Replication of Human Norovirus in Human Intestinal Enteroids Is Affected by Fecal Sample Processing
Abstract
:1. Introduction
2. Materials and Methods
2.1. HIE Maintenance and Propagation
2.2. Establishing and Differentiation of HIE Monolayers
2.3. HuNoV Infection of Differentiated HIE Monolayers
2.4. Fecal Processing by Serial Centrifugation and Serial Filtration
2.5. RNA Extraction Using QIAamp and MagMAX Kits
2.6. Two Intervals of HIE Passages
2.7. HuNoV-Specific RT-qPCR
2.8. Statistics
3. Results
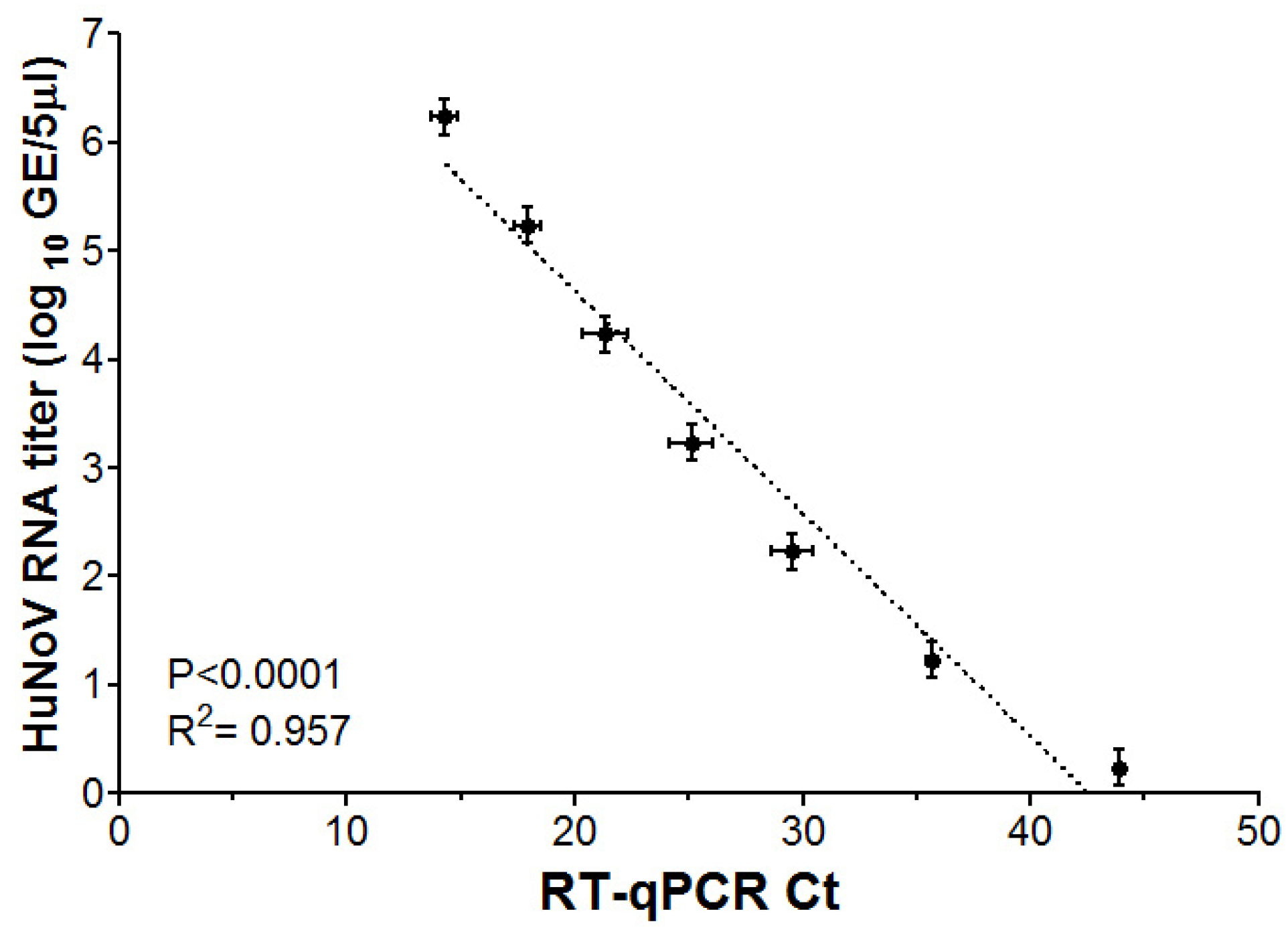
3.1. RT-qPCR Limit of Detection for HuNoV GII RNA
3.2. Effect of Fecal Sample Processing on HuNoV Replication in HIE
3.3. Effect of RNA Extraction Kit on Serially Centrifuged HuNoV Replication in HIE
3.4. Effect of HIE Age on Serially Centrifuged HuNoV Replication in HIE
3.5. Overall Comparisons of HuNoV Genotypes among Various Treatments
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Hoa Tran, T.N.; Trainor, E.; Nakagomi, T.; Cunliffe, N.A.; Nakagomi, O. Molecular epidemiology of noroviruses associated with acute sporadic gastroenteritis in children: Global distribution of genogroups, genotypes and GII.4 variants. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2013, 56, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Lopman, B.A.; Steele, D.; Kirkwood, C.D.; Parashar, U.D. The Vast and Varied Global Burden of Norovirus: Prospects for Prevention and Control. PLoS Med. 2016, 13, e1001999. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; Lopman, B.A.; Ozawa, S.; Hall, A.J.; Lee, B.Y. Global Economic Burden of Norovirus Gastroenteritis. PLoS ONE 2016, 11, e0151219. [Google Scholar] [CrossRef] [PubMed]
- Burke, R.M.; Mattison, C.P.; Marsh, Z.; Shioda, K.; Donald, J.; Salas, S.B.; Naleway, A.L.; Biggs, C.; Schmidt, M.A.; Hall, A.J. Norovirus and Other Viral Causes of Medically Attended Acute Gastroenteritis Across the Age Spectrum: Results from the Medically Attended Acute Gastroenteritis Study in the United States. Clin. Infect. Dis. 2021, 73, e913–e920. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, W.; D’Souza, D.H.; Sanchez, G. Norovirus: The Burden of the Unknown. Adv. Food Nutr. Res. 2018, 86, 13–53. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Kroneman, A.; Verhoef, L.; Harris, J.; Vennema, H.; Duizer, E.; van Duynhoven, Y.; Gray, J.; Iturriza, M.; Bottiger, B.; Falkenhorst, G.; et al. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. J. Clin. Microbiol. 2008, 46, 2959–2965. [Google Scholar] [CrossRef]
- Straub, T.M.; zu Bentrup, K.H.; Orosz-Coghlan, P.; Dohnalkova, A.; Mayer, B.K.; Bartholomew, R.A.; Valdez, C.O.; Bruckner-Lea, C.J.; Gerba, C.P.; Abbaszadegan, M.; et al. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect. Dis. 2007, 13, 396–403. [Google Scholar] [CrossRef]
- Straub, T.M.; Bartholomew, R.A.; Valdez, C.O.; Valentine, N.B.; Dohnalkova, A.; Ozanich, R.M.; Bruckner-Lea, C.J.; Call, D.R. Human norovirus infection of caco-2 cells grown as a three-dimensional tissue structure. J. Water Health 2011, 9, 225–240. [Google Scholar] [CrossRef]
- Papafragkou, E.; Hewitt, J.; Park, G.W.; Greening, G.; Vinje, J. Challenges of culturing human norovirus in three-dimensional organoid intestinal cell culture models. PLoS ONE 2014, 8, e63485. [Google Scholar] [CrossRef]
- Takanashi, S.; Saif, L.J.; Hughes, J.H.; Meulia, T.; Jung, K.; Scheuer, K.A.; Wang, Q. Failure of propagation of human norovirus in intestinal epithelial cells with microvilli grown in three-dimensional cultures. Arch. Virol. 2014, 159, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Costantini, V.; Morantz, E.K.; Browne, H.; Ettayebi, K.; Zeng, X.L.; Atmar, R.L.; Estes, M.K.; Vinje, J. Human Norovirus Replication in Human Intestinal Enteroids as Model to Evaluate Virus Inactivation. Emerg. Infect. Dis. 2018, 24, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: Mechanism and applications. Science 2013, 340, 1190–1194. [Google Scholar] [CrossRef] [PubMed]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R.; et al. Human Intestinal Enteroids: A New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J. Virol. 2016, 90, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Santiana, M.; Ghosh, S.; Ho, B.A.; Rajasekaran, V.; Du, W.L.; Mutsafi, Y.; De Jesus-Diaz, D.A.; Sosnovtsev, S.V.; Levenson, E.A.; Parra, G.I.; et al. Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-organismal Viral Transmission. Cell Host Microbe 2018, 24, 208–220.e8. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.Y.; Blutt, S.E.; Crawford, S.E.; Ettayebi, K.; Zeng, X.L.; Saxena, K.; Ramani, S.; Karandikar, U.C.; Zachos, N.C.; Estes, M.K. Human Intestinal Enteroids: New Models to Study Gastrointestinal Virus Infections. Methods Mol. Biol. 2019, 1576, 229–247. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Tenge, V.R.; Cortes-Penfield, N.W.; Crawford, S.E.; Neill, F.H.; Zeng, X.L.; Yu, X.; Ayyar, B.V.; Burrin, D.; Ramani, S.; et al. New Insights and Enhanced Human Norovirus Cultivation in Human Intestinal Enteroids. mSphere 2021, 6, e01136-20. [Google Scholar] [CrossRef]
- Overbey, K.N.; Zachos, N.C.; Coulter, C.; Schwab, K.J. Optimizing Human Intestinal Enteroids for Environmental Monitoring of Human Norovirus. Food Environ. Virol. 2021, 13, 470–484. [Google Scholar] [CrossRef]
- Ford-Siltz, L.A.; Wales, S.; Tohma, K.; Gao, Y.; Parra, G.I. Genotype-Specific Neutralization of Norovirus Is Mediated by Antibodies Against the Protruding Domain of the Major Capsid Protein. J. Infect. Dis. 2022, 225, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Mammel, M.; Wales, S.Q. Near-Complete Genome Sequence of a Human Norovirus GII.1[Pg] Strain Associated with Acute Gastroenteritis, Determined Using Long-Read Sequencing. Microbiol. Resour. Announc. 2021, 10, e0040121. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Vinje, J.; Elkins, C.A.; Kulka, M. Complete Genome Sequence of Human Norovirus Strain GII.P7-GII.6 Detected in a Patient in the United States in 2014. Genome Announc. 2016, 4, e01211-16. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, T.; Kojima, S.; Shinohara, M.; Uchida, K.; Fukushi, S.; Hoshino, F.B.; Takeda, N.; Katayama, K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003, 41, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.; Cheung, S.K.C.; Mohammad, K.N.; Chan, J.C.M.; Estes, M.K.; Chan, P.K.S. Use of Human Intestinal Enteroids to Detect Human Norovirus Infectivity. Emerg. Infect. Dis. 2019, 25, 1730–1735. [Google Scholar] [CrossRef] [PubMed]
- Overbey, K.N.; Zachos, N.C.; Coulter, C.; Jacangelo, J.; Schwab, K.J. Recovery of Infectious Human Norovirus GII.4 Sydney From Fomites via Replication in Human Intestinal Enteroids. Front. Cell. Infect. Microbiol. 2021, 11, 693090. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, W.; Costantini, V.; Morantz, E.K.; Vinje, J. Human Intestinal Enteroids to Evaluate Human Norovirus GII.4 Inactivation by Aged-Green Tea. Front. Microbiol. 2020, 11, 1917. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Murakami, K.; Hirano, J.; Fujii, Y.; Yamaoka, Y.; Ohashi, H.; Watashi, K.; Estes, M.K.; Muramatsu, M. Dasabuvir Inhibits Human Norovirus Infection in Human Intestinal Enteroids. mSphere 2021, 6, e0062321. [Google Scholar] [CrossRef]
- Fumian, T.M.; Leite, J.P.; Marin, V.A.; Miagostovich, M.P. A rapid procedure for detecting noroviruses from cheese and fresh lettuce. J. Virol. Methods 2009, 155, 39–43. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kwak, I.S.; Hwang, I.G.; Ko, G. Optimization of methods for detecting norovirus on various fruit. J. Virol. Methods 2008, 153, 104–110. [Google Scholar] [CrossRef]
- Kim, K.; Katayama, H.; Kitajima, M.; Tohya, Y.; Ohgaki, S. Development of a real-time RT-PCR assay combined with ethidium monoazide treatment for RNA viruses and its application to detect viral RNA after heat exposure. Water Sci. Technol. 2011, 63, 502–507. [Google Scholar] [CrossRef]
- Hagbom, M.; Lin, J.; Falkeborn, T.; Serrander, L.; Albert, J.; Nordgren, J.; Sharma, S. Replication in Human Intestinal Enteroids of Infectious Norovirus from Vomit Samples. Emerg. Infect. Dis. 2021, 27, 2212–2214. [Google Scholar] [CrossRef]




| Genotype | P-Type | GenBank Accession Number | Serial Filtration Titer (Log GE/Well) | Serial Centrifugation Titer (Log GE/Well) | Reference |
|---|---|---|---|---|---|
| GII.1 | [Pg] | MW854326 | 4.7 ± 0.03 Aa | 4.6 ± 0.5 Aa | [22] |
| GII.4 | [P16] | MN782359 | 4.8 ± 0.2 Aa | 5.0 ± 0.3 Aa | [21] |
| GII.6 | [P7] | KX268709 | 4.5 ± 0.08 Aa | 4.4 ± 0.6 Aa | [23] |
| Fecal Processing | Serial Filtration | Serial Centrifugation | ||||
| RNA Extraction Kit | QIAamp | QIAamp | MagMAX | MagMAX | MagMAX | |
| HIE passages # | 7–28 | 7–28 | 7–28 | 5–19 | 20–34 | |
| GII.1 (log GE/well) | 1 h | 1.5 ± 0.5 Aa | 3.9 ± 0.0 Ab | 2.8 ± 0.5 Aab | 2.0 ± 0.6 Aab | 3.7 ± 0.2 Ab |
| 72 h | 3.7 ± 0.3 Ba | 4.8 ± 0.2 Bb | 3.7 ± 0.1 Aac | 3.7 ± 0.2 Bac | 4.3 ± 0.2 Ab | |
| GII.4 (log GE/well) | 1 h | 1.8 ± 0.5 Aa | 4.1 ± 0.08 Ab | 1.4 ± 0.4 Ac | 0.6 ± 0.0 Aac | 1.9 ± 0.5 Aac |
| 72 h | 3.9 ± 0.5 Bab | 4.5 ± 0.3 Aa | 3.1 ± 0.3 Bb | 2.9 ± 0.4 Bab | 3.9 ± 0.1 Bab | |
| GII.6 (log GE/well | 1 h | 0.6 ± 0.0 Aa | 2.8 ± 0.5 Ab | 1.7± 0.5 Aab | 0.7 ± 0.1 Aac | 2.7 ± 0.4 Ab |
| 72 h | 1.7 ± 0.4 Ba | 3.6 ± 0.0 Ab | 2.9± 0.5 Aab | 2.4 ±0.7 Bab | 2.7 ± 0.5 Aab | |
| Fecal Processing | Serial Filtration | Serial Centrifugation | |||
| RNA Extraction Kit | QIAamp | QIAamp | MagMAX | MagMAX | MagMAX |
| HIE passages # | 7–28 | 7–28 | 7–28 | 5–19 | 20–34 |
| GII.1 | 2.2 ± 0.5 Aa | 0.9 ± 0.2 Ab | 1.0 ± 0.4 Aab | 1.6 ± 0.5 Aab | 0.8 ± 0.4 ABab |
| GII.4 | 2.0 ± 0.6 Aa | 0.4 ± 0.2 Ab | 1.7 ± 0.4 Aac | 2.3 ± 0.4 Aa | 2.0 ± 0.5 Aa |
| GII.6 | 1.2 ± 0.4 Aa | 0.9 ±0.4 Aa | 1.3 ± 0.5 Aa | 1.8 ± 0.6 Aa | 0.4 ± 0.2 Ba |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narwankar, R.; Esseili, M.A. Replication of Human Norovirus in Human Intestinal Enteroids Is Affected by Fecal Sample Processing. Viruses 2024, 16, 241. https://doi.org/10.3390/v16020241
Narwankar R, Esseili MA. Replication of Human Norovirus in Human Intestinal Enteroids Is Affected by Fecal Sample Processing. Viruses. 2024; 16(2):241. https://doi.org/10.3390/v16020241
Chicago/Turabian StyleNarwankar, Revati, and Malak A. Esseili. 2024. "Replication of Human Norovirus in Human Intestinal Enteroids Is Affected by Fecal Sample Processing" Viruses 16, no. 2: 241. https://doi.org/10.3390/v16020241





