Identifies Multiple Novel Rare Variants to Predict Common Human Infectious Diseases Risk. Int. J. Mol. Sci. 2023, 24, 7006. [Google Scholar] [CrossRef]
- Consortium CO-M-oBA. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell 2022, 185, 916–938.e58. [Google Scholar] [CrossRef]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Glotov, A.S.; Zelenkova, I.E.; Vashukova, E.S.; Shuvalova, A.R.; Zolotareva, A.D.; Polev, D.E.; Barbitoff, Y.A.; Glotov, O.S.; Sarana, A.M.; Shcherbak, S.G.; et al. RNA Sequencing of Whole Blood Defines the Signature of High Intensity Exercise at Altitude in Elite Speed Skaters. Genes 2022, 13, 574. [Google Scholar] [CrossRef]
- Manella, G.; Ezagouri, S.; Champigneulle, B.; Gaucher, J.; Mendelson, M.; Lemarie, E.; Stauffer, E.; Pichon, A.; Howe, C.A.; Doutreleau, S.; et al. The human blood transcriptome exhibits time-of-day-dependent response to hypoxia: Lessons from the highest city in the world. Cell Rep. 2022, 40, 111213. [Google Scholar] [CrossRef] [PubMed]
- Pera, J.; Korostynski, M.; Golda, S.; Piechota, M.; Dzbek, J.; Krzyszkowski, T.; Dziedzic, T.; Moskala, M.; Przewlocki, R.; Szczudlik, A.; et al. Gene expression profiling of blood in ruptured intracranial aneurysms: In search of biomarkers. J. Cereb. Blood Flow Metab. 2013, 33, 1025–1031. [Google Scholar] [CrossRef] [PubMed]
- Stamova, B.; Jickling, G.C.; Ander, B.P.; Zhan, X.; Liu, D.; Turner, R.; Ho, C.; Khoury, J.C.; Bushnell, C.; Pancioli, A.; et al. Gene expression in peripheral immune cells following cardioembolic stroke is sexually dimorphic. PLoS ONE 2014, 9, e102550. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2021, 12, 23–40.e7. [Google Scholar] [CrossRef] [PubMed]
- LaSalle, T.J.; Gonye, A.L.; Freeman, S.S.; Kaplonek, P.; Gushterova, I.; Kays, K.R.; Manakongtreecheep, K.; Tantivit, J.; Rojas-Lopez, M.; Russo, B.C.; et al. Longitudinal characterization of circulating neutrophils uncovers phenotypes associated with severity in hospitalized COVID-19 patients. Cell Rep. Med. 2022, 3, 100779. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, A.C.; Mouktaroudi, M.; Krämer, B.; Oestreich, M.; Antonakos, N.; Nuesch-Germano, M.; Gkizeli, K.; Bonaguro, L.; Reusch, N.; Baßler, K.; et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021, 13, 7. [Google Scholar] [CrossRef]
- Dunning, J.; MOSAIC Investigators; Blankley, S.; Hoang, L.T.; Cox, M.; Graham, C.M.; James, P.L.; Bloom, C.I.; Chaussabel, D.; Banchereau, J.; et al. Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza. Nat. Immunol. 2018, 19, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Yang, Z.; Wu, N.C.; Lee, H.H.Y.; Li, Y.; Jiang, W.; Shen, L.; Wu, D.C.; Chen, R.; Zhong, N.; et al. Clinical Correlations of Transcriptional Profile in Patients Infected With Avian Influenza H7N9 Virus. J. Infect. Dis. 2018, 218, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.M.; Shojaei, M.; Teoh, S.; Meyers, A.; Ho, J.; Ball, T.B.; Keynan, Y.; Pisipati, A.; Kumar, A.; Eisen, D.P.; et al. Neutrophils-related host factors associated with severe disease and fatality in patients with influenza infection. Nat. Commun. 2019, 10, 3422. [Google Scholar] [CrossRef] [PubMed]
- de Steenhuijsen Piters, W.A.A.; Heinonen, S.; Hasrat, R.; Bunsow, E.; Smith, B.; Suarez-Arrabal, M.-C.; Chaussabel, D.; Cohen, D.M.; Sanders, E.A.M.; Ramilo, O.; et al. Nasopharyngeal Microbiota, Host Transcriptome, and Disease Severity in Children with Respiratory Syncytial Virus Infection. Am. J. Respir. Crit. Care Med. 2016, 194, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Sun, N.; Tang, H.; Ye, J.; Liu, Y.; He, Q.; Fu, Y.; Zhu, H.; Jiang, C.; et al. NETosis is critical in patients with severe community-acquired pneumonia. Front. Immunol. 2022, 13, 1051140. [Google Scholar] [CrossRef] [PubMed]
- Lodge, K.M.; Cowburn, A.S.; Li, W.; Condliffe, A.M. The Impact of Hypoxia on Neutrophil Degranulation and Consequences for the Host. Int. J. Mol. Sci. 2020, 21, 1183. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Andreakos, E.; Tsiodras, S. COVID-19: Lambda interferon against viral load and hyperinflammation. EMBO Mol. Med. 2020, 12, e12465. [Google Scholar] [CrossRef] [PubMed]
- RReis, G.; Silva, E.A.M.; Silva, D.C.M.; Thabane, L.; Campos, V.H.; Ferreira, T.S.; Santos, C.V.; Nogueira, A.M.; Almeida, A.P.; Savassi, L.C.; et al. Early Treatment with Pegylated Interferon Lambda for COVID-19. N. Engl. J. Med. 2023, 388, 518–528. [Google Scholar] [CrossRef]
- Mousavi, S.R.; Lotfi, H.; Salmanizadeh, S.; Feizbakhshan, S.; Khosravian, F.; Sajjadi, M.S.; Komachali, S.R.; Beni, F.A.; Torkan, B.; Kazemi, M.; et al. An experimental in silico study on COVID-19: Response of neutrophil-related genes to antibiotics. Health Sci. Rep. 2022, 5, e548. [Google Scholar] [CrossRef] [PubMed]
- Rosa, B.A.; Ahmed, M.; Singh, D.K.; Choreño-Parra, J.A.; Cole, J.; Jiménez-Álvarez, L.A.; Rodríguez-Reyna, T.S.; Singh, B.; Gonzalez, O.; Carrion, R., Jr.; et al. IFN signaling and neutrophil degranulation transcriptional signatures are induced during SARS-CoV-2 infection. Commun. Biol. 2021, 4, 290. [Google Scholar] [CrossRef]
- Bibert, S.; Guex, N.; Lourenco, J.; Brahier, T.; Papadimitriou-Olivgeris, M.; Damonti, L.; Manuel, O.; Liechti, R.; Götz, L.; Tschopp, J.; et al. Transcriptomic Signature Differences Between SARS-CoV-2 and Influenza Virus Infected Patients. Front. Immunol. 2021, 12, 666163. [Google Scholar] [CrossRef] [PubMed]
- Ciccosanti, F.; Antonioli, M.; Sacchi, A.; Notari, S.; Farina, A.; Beccacece, A.; Fusto, M.; Vergori, A.; D’offizi, G.; Taglietti, F.; et al. Proteomic analysis identifies a signature of disease severity in the plasma of COVID-19 pneumonia patients associated to neutrophil, platelet and complement activation. Clin. Proteom. 2022, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Pachl, E.; Hellmuth, J.C.; Kneidinger, N.; Heydarian, M.; Frankenberger, M.; Stubbe, H.C.; Ryffel, B.; Petrera, A.; Hauck, S.M.; et al. Proteomics reveals antiviral host response and NETosis during acute COVID-19 in high-risk patients. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166592. [Google Scholar] [CrossRef] [PubMed]
- Meizlish, M.L.; Pine, A.B.; Bishai, J.D.; Goshua, G.; Nadelmann, E.R.; Simonov, M.; Chang, C.-H.; Zhang, H.; Shallow, M.; Bahel, P.; et al. A neutrophil activation signature predicts critical illness and mortality in COVID-19. Blood Adv. 2021, 5, 1164–1177. [Google Scholar] [CrossRef] [PubMed]
- Akgun, E.; Tuzuner, M.B.; Sahin, B.; Kilercik, M.; Kulah, C.; Cakiroglu, H.N.; Serteser, M.; Unsal, I.; Baykal, A.T. Proteins associated with neutrophil degranulation are upregulated in nasopharyngeal swabs from SARS-CoV-2 patients. PLoS ONE 2020, 15, e0240012. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Xu, X.; Wang, C.; Xue, D.; Wang, C.; Chen, J. A host-based two-gene model for the identification of bacterial infection in general clinical settings. Int. J. Infect. Dis. 2021, 105, 662–667. [Google Scholar] [CrossRef]
- Lei, H.; Wang, C.; Wang, Y.; Wang, C. Single-cell RNA-Seq revealed profound immune alteration in the peripheral blood of patients with bacterial infection. Int. J. Infect. Dis. 2021, 103, 527–535. [Google Scholar] [CrossRef]
- Song, F.; Qian, Y.; Peng, X.; Li, X.; Xing, P.; Ye, D.; Lei, H. The frontline of immune response in peripheral blood. PLoS ONE 2017, 12, e0182294. [Google Scholar] [CrossRef]
- Alqutami, F.; Senok, A.; Hachim, M. COVID-19 Transcriptomic Atlas: A Comprehensive Analysis of COVID-19 Related Transcriptomics Datasets. Front. Genet. 2021, 12, 755222. [Google Scholar] [CrossRef]
- Daamen, A.R.; Bachali, P.; Owen, K.A.; Kingsmore, K.M.; Hubbard, E.L.; Labonte, A.C.; Robl, R.; Shrotri, S.; Grammer, A.C.; Lipsky, P.E. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 2021, 11, 7052. [Google Scholar] [CrossRef]
- Islam, A.B.M.M.K.; Khan, M.A.-A.-K.; Ahmed, R.; Hossain, M.S.; Kabir, S.M.T.; Islam, M.S.; Siddiki, A.M.A.M.Z. Transcriptome of nasopharyngeal samples from COVID-19 patients and a comparative analysis with other SARS-CoV-2 infection models reveal disparate host responses against SARS-CoV-2. J. Transl. Med. 2021, 19, 32. [Google Scholar] [CrossRef] [PubMed]


| Accession ID | Sources | Case Number | Control Number | Reference |
|---|---|---|---|---|
| GSE152641 | Whole blood | 62 | 24 | [9] |
| GSE161731 | Whole blood | 77 | 19 | [10] |
| GSE161777 | Whole blood | 11 | 14 | [11] |
| GSE171110 | Whole blood | 44 | 10 | [12] |
| GSE179850 | Whole blood | 31 | 16 | [13] |
| GSE185557 | Whole blood | 11 | 10 | [14] |
| GSE189990 | Whole blood | 20 | 4 | [15] |
| GSE196822 | Whole blood | 40 | 9 | [16] |
| GSE213313 | Whole blood | 83 | 11 | [17] |
| GSE217948 | Whole blood | 395 | 72 | [18] |
| GSE223885 | Whole blood | 10 | 10 | [19] |
| Zenodo 6120249 | Whole blood | 78 | 10 | [20] |
| HRA000238 | Whole blood | 12 | 4 | [21] |
| Gene Symbol | Frequency (logFC > 1) | Median logFC | Correlation w/Severity |
|---|---|---|---|
| ARG1 | 13 | 2.87 | 0.68 |
| CEACAM8 | 13 | 2.93 | 0.67 |
| RNASE2 | 13 | 2.01 | 0.77 |
| CD177 | 12 | 4.98 | 0.61 |
| CKAP4 | 12 | 1.43 | 0.70 |
| ELANE | 12 | 2.92 | 0.65 |
| HP | 12 | 3.13 | 0.75 |
| MCEMP1 | 12 | 2.44 | 0.69 |
| MMP9 | 12 | 2.9 | 0.67 |
| MPO | 12 | 2.27 | 0.70 |
| S100A12 | 12 | 2.02 | 0.69 |
| S100A9 | 12 | 1.63 | 0.72 |
| TCN1 | 12 | 2.18 | 0.69 |
| Accession ID | Sources | Condition | Sample Numbers | Ref |
|---|---|---|---|---|
| GSE164890 | Whole blood | Training at high altitude | 7 athletes Before and after training | [23] |
| GSE196728 | Whole blood | High altitude | 8 subjects, 3 altitudes, 6 time points | [24] |
| GSE36791 | Whole blood | Stroke | 18 controls, 43 patients | [25] |
| GSE58294 | Whole blood | Stroke | 23 controls, 23 patients, 3 time points | [26] |
| Zenodo 6120249 | Whole blood | COVID-19 | 10 controls, 91 patients | [20] |
| GSE157103 | Whole blood | COVID-19 | 50 ICU patients, 50 non-ICU patients | [27] |
| GSE212041 | Neutrophils | COVID-19 | 8 controls, 299 patients | [28] |
| EGAS00001004503 | Neutrophils | COVID-19 | 10 controls, 39 patients | [29] |
| GSE111368 | Whole blood | H1N1 | 130 controls, 198 patients | [30] |
| GSE114466 | Whole blood | H7N9 | 15 controls, 8 patients | [31] |
| GSE101702 | Whole blood | Influenza A | 52 controls, 107 patients | [32] |
| GSE77087 | Whole blood | RSV | 23 controls, 81 patients | [33] |
| GSE196399 | Whole blood | SCAP | 21 controls, 56 patients | [34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, H. Hypoxia and Activation of Neutrophil Degranulation-Related Genes in the Peripheral Blood of COVID-19 Patients. Viruses 2024, 16, 201. https://doi.org/10.3390/v16020201
Lei H. Hypoxia and Activation of Neutrophil Degranulation-Related Genes in the Peripheral Blood of COVID-19 Patients. Viruses. 2024; 16(2):201. https://doi.org/10.3390/v16020201
Chicago/Turabian StyleLei, Hongxing. 2024. "Hypoxia and Activation of Neutrophil Degranulation-Related Genes in the Peripheral Blood of COVID-19 Patients" Viruses 16, no. 2: 201. https://doi.org/10.3390/v16020201
APA StyleLei, H. (2024). Hypoxia and Activation of Neutrophil Degranulation-Related Genes in the Peripheral Blood of COVID-19 Patients. Viruses, 16(2), 201. https://doi.org/10.3390/v16020201






