The Adaptive Immune Response against Bunyavirales
Abstract
:1. Introduction
2. Bunyavirales Structure and Life Cycle
3. T Cell Responses against Bunyavirales
3.1. Peribunyaviridae
3.2. Phenuiviridae
3.3. Hantaviridae
3.4. Nairoviridae
3.5. Arenaviridae
4. Antibody Responses against Bunyavirales
4.1. Peribunyaviridea
4.2. Phenuiviridae
4.3. Hantaviridae
4.4. Nairoviridea
4.5. Arenaviridae
5. Bunyavirales Vaccines and Therapeutic Strategies
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch. Virol. 2017, 162, 2505–2538. [Google Scholar] [CrossRef] [PubMed]
- Abudurexiti, A.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Avšič-Županc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, É.; Blair, C.D.; et al. Taxonomy of the order Bunyavirales: Update 2019. Arch. Virol. 2019, 164, 1949–1965. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef] [PubMed]
- Orba, Y.; Abu, Y.E.; Chambaro, H.M.; Lundu, T.; Muleya, W.; Eshita, Y.; Qiu, Y.; Harima, H.; Kajihara, M.; Mori-Kajihara, A.; et al. Expanding diversity of bunyaviruses identified in mosquitoes. Sci. Rep. 2023, 13, 18165. [Google Scholar] [CrossRef] [PubMed]
- Horne, K.M.; Vanlandingham, D.L. Bunyavirus-vector interactions. Viruses 2014, 6, 4373–4397. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.M. Bunyaviruses and climate change. Clin. Microbiol. Infect. 2009, 15, 510–517. [Google Scholar] [CrossRef]
- Soldan, S.S.; González-Scarano, F. Emerging infectious diseases: The Bunyaviridae. J. NeuroVirol. 2005, 11, 412–423. [Google Scholar] [CrossRef]
- Elliott, R.M. Emerging Viruses: The Bunyaviridae. Mol. Med. 1997, 3, 572–577. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Family-Bunyaviridae. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 725–741. [Google Scholar]
- Hastie, K.M.; Melnik, L.I.; Cross, R.W.; Klitting, R.M.; Andersen, K.G.; Saphire, E.O.; Garry, R.F. The Arenaviridae Family: Knowledge Gaps, Animal Models, Countermeasures, and Prototype Pathogens. J. Infect. Dis. 2023, 228 (Suppl. S6), S359–S375. [Google Scholar] [CrossRef]
- Leventhal, S.S.; Wilson, D.; Feldmann, H.; Hawman, D.W. A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins. Viruses 2021, 13, 314. [Google Scholar] [CrossRef]
- Hulswit, R.J.G.; Paesen, G.C.; Bowden, T.A.; Shi, X. Recent Advances in Bunyavirus Glycoprotein Research: Precursor Processing, Receptor Binding and Structure. Viruses 2021, 13, 353. [Google Scholar] [CrossRef]
- Fontana, J.; López-Montero, N.; Elliott, R.M.; Fernández, J.J.; Risco, C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cell. Microbiol. 2008, 10, 2012–2028. [Google Scholar] [CrossRef]
- Won, S.; Ikegami, T.; Peters, C.J.; Makino, S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J. Virol. 2007, 81, 13335–13345. [Google Scholar] [CrossRef] [PubMed]
- Ferron, F.; Weber, F.; de la Torre, J.C.; Reguera, J. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 2017, 234, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Suda, Y.; Fukushi, S.; Tani, H.; Murakami, S.; Saijo, M.; Horimoto, T.; Shimojima, M. Analysis of the entry mechanism of Crimean-Congo hemorrhagic fever virus, using a vesicular stomatitis virus pseudotyping system. Arch. Virol. 2016, 161, 1447–1454. [Google Scholar] [CrossRef]
- Léger, P.; Tetard, M.; Youness, B.; Cordes, N.; Rouxel, R.N.; Flamand, M.; Lozach, P.Y. Differential use of the C-type lectins L-SIGN and DC-SIGN for phlebovirus endocytosis. Traffic 2016, 17, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Shimojima, M.; Kawaoka, Y. Cell surface molecules involved in infection mediated by lymphocytic choriomeningitis virus glycoprotein. J. Vet. Med. Sci. 2012, 74, 1363–1366. [Google Scholar] [CrossRef] [PubMed]
- Khoo, U.S.; Chan, K.Y.; Chan, V.S.; Lin, C.L. DC-SIGN and L-SIGN: The SIGNs for infection. J. Mol. Med. 2008, 86, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Albornoz, A.; Hoffmann, A.B.; Lozach, P.Y.; Tischler, N.D. Early Bunyavirus-Host Cell Interactions. Viruses 2016, 8, 143. [Google Scholar] [CrossRef]
- Garrison, A.R.; Radoshitzky, S.R.; Kota, K.P.; Pegoraro, G.; Ruthel, G.; Kuhn, J.H.; Altamura, L.A.; Kwilas, S.A.; Bavari, S.; Haucke, V.; et al. Crimean-Congo hemorrhagic fever virus utilizes a clathrin- and early endosome-dependent entry pathway. Virology 2013, 444, 45–54. [Google Scholar] [CrossRef]
- Boshra, H. An Overview of the Infectious Cycle of Bunyaviruses. Viruses 2022, 14, 2139. [Google Scholar] [CrossRef]
- Barker, J.; daSilva, L.L.P.; Crump, C.M. Mechanisms of bunyavirus morphogenesis and egress. J. Gen. Virol. 2023, 104, 001845. [Google Scholar] [CrossRef]
- Urata, S.; Yasuda, J. Molecular Mechanism of Arenavirus Assembly and Budding. Viruses 2012, 4, 2049–2079. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Elliott, L.H.; Peters, C.J.; Zaki, S.R. Ultrastructural characteristics of Sin Nombre virus, causative agent of hantavirus pulmonary syndrome. Arch. Virol. 1995, 140, 2107–2122. [Google Scholar] [CrossRef] [PubMed]
- Ravkov, E.V.; Nichol, S.T.; Compans, R.W. Polarized entry and release in epithelial cells of Black Creek Canal virus, a New World hantavirus. J. Virol. 1997, 71, 1147–1154. [Google Scholar] [CrossRef]
- Ter Horst, S.; Conceição-Neto, N.; Neyts, J.; Rocha-Pereira, J. Structural and functional similarities in bunyaviruses: Perspectives for pan-bunya antivirals. Rev. Med. Virol. 2019, 29, e2039. [Google Scholar] [CrossRef]
- Dutuze, M.F.; Nzayirambaho, M.; Mores, C.N.; Christofferson, R.C. A Review of Bunyamwera, Batai, and Ngari Viruses: Understudied Orthobunyaviruses with Potential One Health Implications. Front. Vet. Sci. 2018, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.M. Orthobunyaviruses: Recent genetic and structural insights. Nat. Rev. Microbiol. 2014, 12, 673–685. [Google Scholar] [CrossRef]
- Adhikari, U.K.; Tayebi, M.; Rahman, M.M. Immunoinformatics Approach for Epitope-Based Peptide Vaccine Design and Active Site Prediction against Polyprotein of Emerging Oropouche Virus. J. Immunol. Res. 2018, 2018, 6718083. [Google Scholar] [CrossRef]
- Shahab, M.; Aiman, S.; Alshammari, A.; Alasmari, A.F.; Alharbi, M.; Khan, A.; Wei, D.Q.; Zheng, G. Immunoinformatics-based potential multi-peptide vaccine designing against Jamestown Canyon Virus (JCV) capable of eliciting cellular and humoral immune responses. Int. J. Biol. Macromol. 2023, 253 Pt 2, 126678. [Google Scholar] [CrossRef] [PubMed]
- Nelluri, K.D.D.; Ammulu, M.A.; Durga, M.L.; Sravani, M.; Kumar, V.P.; Poda, S. In silico multi-epitope Bunyumwera virus vaccine to target virus nucleocapsid N protein. J. Genet. Eng. Biotechnol. 2022, 20, 89. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Bhattacharya, M.; Patra, P.; Sharma, G.; Patra, B.C.; Lee, S.S.; Sharma, A.R.; Chakraborty, C. Evaluation and Designing of Epitopic-Peptide Vaccine Against Bunyamwera orthobunyavirus Using M-Polyprotein Target Sequences. Int. J. Pept. Res. Ther. 2022, 28, 5. [Google Scholar] [CrossRef]
- Boshra, H.Y.; Charro, D.; Lorenzo, G.; Sánchez, I.; Lazaro, B.; Brun, A.; Abrescia, N.G. DNA vaccination regimes against Schmallenberg virus infection in IFNAR(−/−) mice suggest two targets for immunization. Antivir. Res. 2017, 141, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Schuh, T.; Schultz, J.; Moelling, K.; Pavlovic, J. DNA-based vaccine against La Crosse virus: Protective immune response mediated by neutralizing antibodies and CD4+ T cells. Hum. Gene Ther. 1999, 10, 1649–1658. [Google Scholar] [CrossRef]
- Boshra, H.; Lorenzo, G.; Charro, D.; Moreno, S.; Guerra, G.S.; Sanchez, I.; Garrido, J.M.; Geijo, M.; Brun, A.; Abrescia, N.G.A. A novel Schmallenberg virus subunit vaccine candidate protects IFNAR(−/−) mice against virulent SBV challenge. Sci. Rep. 2020, 10, 18725. [Google Scholar] [CrossRef]
- Jain, A.; Tripathi, P.; Shrotriya, A.; Chaudhary, R.; Singh, A. In silico analysis and modeling of putative T cell epitopes for vaccine design of Toscana virus. 3 Biotech 2015, 5, 497–503. [Google Scholar] [CrossRef]
- Suleman, M.; Asad, U.; Arshad, S.; Rahman, A.U.; Akbar, F.; Khan, H.; Hussain, Z.; Ali, S.S.; Mohammad, A.; Khan, A.; et al. Screening of immune epitope in the proteome of the Dabie bandavirus, SFTS, to design a protein-specific and proteome-wide vaccine for immune response instigation using an immunoinformatics approaches. Comput. Biol. Med. 2022, 148, 105893. [Google Scholar] [CrossRef]
- Adhikari, U.K.; Rahman, M.M. Overlapping CD8+ and CD4+ T-cell epitopes identification for the progression of epitope-based peptide vaccine from nucleocapsid and glycoprotein of emerging Rift Valley fever virus using immunoinformatics approach. Infect. Genet. Evol. 2017, 56, 75–91. [Google Scholar] [CrossRef]
- Harmon, J.R.; Barbeau, D.J.; Nichol, S.T.; Spiropoulou, C.F.; McElroy, A.K. Rift Valley fever virus vaccination induces long-lived, antigen-specific human T cell responses. NPJ Vaccines 2020, 5, 17. [Google Scholar] [CrossRef]
- Xu, W.; Watts, D.M.; Costanzo, M.C.; Tang, X.; Venegas, L.A.; Jiao, F.; Sette, A.; Sidney, J.; Sewell, A.K.; Wooldridge, L.; et al. The nucleocapsid protein of Rift Valley fever virus is a potent human CD8+ T cell antigen and elicits memory responses. PLoS ONE 2013, 8, e59210. [Google Scholar] [CrossRef]
- Barbeau, D.J.; Cartwright, H.N.; Harmon, J.R.; Spengler, J.R.; Spiropoulou, C.F.; Sidney, J.; Sette, A.; McElroy, A.K. Identification and Characterization of Rift Valley Fever Virus-Specific T Cells Reveals a Dependence on CD40/CD40L Interactions for Prevention of Encephalitis. J. Virol. 2021, 95, e0150621. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, F.; Nain, Z.; Hossain, M.M.; Syed, S.B.; Ahmed Khan, M.S.; Adhikari, U.K. A comprehensive screening of the whole proteome of hantavirus and designing a multi-epitope subunit vaccine for cross-protection against hantavirus: Structural vaccinology and immunoinformatics study. Microb. Pathog. 2021, 150, 104705. [Google Scholar] [CrossRef]
- Van Epps, H.L.; Schmaljohn, C.S.; Ennis, F.A. Human memory cytotoxic T-lymphocyte (CTL) responses to Hantaan virus infection: Identification of virus-specific and cross-reactive CD8(+) CTL epitopes on nucleocapsid protein. J. Virol. 1999, 73, 5301–5308. [Google Scholar] [CrossRef]
- Wang, M.; Wang, J.; Kang, Z.; Zhao, Q.; Wang, X.; Hui, L. Kinetics and Immunodominance of Virus-Specific T Cell Responses During Hantaan Virus Infection. Viral Immunol. 2015, 28, 265–271. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, Y.; Wang, J.; Lv, T.; Jin, B. Identification of three novel CTL epitopes within nucleocapsid protein of Hantaan virus. Viral Immunol. 2011, 24, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Chun, E.; Kim, N.Y.; Seong, B.L. Characterization of HLA-A2.1-restricted epitopes, conserved in both Hantaan and Sin Nombre viruses, in Hantaan virus-infected patients. J. Gen. Virol. 2002, 83 Pt 5, 1131–1136. [Google Scholar] [CrossRef]
- Ennis, F.A.; Cruz, J.; Spiropoulou, C.F.; Waite, D.; Peters, C.J.; Nichol, S.T.; Kariwa, H.; Koster, F.T. Hantavirus pulmonary syndrome: CD8+ and CD4+ cytotoxic T lymphocytes to epitopes on Sin Nombre virus nucleocapsid protein isolated during acute illness. Virology 1997, 238, 380–390. [Google Scholar] [CrossRef]
- Tang, K.; Cheng, L.; Zhang, C.; Zhang, Y.; Zheng, X.; Zhang, Y.; Zhuang, R.; Jin, B.; Zhang, F.; Ma, Y. Novel Identified HLA-A*0201-Restricted Hantaan Virus Glycoprotein Cytotoxic T-Cell Epitopes Could Effectively Induce Protective Responses in HLA-A2.1/K(b) Transgenic Mice May Associate with the Severity of Hemorrhagic Fever with Renal Syndrome. Front. Immunol. 2017, 8, 1797. [Google Scholar] [CrossRef]
- Manigold, T.; Mori, A.; Graumann, R.; Llop, E.; Simon, V.; Ferrés, M.; Valdivieso, F.; Castillo, C.; Hjelle, B.; Vial, P. Highly differentiated, resting gn-specific memory CD8+ T cells persist years after infection by andes hantavirus. PLoS Pathog. 2010, 6, e1000779. [Google Scholar] [CrossRef]
- Terajima, M.; Van Epps, H.L.; Li, D.; Leporati, A.M.; Juhlin, S.E.; Mustonen, J.; Vaheri, A.; Ennis, F.A. Generation of recombinant vaccinia viruses expressing Puumala virus proteins and use in isolating cytotoxic T cells specific for Puumala virus. Virus Res. 2002, 84, 67–77. [Google Scholar] [CrossRef]
- Nosrati, M.; Behbahani, M.; Mohabatkar, H. Towards the first multi-epitope recombinant vaccine against Crimean-Congo hemorrhagic fever virus: A computer-aided vaccine design approach. J. Biomed. Inf. 2019, 93, 103160. [Google Scholar] [CrossRef]
- Shrivastava, N.; Verma, A.; Dash, P.K. Identification of functional epitopes of structural proteins and in-silico designing of dual acting multiepitope anti-tick vaccine against emerging Crimean-Congo hemorrhagic fever virus. Eur. J. Pharm. Sci. 2020, 151, 105396. [Google Scholar] [CrossRef]
- Oany, A.R.; Ahmad, S.A.; Hossain, M.U.; Jyoti, T.P. Identification of highly conserved regions in L-segment of Crimean-Congo hemorrhagic fever virus and immunoinformatic prediction about potential novel vaccine. Adv. Appl. Bioinform. Chem. 2015, 8, 1–10. [Google Scholar] [CrossRef]
- Maotoana, M.G.; Burt, F.J.; Goedhals, D. Identification of T cell responses to the nonstructural glycoproteins in survivors of Crimean-Congo hemorrhagic fever in South Africa. J. Med. Virol. 2023, 95, e29154. [Google Scholar] [CrossRef]
- Goedhals, D.; Paweska, J.T.; Burt, F.J. Long-lived CD8+ T cell responses following Crimean-Congo haemorrhagic fever virus infection. PLoS Negl. Trop. Dis. 2017, 11, e0006149. [Google Scholar] [CrossRef]
- Rao, D.; Meade-White, K.; Leventhal, S.; Mihalakakos, E.; Carmody, A.; Feldmann, H.; Hawman, D.W. CD8(+) T-cells target the Crimean-Congo haemorrhagic fever virus Gc protein to control the infection in wild-type mice. EBioMedicine 2023, 97, 104839. [Google Scholar] [CrossRef]
- Abass, O.A.; Timofeev, V.I.; Sarkar, B.; Onobun, D.O.; Ogunsola, S.O.; Aiyenuro, A.E.; Aborode, A.T.; Aigboje, A.E.; Omobolanle, B.N.; Imolele, A.G.; et al. Immunoinformatics analysis to design novel epitope based vaccine candidate targeting the glycoprotein and nucleoprotein of Lassa mammarenavirus (LASMV) using strains from Nigeria. J. Biomol. Struct. Dyn. 2022, 40, 7283–7302. [Google Scholar] [CrossRef]
- Sakabe, S.; Hartnett, J.N.; Ngo, N.; Goba, A.; Momoh, M.; Sandi, J.D.; Kanneh, L.; Cubitt, B.; Garcia, S.D.; Ware, B.C.; et al. Identification of Common CD8(+) T Cell Epitopes from Lassa Fever Survivors in Nigeria and Sierra Leone. J. Virol. 2020, 94, e00153-20. [Google Scholar] [CrossRef]
- Ugwu, C.; Olumade, T.; Nwakpakpa, E.; Onyia, V.; Odeh, E.; Duruiheoma, R.O.; Ojide, C.K.; Eke, M.A.; Nwafor, I.E.; Chika-Igwenyi, N.; et al. Humoral and cellular immune responses to Lassa fever virus in Lassa fever survivors and their exposed contacts in Southern Nigeria. Sci. Rep. 2022, 12, 22330. [Google Scholar] [CrossRef] [PubMed]
- Meulen, J.; Badusche, M.; Satoguina, J.; Strecker, T.; Lenz, O.; Loeliger, C.; Sakho, M.; Koulemou, K.; Koivogui, L.; Hoerauf, A. Old and New World arenaviruses share a highly conserved epitope in the fusion domain of the glycoprotein 2, which is recognized by Lassa virus-specific human CD4+ T-cell clones. Virology 2004, 321, 134–143. [Google Scholar] [CrossRef] [PubMed]
- ter Meulen, J.; Badusche, M.; Kuhnt, K.; Doetze, A.; Satoguina, J.; Marti, T.; Loeliger, C.; Koulemou, K.; Koivogui, L.; Schmitz, H.; et al. Characterization of human CD4(+) T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J. Virol. 2000, 74, 2186–2192. [Google Scholar] [CrossRef]
- La Posta, V.J.; Auperin, D.D.; Kamin-Lewis, R.; Cole, G.A. Cross-protection against lymphocytic choriomeningitis virus mediated by a CD4+ T-cell clone specific for an envelope glycoprotein epitope of Lassa virus. J. Virol. 1993, 67, 3497–3506. [Google Scholar] [CrossRef]
- Vahey, G.M.; Lindsey, N.P.; Staples, J.E.; Hills, S.L. La Crosse Virus Disease in the United States, 2003–2019. Am. J. Trop. Med. Hyg. 2021, 105, 807–812. [Google Scholar] [CrossRef]
- Winkler, C.W.; Myers, L.M.; Woods, T.A.; Carmody, A.B.; Taylor, K.G.; Peterson, K.E. Lymphocytes have a role in protection, but not in pathogenesis, during La Crosse Virus infection in mice. J. Neuroinflamm. 2017, 14, 62. [Google Scholar] [CrossRef]
- Sun, M.-H.; Ji, Y.-F.; Li, G.-H.; Shao, J.-W.; Chen, R.-X.; Gong, H.-Y.; Chen, S.-Y.; Chen, J.-M. Highly adaptive Phenuiviridae with biomedical importance in multiple fields. J. Med. Virol. 2022, 94, 2388–2401. [Google Scholar] [CrossRef]
- Mehand, M.S.; Al-Shorbaji, F.; Millett, P.; Murgue, B. The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antivir. Res. 2018, 159, 63–67. [Google Scholar] [CrossRef]
- Kwaśnik, M.; Rożek, W.; Rola, J. Rift Valley Fever—A Growing Threat To Humans and Animals. J. Vet. Res. 2021, 65, 7–14. [Google Scholar] [CrossRef]
- Bron, G.M.; Strimbu, K.; Cecilia, H.; Lerch, A.; Moore, S.M.; Tran, Q.; Perkins, T.A.; Ten Bosch, Q.A. Over 100 Years of Rift Valley Fever: A Patchwork of Data on Pathogen Spread and Spillover. Pathogens 2021, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Huang, H.; Jiang, L.; Li, J. Overview of the immunological mechanism underlying severe fever with thrombocytopenia syndrome (Review). Int. J. Mol. Med. 2022, 50, 118. [Google Scholar] [CrossRef] [PubMed]
- Ayhan, N.; Charrel, R.N. An update on Toscana virus distribution, genetics, medical and diagnostic aspects. Clin. Microbiol. Infect. 2020, 26, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Ayari-Fakhfakh, E.; Ghram, A.; Albina, E.; Cêtre-Sossah, C. Expression of cytokines following vaccination of goats with a recombinant capripoxvirus vaccine expressing Rift Valley fever virus proteins. Vet. Immunol. Immunopathol. 2018, 197, 15–20. [Google Scholar] [CrossRef]
- López-Gil, E.; Lorenzo, G.; Hevia, E.; Borrego, B.; Eiden, M.; Groschup, M.; Gilbert, S.C.; Brun, A. A single immunization with MVA expressing GnGc glycoproteins promotes epitope-specific CD8+-T cell activation and protects immune-competent mice against a lethal RVFV infection. PLoS Negl. Trop. Dis. 2013, 7, e2309. [Google Scholar] [CrossRef]
- Pavulraj, S.; Stout, R.W.; Barras, E.D.; Paulsen, D.B.; Chowdhury, S.I. A Novel Quadruple Gene-Deleted BoHV-1-Vectored RVFV Subunit Vaccine Induces Humoral and Cell-Mediated Immune Response against Rift Valley Fever in Calves. Viruses 2023, 15, 2183. [Google Scholar] [CrossRef]
- Kwak, J.E.; Kim, Y.I.; Park, S.J.; Yu, M.A.; Kwon, H.I.; Eo, S.; Kim, T.S.; Seok, J.; Choi, W.S.; Jeong, J.H.; et al. Development of a SFTSV DNA vaccine that confers complete protection against lethal infection in ferrets. Nat. Commun. 2019, 10, 3836. [Google Scholar] [CrossRef]
- Kang, J.G.; Jeon, K.; Choi, H.; Kim, Y.; Kim, H.I.; Ro, H.J.; Seo, Y.B.; Shin, J.; Chung, J.; Jeon, Y.K.; et al. Vaccination with single plasmid DNA encoding IL-12 and antigens of severe fever with thrombocytopenia syndrome virus elicits complete protection in IFNAR knockout mice. PLoS Negl. Trop. Dis. 2020, 14, e0007813. [Google Scholar] [CrossRef]
- Kim, J.Y.; Jeon, K.; Hong, J.J.; Park, S.I.; Cho, H.; Park, H.J.; Kwak, H.W.; Park, H.J.; Bang, Y.J.; Lee, Y.S.; et al. Heterologous vaccination utilizing viral vector and protein platforms confers complete protection against SFTSV. Sci. Rep. 2023, 13, 8189. [Google Scholar] [CrossRef]
- Park, J.Y.; Hewawaduge, C.; Sivasankar, C.; Lloren, K.K.S.; Oh, B.; So, M.Y.; Lee, J.H. An mRNA-Based Multiple Antigenic Gene Expression System Delivered by Engineered Salmonella for Severe Fever with Thrombocytopenia Syndrome and Assessment of Its Immunogenicity and Protection Using a Human DC-SIGN-Transduced Mouse Model. Pharmaceutics 2023, 15, 1339. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Taniguchi, S.; Kato, H.; Iwata-Yoshikawa, N.; Tani, H.; Kurosu, T.; Fujii, H.; Omura, N.; Shibamura, M.; Watanabe, S.; et al. A highly attenuated vaccinia virus strain LC16m8-based vaccine for severe fever with thrombocytopenia syndrome. PLoS Pathog. 2021, 17, e1008859. [Google Scholar] [CrossRef] [PubMed]
- Gori Savellini, G.; Di Genova, G.; Terrosi, C.; Di Bonito, P.; Giorgi, C.; Valentini, M.; Docquier, J.D.; Cusi, M.G. Immunization with Toscana virus N-Gc proteins protects mice against virus challenge. Virology 2008, 375, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Harmon, J.R.; Spengler, J.R.; Coleman-McCray, J.D.; Nichol, S.T.; Spiropoulou, C.F.; McElroy, A.K. CD4 T Cells, CD8 T Cells, and Monocytes Coordinate to Prevent Rift Valley Fever Virus Encephalitis. J. Virol. 2018, 92, e01270-18. [Google Scholar] [CrossRef] [PubMed]
- Dodd, K.A.; McElroy, A.K.; Jones, M.E.; Nichol, S.T.; Spiropoulou, C.F. Rift Valley fever virus clearance and protection from neurologic disease are dependent on CD4+ T cell and virus-specific antibody responses. J. Virol. 2013, 87, 6161–6171. [Google Scholar] [CrossRef] [PubMed]
- Dodd, K.A.; McElroy, A.K.; Jones, T.L.; Zaki, S.R.; Nichol, S.T.; Spiropoulou, C.F. Rift valley Fever virus encephalitis is associated with an ineffective systemic immune response and activated T cell infiltration into the CNS in an immunocompetent mouse model. PLoS Negl. Trop. Dis. 2014, 8, e2874. [Google Scholar] [CrossRef]
- Michaely, L.M.; Rissmann, M.; Keller, M.; König, R.; von Arnim, F.; Eiden, M.; Rohn, K.; Baumgärtner, W.; Groschup, M.; Ulrich, R. NSG-Mice Reveal the Importance of a Functional Innate and Adaptive Immune Response to Overcome RVFV Infection. Viruses 2022, 14, 350. [Google Scholar] [CrossRef]
- Nair, N.; Osterhaus, A.; Rimmelzwaan, G.F.; Prajeeth, C.K. Rift Valley Fever Virus-Infection, Pathogenesis and Host Immune Responses. Pathogens 2023, 12, 1174. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Cao, K.; Shen, X.; Zhang, B.; Chen, M.; Yu, W. Clinical Characteristics and Immune Status of Patients with Severe Fever with Thrombocytopenia Syndrome. Viral Immunol. 2022, 35, 465–473. [Google Scholar] [CrossRef]
- Sun, L.; Hu, Y.; Niyonsaba, A.; Tong, Q.; Lu, L.; Li, H.; Jie, S. Detection and evaluation of immunofunction of patients with severe fever with thrombocytopenia syndrome. Clin. Exp. Med. 2014, 14, 389–395. [Google Scholar] [CrossRef]
- Li, M.M.; Zhang, W.J.; Weng, X.F.; Li, M.Y.; Liu, J.; Xiong, Y.; Xiong, S.E.; Zou, C.C.; Wang, H.; Lu, M.J.; et al. CD4 T cell loss and Th2 and Th17 bias are associated with the severity of severe fever with thrombocytopenia syndrome (SFTS). Clin. Immunol. 2018, 195, 8–17. [Google Scholar] [CrossRef]
- Yi, X.; Li, W.; Li, H.; Jie, S. Circulating regulatory T cells in patients with severe fever with thrombocytopenia syndrome. Infect. Dis. 2015, 47, 294–301. [Google Scholar] [CrossRef]
- Kim, M.; Heo, S.T.; Seong, G.M.; Lee, K.H.; Yoo, J.R. Severe fever with thrombocytopenia syndrome (SFTS) associated with invasive pulmonary Aspergillosis in a patient with a low CD4+ T-cell count: A case report. Int. J. Crit. Illn. Inj. Sci. 2020, 10 (Suppl. S1), 53–56. [Google Scholar] [CrossRef]
- Li, J.; Han, Y.; Xing, Y.; Li, S.; Kong, L.; Zhang, Y.; Zhang, L.; Liu, N.; Wang, Q.; Wang, S.; et al. Concurrent measurement of dynamic changes in viral load, serum enzymes, T cell subsets, and cytokines in patients with severe fever with thrombocytopenia syndrome. PLoS ONE 2014, 9, e91679. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; Zhang, W.J.; Liu, J.; Li, M.Y.; Zhang, Y.F.; Xiong, Y.; Xiong, S.E.; Zou, C.C.; Xiong, L.Q.; Liang, B.Y.; et al. Dynamic changes in the immunological characteristics of T lymphocytes in surviving patients with severe fever with thrombocytopenia syndrome (SFTS). Int. J. Infect. Dis. 2018, 70, 72–80. [Google Scholar] [CrossRef]
- Zong, L.; Yang, F.; Liu, S.; Gao, Y.; Xia, F.; Zheng, M.; Xu, Y. CD8(+) T cells mediate antiviral response in severe fever with thrombocytopenia syndrome. FASEB J. 2023, 37, e22722. [Google Scholar] [CrossRef]
- Jonsson, C.B.; Figueiredo, L.T.; Vapalahti, O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010, 23, 412–441. [Google Scholar] [CrossRef]
- Schönrich, G.; Rang, A.; Lütteke, N.; Raftery, M.J.; Charbonnel, N.; Ulrich, R.G. Hantavirus-induced immunity in rodent reservoirs and humans. Immunol. Rev. 2008, 225, 163–189. [Google Scholar] [CrossRef] [PubMed]
- Easterbrook, J.D.; Klein, S.L. Immunological mechanisms mediating hantavirus persistence in rodent reservoirs. PLoS Pathog. 2008, 4, e1000172. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, F.; Díaz, F.E.; Retamal-Díaz, A.; Covián, C.; González, P.A.; Kalergis, A.M. Immune response during hantavirus diseases: Implications for immunotherapies and vaccine design. Immunology 2021, 163, 262–277. [Google Scholar] [CrossRef] [PubMed]
- Klingström, J.; Smed-Sörensen, A.; Maleki, K.T.; Solà-Riera, C.; Ahlm, C.; Björkström, N.K.; Ljunggren, H.G. Innate and adaptive immune responses against human Puumala virus infection: Immunopathogenesis and suggestions for novel treatment strategies for severe hantavirus-associated syndromes. J. Intern. Med. 2019, 285, 510–523. [Google Scholar] [CrossRef]
- Ma, R.X.; Cheng, L.F.; Ying, Q.K.; Liu, R.R.; Ma, T.J.; Zhang, X.X.; Liu, Z.Y.; Zhang, L.; Ye, W.; Zhang, F.L.; et al. Screening and Identification of an H-2Kb-Restricted CTL Epitope within the Glycoprotein of Hantaan Virus. Front. Cell Infect. Microbiol. 2016, 6, 151. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.L.; Zhu, Y.; Wang, J.P.; Liu, J.M.; Fang, L.; Jin, B.Q. Identification of HTNV-NP-specific T lymphocyte epitopes and analysis of the epitope-specific T cell response. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2005, 21, 704–706. [Google Scholar]
- Maeda, K.; West, K.; Toyosaki-Maeda, T.; Rothman, A.L.; Ennis, F.A.; Terajima, M. Identification and analysis for cross-reactivity among hantaviruses of H-2b-restricted cytotoxic T-lymphocyte epitopes in Sin Nombre virus nucleocapsid protein. J. Gen. Virol. 2004, 85, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Dong, Y.; Zhou, Y.; Ren, H.; Ji, Y.; Lv, S. Levels of HTNV-specific CD8+ T lymphocytes in PBMC from the patients with hemorrhagic fever with renal syndrome. Intern. Emerg. Med. 2013, 8, 503–508. [Google Scholar] [CrossRef]
- Liu, B.; Ma, Y.; Zhang, Y.; Zhang, C.; Yi, J.; Zhuang, R.; Yu, H.; Yang, A.; Zhang, Y.; Jin, B. CD8low CD100- T Cells Identify a Novel CD8 T Cell Subset Associated with Viral Control during Human Hantaan Virus Infection. J. Virol. 2015, 89, 11834–11844. [Google Scholar] [CrossRef]
- Tang, K.; Zhang, Y.; Li, X.; Zhang, C.; Jia, X.; Hu, H.; Chen, L.; Zhuang, R.; Zhang, Y.; Jin, B.; et al. HLA-E-restricted Hantaan virus-specific CD8+ T cell responses enhance the control of infection in hemorrhagic fever with renal syndrome. Biosaf. Health 2023, 5, 289–299. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, J.; Yuan, B.; Wang, M.; Zhang, Y.; Xu, Z.; Zhang, C.; Zhang, Y.; Liu, B.; Yi, J.; et al. HLA-A2 and B35 restricted hantaan virus nucleoprotein CD8+ T-cell epitope-specific immune response correlates with milder disease in hemorrhagic fever with renal syndrome. PLoS Negl. Trop. Dis. 2013, 7, e2076. [Google Scholar] [CrossRef]
- Iglesias, A.A.; Períolo, N.; Bellomo, C.M.; Lewis, L.C.; Olivera, C.P.; Anselmo, C.R.; García, M.; Coelho, R.M.; Alonso, D.O.; Dighero-Kemp, B.; et al. Delayed viral clearance despite high number of activated T cells during the acute phase in Argentinean patients with hantavirus pulmonary syndrome. eBioMedicine 2022, 75, 103765. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ma, R.; Liu, Z.; Hu, H.; Shu, J.; Hu, P.; Kang, J.; Zhang, Y.; Han, M.; Zhang, X.; et al. HTNV infection of CD8+ T cells is associated with disease progression in HFRS patients. Commun. Biol. 2021, 4, 652. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.A.; Campos, G.M.; Moreli, M.L.; Moro Souza, R.L.; Saggioro, F.P.; Figueiredo, G.G.; Livonesi, M.C.; Moraes Figueiredo, L.T. Role of mixed Th1 and Th2 serum cytokines on pathogenesis and prognosis of hantavirus pulmonary syndrome. Microbes Infect. 2008, 10, 1150–1157. [Google Scholar] [CrossRef]
- Ma, Y.; Yuan, B.; Zhuang, R.; Zhang, Y.; Liu, B.; Zhang, C.; Zhang, Y.; Yu, H.; Yi, J.; Yang, A.; et al. Hantaan virus infection induces both Th1 and ThGranzyme B+ cell immune responses that associated with viral control and clinical outcome in humans. PLoS Pathog. 2015, 11, e1004788. [Google Scholar] [CrossRef]
- de Carvalho Nicacio, C.; Sällberg, M.; Hultgren, C.; Lundkvist, Å. T-helper and humoral responses to Puumala hantavirus nucleocapsid protein: Identification of T-helper epitopes in a mouse model. J. Gen. Virol. 2001, 82 Pt 1, 129–138. [Google Scholar] [CrossRef]
- Ma, Y.; Cheng, L.; Yuan, B.; Zhang, Y.; Zhang, C.; Zhang, Y.; Tang, K.; Zhuang, R.; Chen, L.; Yang, K.; et al. Structure and Function of HLA-A*02-Restricted Hantaan Virus Cytotoxic T-Cell Epitope That Mediates Effective Protective Responses in HLA-A2.1/K(b) Transgenic Mice. Front. Immunol. 2016, 7, 298. [Google Scholar] [CrossRef]
- Ma, Y.; Tang, K.; Zhang, Y.; Zhang, C.; Zhang, Y.; Jin, B.; Ma, Y. Design and synthesis of HLA-A*02-restricted Hantaan virus multiple-antigenic peptide for CD8(+) T cells. Virol. J. 2020, 17, 15. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Tang, K.; Zhang, Y.; Zhang, C.; Cheng, L.; Zhang, F.; Zhuang, R.; Jin, B.; Zhang, Y. Protective CD8(+) T-cell response against Hantaan virus infection induced by immunization with designed linear multi-epitope peptides in HLA-A2.1/K(b) transgenic mice. Virol. J. 2020, 17, 146. [Google Scholar] [CrossRef] [PubMed]
- Kallio-Kokko, H.; Leveelahti, R.; Brummer-Korvenkontio, M.; Lundkvist, A.; Vaheri, A.; Vapalahti, O. Human immune response to Puumala virus glycoproteins and nucleocapsid protein expressed in mammalian cells. J. Med. Virol. 2001, 65, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Lindkvist, M.; Lahti, K.; Lilliehöök, B.; Holmström, A.; Ahlm, C.; Bucht, G. Cross-reactive immune responses in mice after genetic vaccination with cDNA encoding hantavirus nucleocapsid proteins. Vaccine 2007, 25, 1690–1699. [Google Scholar] [CrossRef]
- Lundkvist, A.; Meisel, H.; Koletzki, D.; Lankinen, H.; Cifire, F.; Geldmacher, A.; Sibold, C.; Gött, P.; Vaheri, A.; Krüger, D.H.; et al. Mapping of B-cell epitopes in the nucleocapsid protein of Puumala hantavirus. Viral Immunol. 2002, 15, 177–192. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho Nicacio, C.; Gonzalez Della Valle, M.; Padula, P.; Björling, E.; Plyusnin, A.; Lundkvist, A. Cross-protection against challenge with Puumala virus after immunization with nucleocapsid proteins from different hantaviruses. J. Virol. 2002, 76, 6669–6677. [Google Scholar] [CrossRef] [PubMed]
- Garrison, A.R.; Alkhovsky, S.V.; Avšič-Županc, T.; Bente, D.A.; Bergeron, É.; Burt, F.; Di Paola, N.; Ergünay, K.; Hewson, R.; Kuhn, J.H.; et al. ICTV Virus Taxonomy Profile: Nairoviridae. J. Gen. Virol. 2020, 101, 798–799. [Google Scholar] [CrossRef]
- Krasteva, S.; Jara, M.; Frias-De-Diego, A.; Machado, G. Nairobi Sheep Disease Virus: A Historical and Epidemiological Perspective. Front. Vet. Sci. 2020, 7, 419. [Google Scholar] [CrossRef]
- Rodriguez, S.E.; Hawman, D.W.; Sorvillo, T.E.; O’Neal, T.J.; Bird, B.H.; Rodriguez, L.L.; Bergeron, É.; Nichol, S.T.; Montgomery, J.M.; Spiropoulou, C.F.; et al. Immunobiology of Crimean-Congo hemorrhagic fever. Antivir. Res. 2022, 199, 105244. [Google Scholar] [CrossRef]
- Khan, M.S.A.; Nain, Z.; Syed, S.B.; Abdulla, F.; Moni, M.A.; Sheam, M.M.; Karim, M.M.; Adhikari, U.K. Computational formulation and immune dynamics of a multi-peptide vaccine candidate against Crimean-Congo hemorrhagic fever virus. Mol. Cell Probes 2021, 55, 101693. [Google Scholar] [CrossRef]
- Golden, J.W.; Fitzpatrick, C.J.; Suschak, J.J.; Clements, T.L.; Ricks, K.M.; Sanchez-Lockhart, M.; Garrison, A.R. Induced protection from a CCHFV-M DNA vaccine requires CD8(+) T cells. Virus Res. 2023, 334, 199173. [Google Scholar] [CrossRef] [PubMed]
- Appelberg, S.; John, L.; Pardi, N.; Végvári, Á.; Bereczky, S.; Ahlén, G.; Monteil, V.; Abdurahman, S.; Mikaeloff, F.; Beattie, M.; et al. Nucleoside-Modified mRNA Vaccines Protect IFNAR(−/−) Mice against Crimean-Congo Hemorrhagic Fever Virus Infection. J. Virol. 2022, 96, e0156821. [Google Scholar] [CrossRef] [PubMed]
- Kortekaas, J.; Vloet, R.P.; McAuley, A.J.; Shen, X.; Bosch, B.J.; De Vries, L.; Moormann, R.J.; Bente, D.A. Crimean-Congo Hemorrhagic Fever Virus Subunit Vaccines Induce High Levels of Neutralizing Antibodies But No Protection in STAT1 Knockout Mice. Vector-Borne Zoonotic Dis. 2015, 15, 759–764. [Google Scholar] [CrossRef]
- Dowall, S.D.; Graham, V.A.; Rayner, E.; Hunter, L.; Watson, R.; Taylor, I.; Rule, A.; Carroll, M.W.; Hewson, R. Protective effects of a Modified Vaccinia Ankara-based vaccine candidate against Crimean-Congo Haemorrhagic Fever virus require both cellular and humoral responses. PLoS ONE 2016, 11, e0156637. [Google Scholar] [CrossRef]
- Hawman, D.W.; Meade-White, K.; Leventhal, S.; Feldmann, F.; Okumura, A.; Smith, B.; Scott, D.; Feldmann, H. Immunocompetent mouse model for Crimean-Congo hemorrhagic fever virus. eLife 2021, 10, e63906. [Google Scholar] [CrossRef] [PubMed]
- Akinci, E.; Yilmaz, M.; Bodur, H.; Ongürü, P.; Bayazit, F.N.; Erbay, A.; Ozet, G. Analysis of lymphocyte subgroups in Crimean-Congo hemorrhagic fever. Int. J. Infect. Dis. 2009, 13, 560–563. [Google Scholar] [CrossRef]
- Salvati, M.V.; Salaris, C.; Monteil, V.; Del Vecchio, C.; Palù, G.; Parolin, C.; Calistri, A.; Bell-Sakyi, L.; Mirazimi, A.; Salata, C. Virus-Derived DNA Forms Mediate the Persistent Infection of Tick Cells by Hazara Virus and Crimean-Congo Hemorrhagic Fever Virus. J. Virol. 2021, 95, e0163821. [Google Scholar] [CrossRef] [PubMed]
- Ohta, K.; Saka, N.; Nishio, M. Hazara Orthonairovirus Nucleoprotein Antagonizes Type I Interferon Production by Inhibition of RIG-I Ubiquitination. Viruses 2022, 14, 1965. [Google Scholar] [CrossRef]
- Tapia-Ramírez, G.; Lorenzo, C.; Navarrete, D.; Carrillo-Reyes, A.; Retana, Ó.; Carrasco-Hernández, R. A Review of Mammarenaviruses and Rodent Reservoirs in the Americas. Ecohealth 2022, 19, 22–39. [Google Scholar] [CrossRef]
- Briese, T.; Paweska, J.T.; McMullan, L.K.; Hutchison, S.K.; Street, C.; Palacios, G.; Khristova, M.L.; Weyer, J.; Swanepoel, R.; Egholm, M.; et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009, 5, e1000455. [Google Scholar] [CrossRef]
- Abdel-Hakeem, M.S. Viruses Teaching Immunology: Role of LCMV Model and Human Viral Infections in Immunological Discoveries. Viruses 2019, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Oldstone, M.B.; Lewicki, H.; Homann, D.; Nguyen, C.; Julien, S.; Gairin, J.E. Common antiviral cytotoxic t-lymphocyte epitope for diverse arenaviruses. J. Virol. 2001, 75, 6273–6278. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Muzaffar, A.; Shoaib, R.M.; Khan, A.; Waheed, Y.; Wei, D.Q. Towards specie-specific ensemble vaccine candidates against mammarenaviruses using optimized structural vaccinology pipeline and molecular modelling approaches. Microb. Pathog. 2022, 172, 105793. [Google Scholar] [CrossRef] [PubMed]
- Azim, K.F.; Lasker, T.; Akter, R.; Hia, M.M.; Bhuiyan, O.F.; Hasan, M.; Hossain, M.N. Combination of highly antigenic nucleoproteins to inaugurate a cross-reactive next generation vaccine candidate against Arenaviridae family. Heliyon 2021, 7, e07022. [Google Scholar] [CrossRef] [PubMed]
- Botten, J.; Alexander, J.; Pasquetto, V.; Sidney, J.; Barrowman, P.; Ting, J.; Peters, B.; Southwood, S.; Stewart, B.; Rodriguez-Carreno, M.P.; et al. Identification of protective Lassa virus epitopes that are restricted by HLA-A2. J. Virol. 2006, 80, 8351–8361. [Google Scholar] [CrossRef] [PubMed]
- Boesen, A.; Sundar, K.; Coico, R. Lassa fever virus peptides predicted by computational analysis induce epitope-specific cytotoxic-T-lymphocyte responses in HLA-A2.1 transgenic mice. Clin. Diagn. Lab. Immunol. 2005, 12, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Clegg, J.C.S. Current progress towards vaccines for arenavirus-caused diseases. Vaccine 1992, 10, 89–95. [Google Scholar] [CrossRef]
- Fisher-Hoch, S.P.; McCormick, J.B. Towards a human Lassa fever vaccine. Rev. Med. Virol. 2001, 11, 331–341. [Google Scholar] [CrossRef]
- Jahrling, P.B.; Frame, J.D.; Rhoderick, J.B.; Monson, M.H. Endemic Lassa fever in Liberia. IV. Selection of optimally effective plasma for treatment by passive immunization. Trans. R. Soc. Trop. Med. Hyg. 1985, 79, 380–384. [Google Scholar] [CrossRef]
- Port, J.R.; Wozniak, D.M.; Oestereich, L.; Pallasch, E.; Becker-Ziaja, B.; Müller, J.; Rottstegge, M.; Olal, C.; Gómez-Medina, S.; Oyakhliome, J.; et al. Severe Human Lassa Fever Is Characterized by Nonspecific T-Cell Activation and Lymphocyte Homing to Inflamed Tissues. J. Virol. 2020, 94, e01367-20. [Google Scholar] [CrossRef] [PubMed]
- Flatz, L.; Rieger, T.; Merkler, D.; Bergthaler, A.; Regen, T.; Schedensack, M.; Bestmann, L.; Verschoor, A.; Kreutzfeldt, M.; Brück, W.; et al. T cell-dependence of Lassa fever pathogenesis. PLoS Pathog. 2010, 6, e1000836. [Google Scholar] [CrossRef]
- Carballal, G.; Oubiña, J.R.; Rondinone, S.N.; Elsner, B.; Frigerio, M.J. Cell-mediated immunity and lymphocyte populations in experimental Argentine hemorrhagic fever (Junín Virus). Infect. Immun. 1981, 34, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Barrios, H.A.; Rondinone, S.N.; Blejer, J.L.; Giovanniello, O.A.; Nota, N.R. Development of specific immune response in mice infected with Junin virus. Acta Virol. 1982, 26, 156–164. [Google Scholar]
- Evans, A.B.; Peterson, K.E. Cross reactivity of neutralizing antibodies to the encephalitic California Serogroup orthobunyaviruses varies by virus and genetic relatedness. Sci. Rep. 2021, 11, 16424. [Google Scholar] [CrossRef]
- Wernike, K.; Aebischer, A.; Sick, F.; Szillat, K.P.; Beer, M. Differentiation of Antibodies against Selected Simbu Serogroup Viruses by a Glycoprotein Gc-Based Triplex ELISA. Vet. Sci. 2021, 8, 12. [Google Scholar] [CrossRef]
- Skinner, B.; Mikula, S.; Davis, B.S.; Powers, J.A.; Hughes, H.R.; Calvert, A.E. Monoclonal antibodies to Cache Valley virus for serological diagnosis. PLoS Negl. Trop. Dis. 2022, 16, e0010156. [Google Scholar] [CrossRef]
- Srihongse, S.; Grayson, M.A.; Deibel, R. California serogroup viruses in New York State: The role of subtypes in human infections. Am. J. Trop. Med. Hyg. 1984, 33, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Heinz, F.; Asera, J. Presence of viruse-neutralizing antibodies ot the Tahyna virus in the inhabitants of North Moravia. Folia Parasitol. 1972, 19, 315–320. [Google Scholar]
- Blitvich, B.J.; Saiyasombat, R.; Talavera-Aguilar, L.G.; Garcia-Rejon, J.E.; Farfan-Ale, J.A.; Machain-Williams, C.; Loroño-Pino, M.A. Orthobunyavirus Antibodies in Humans, Yucatan Peninsula, Mexico. Emerg. Infect. Dis. 2012, 18, 1629–1632. [Google Scholar] [CrossRef] [PubMed]
- Grimstad, P.R.; Schmitt, S.M.; Williams, D.G. Prevalence of neutralizing antibody to Jamestown Canyon virus (California group) in populations of elk and moose in northern Michigan and Ontario, Canada. J. Wildl. Dis. 1986, 22, 453–458. [Google Scholar] [CrossRef]
- Putkuri, N.; Vaheri, A.; Vapalahti, O. Prevalence and protein specificity of human antibodies to Inkoo virus infection. Clin. Vaccine Immunol. 2007, 14, 1555–1562. [Google Scholar] [CrossRef]
- Gonzalez-Scarano, F.; Shope, R.E.; Calisher, C.E.; Nathanson, N. Characterization of monoclonal antibodies against the G1 and N proteins of LaCrosse and Tahyna, two California serogroup bunyaviruses. Virology 1982, 120, 42–53. [Google Scholar] [CrossRef]
- Wernike, K.; Brocchi, E.; Cordioli, P.; Sénéchal, Y.; Schelp, C.; Wegelt, A.; Aebischer, A.; Roman-Sosa, G.; Reimann, I.; Beer, M. A novel panel of monoclonal antibodies against Schmallenberg virus nucleoprotein and glycoprotein Gc allows specific orthobunyavirus detection and reveals antigenic differences. Vet. Res. 2015, 46, 27. [Google Scholar] [CrossRef]
- Kingsford, L.; Hill, D.W. The effect of proteolytic cleavage of La Crosse virus G1 glycoprotein on antibody neutralization. J. Gen. Virol. 1983, 64 Pt 10, 2147–2156. [Google Scholar] [CrossRef]
- Powers, J.A.; Boroughs, K.L.; Mikula, S.; Goodman, C.H.; Davis, E.H.; Thrasher, E.M.; Hughes, H.R.; Biggerstaff, B.J.; Calvert, A.E. Characterization of a monoclonal antibody specific to California serogroup orthobunyaviruses and development as a chimeric immunoglobulin M-positive control in human diagnostics. Microbiol. Spectr. 2023, 11, e01966-23. [Google Scholar] [CrossRef] [PubMed]
- Hellert, J.; Aebischer, A.; Wernike, K.; Haouz, A.; Brocchi, E.; Reiche, S.; Guardado-Calvo, P.; Beer, M.; Rey, F.A. Orthobunyavirus spike architecture and recognition by neutralizing antibodies. Nat. Commun. 2019, 10, 879. [Google Scholar] [CrossRef]
- Roman-Sosa, G.; Brocchi, E.; Schirrmeier, H.; Wernike, K.; Schelp, C.; Beer, M. Analysis of the humoral immune response against the envelope glycoprotein Gc of Schmallenberg virus reveals a domain located at the amino terminus targeted by mAbs with neutralizing activity. J. Gen. Virol. 2016, 97, 571–580. [Google Scholar] [CrossRef]
- Kingsford, L. Enhanced neutralization of La Crosse virus by the binding of specific pairs of monoclonal antibodies to the G1 glycoprotein. Virology 1984, 136, 265–273. [Google Scholar] [CrossRef]
- Ogawa, Y.; Eguchi, M.; Shimoji, Y. Two Akabane virus glycoprotein Gc domains induce neutralizing antibodies in mice. J. Vet. Med. Sci. 2022, 84, 538–542. [Google Scholar] [CrossRef]
- Kingsford, L.; Boucquey, K.H. Monoclonal antibodies specific for the G1 glycoprotein of La Crosse virus that react with other California serogroup viruses. J. Gen. Virol. 1990, 71 Pt 3, 523–530. [Google Scholar] [CrossRef]
- Bréard, E.; Lara, E.; Comtet, L.; Viarouge, C.; Doceul, V.; Desprat, A.; Vitour, D.; Pozzi, N.; Cay, A.B.; De Regge, N.; et al. Validation of a commercially available indirect ELISA using a nucleocapside recombinant protein for detection of Schmallenberg virus antibodies. PLoS ONE 2013, 8, e53446. [Google Scholar] [CrossRef] [PubMed]
- Roman-Sosa, G.; Karger, A.; Kraatz, F.; Aebischer, A.; Wernike, K.; Maksimov, P.; Lillig, C.H.; Reimann, I.; Brocchi, E.; Keller, M.; et al. The amino terminal subdomain of glycoprotein Gc of Schmallenberg virus: Disulfide bonding and structural determinants of neutralization. J. Gen. Virol. 2017, 98, 1259–1273. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.S.; Gresko, A.K.; Nelson, J.T.; Murphy, B.R.; Whitehead, S.S. A recombinant chimeric La Crosse virus expressing the surface glycoproteins of Jamestown Canyon virus is immunogenic and protective against challenge with either parental virus in mice or monkeys. J. Virol. 2012, 86, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Operschall, E.; Schuh, T.; Heinzerling, L.; Pavlovic, J.; Moelling, K. Enhanced protection against viral infection by co-administration of plasmid DNA coding for viral antigen and cytokines in mice. J. Clin. Virol. 1999, 13, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Pekosz, A.; Griot, C.; Stillmock, K.; Nathanson, N.; Gonzalez-Scarano, F. Protection from La Crosse virus encephalitis with recombinant glycoproteins: Role of neutralizing anti-G1 antibodies. J. Virol. 1995, 69, 3475–3481. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, S.H.; Cornejo Pontelli, M.; Mishra, N.; Zhou, C.; de Paula Souza, J.; Mendes Viana, R.M.; Lipkin, W.I.; Knipe, D.M.; Arruda, E.; Whelan, S.P.J. Vesicular Stomatitis Virus Chimeras Expressing the Oropouche Virus Glycoproteins Elicit Protective Immune Responses in Mice. mBio 2021, 12, e0046321. [Google Scholar] [CrossRef]
- Hertz, T.; Beatty, P.R.; MacMillen, Z.; Killingbeck, S.S.; Wang, C.; Harris, E. Antibody Epitopes Identified in Critical Regions of Dengue Virus Nonstructural 1 Protein in Mouse Vaccination and Natural Human Infections. J. Immunol. 2017, 198, 4025–4035. [Google Scholar] [CrossRef]
- Sootichote, R.; Puangmanee, W.; Benjathummarak, S.; Kowaboot, S.; Yamanaka, A.; Boonnak, K.; Ampawong, S.; Chatchen, S.; Ramasoota, P.; Pitaksajjakul, P. Potential Protective Effect of Dengue NS1 Human Monoclonal Antibodies against Dengue and Zika Virus Infections. Biomedicines 2023, 11, 227. [Google Scholar] [CrossRef]
- Lai, Y.-C.; Chuang, Y.-C.; Liu, C.-C.; Ho, T.-S.; Lin, Y.-S.; Anderson, R.; Yeh, T.-M. Antibodies Against Modified NS1 Wing Domain Peptide Protect Against Dengue Virus Infection. Sci. Rep. 2017, 7, 6975. [Google Scholar] [CrossRef]
- Wright, D.; Allen, E.R.; Clark, M.H.A.; Gitonga, J.N.; Karanja, H.K.; Hulswit, R.J.G.; Taylor, I.; Biswas, S.; Marshall, J.; Mwololo, D.; et al. Naturally Acquired Rift Valley Fever Virus Neutralizing Antibodies Predominantly Target the Gn Glycoprotein. iScience 2020, 23, 101669. [Google Scholar] [CrossRef]
- Chapman, N.S.; Zhao, H.; Kose, N.; Westover, J.B.; Kalveram, B.; Bombardi, R.; Rodriguez, J.; Sutton, R.; Genualdi, J.; LaBeaud, A.D.; et al. Potent neutralization of Rift Valley fever virus by human monoclonal antibodies through fusion inhibition. Proc. Natl. Acad. Sci. USA 2021, 118, e2025642118. [Google Scholar] [CrossRef]
- Fu, L.; Xu, L.; Qian, J.; Wu, X.; Wang, Z.; Wang, H.; Liu, D.; Deng, F.; Shen, S. The Neutralizing Monoclonal Antibodies against SFTS Group Bandaviruses Suggest New Targets of Specific or Broad-Spectrum Antivirals. Am. J. Trop. Med. Hyg. 2023, 109, 1319–1328. [Google Scholar] [CrossRef]
- Allen, E.R.; Krumm, S.A.; Raghwani, J.; Halldorsson, S.; Elliott, A.; Graham, V.A.; Koudriakova, E.; Harlos, K.; Wright, D.; Warimwe, G.M.; et al. A Protective Monoclonal Antibody Targets a Site of Vulnerability on the Surface of Rift Valley Fever Virus. Cell Rep. 2018, 25, 3750–3758.e3754. [Google Scholar] [CrossRef]
- Besselaar, T.G.; Blackburn, N.K. Topological mapping of antigenic sites on the Rift Valley fever virus envelope glycoproteins using monoclonal antibodies. Arch. Virol. 1991, 121, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhu, Y.; Gao, F.; Jiao, Y.; Oladejo, B.O.; Chai, Y.; Bi, Y.; Lu, S.; Dong, M.; Zhang, C.; et al. Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope. Proc. Natl. Acad. Sci. USA 2017, 114, e7564–e7573. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Zhang, G.; Zhang, S.; Chen, Z.; Chi, X.; Dong, Y.; Fan, P.; Liu, Y.; Chen, Y.; Song, X.; et al. Characterization of Two Neutralizing Antibodies against Rift Valley Fever Virus Gn Protein. Viruses 2020, 12, 259. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, L.; Zhang, W.; Chi, Y.; Zeng, X.; Li, X.; Qi, X.; Jin, Q.; Zhang, X.; Huang, M.; et al. Human antibody neutralizes severe Fever with thrombocytopenia syndrome virus, an emerging hemorrhagic Fever virus. Clin. Vaccine Immunol. 2013, 20, 1426–1432. [Google Scholar] [CrossRef]
- Gandolfo, C.; Prathyumn, S.; Terrosi, C.; Anichini, G.; Gori Savellini, G.; Corti, D.; Bracci, L.; Lanzavecchia, A.; Roman-Sosa, G.; Cusi, M.G. Identification of a Neutralizing Epitope on TOSV Gn Glycoprotein. Vaccines 2021, 9, 924. [Google Scholar] [CrossRef] [PubMed]
- Magurano, F.; Nicoletti, L. Humoral response in Toscana virus acute neurologic disease investigated by viral-protein-specific immunoassays. Clin. Diagn. Lab. Immunol. 1999, 6, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Di Bonito, P.; Bosco, S.; Mochi, S.; Accardi, L.; Ciufolini, M.G.; Nicoletti, L.; Giorgi, C. Human antibody response to Toscana virus glycoproteins expressed by recombinant baculovirus. J. Med. Virol. 2002, 68, 615–619. [Google Scholar] [CrossRef]
- Fernandez, J.C.; Billecocq, A.; Durand, J.P.; Cêtre-Sossah, C.; Cardinale, E.; Marianneau, P.; Pépin, M.; Tordo, N.; Bouloy, M. The nonstructural protein NSs induces a variable antibody response in domestic ruminants naturally infected with Rift Valley fever virus. Clin. Vaccine Immunol. 2012, 19, 5–10. [Google Scholar] [CrossRef]
- Findlay, G.M. The Mechanism of Immunity in Rift Valley Fever. Br. J. Exp. Pathol. 1936, 17, 89–104. [Google Scholar]
- Pierro, A.; Ficarelli, S.; Ayhan, N.; Morini, S.; Raumer, L.; Bartoletti, M.; Mastroianni, A.; Prati, F.; Schivazappa, S.; Cenni, P.; et al. Characterization of antibody response in neuroinvasive infection caused by Toscana virus. Clin. Microbiol. Infect. 2017, 23, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Kong, Q.; Liu, Y.; Li, J.; Bian, T.; Ma, X.; Ye, Y.; Li, J. Time Course of Severe Fever With Thrombocytopenia Syndrome Virus and Antibodies in Patients by Long-Term Follow-Up Study, China. Front. Microbiol. 2021, 12, 744037. [Google Scholar] [CrossRef] [PubMed]
- Mhamadi, M.; Badji, A.; Barry, M.A.; Ndiaye, E.H.; Gaye, A.; Ndiaye, M.; Mhamadi, M.; Touré, C.T.; Ndiaye, O.; Faye, B.; et al. Human and Livestock Surveillance Revealed the Circulation of Rift Valley Fever Virus in Agnam, Northern Senegal, 2021. Trop. Med. Infect. Dis. 2023, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Y.; Huang, B.; Ma, X.; Zhu, L.; Zheng, N.; Xu, S.; Nawaz, W.; Xu, C.; Wu, Z. A single-domain antibody inhibits SFTSV and mitigates virus-induced pathogenesis in vivo. JCI Insight 2020, 5, e136855. [Google Scholar] [CrossRef]
- Li, J.C.; Ding, H.; Wang, G.; Zhang, S.; Yang, X.; Wu, Y.X.; Peng, X.F.; Zhang, X.A.; Yang, Z.D.; Cui, N.; et al. Dynamics of neutralizing antibodies against severe fever with thrombocytopenia syndrome virus. Int. J. Infect. Dis. 2023, 134, 95–98. [Google Scholar] [CrossRef]
- Wang, G.; Chang, H.; Jia, B.; Liu, Y.; Huang, R.; Wu, W.; Hao, Y.; Yan, X.; Xia, J.; Chen, Y.; et al. Nucleocapsid protein-specific IgM antibody responses in the disease progression of severe fever with thrombocytopenia syndrome. Ticks Tick. Borne Dis. 2019, 10, 639–646. [Google Scholar] [CrossRef]
- Salekwa, L.P.; Wambura, P.N.; Matiko, M.K.; Watts, D.M. Circulation of Rift Valley Fever Virus Antibody in Cattle during Inter-Epizootic/Epidemic Periods in Selected Regions of Tanzania. Am. J. Trop. Med. Hyg. 2019, 101, 459–466. [Google Scholar] [CrossRef]
- Nfon, C.K.; Marszal, P.; Zhang, S.; Weingartl, H.M. Innate Immune Response to Rift Valley Fever Virus in Goats. PLoS Negl. Trop. Dis. 2012, 6, e1623. [Google Scholar] [CrossRef] [PubMed]
- Selina, O.; Imatdinov, I.; Balysheva, V.; Akasov, R.; Kryukov, A.; Balyshev, V.; Markvicheva, E. Microencapsulated plasmids expressing Gn and Gc glycoproteins of Rift Valley Fever virus enhance humoral immune response in mice. Biotechnol. Lett. 2020, 42, 529–536. [Google Scholar] [CrossRef]
- Faburay, B.; Lebedev, M.; McVey, D.S.; Wilson, W.; Morozov, I.; Young, A.; Richt, J.A. A glycoprotein subunit vaccine elicits a strong Rift Valley fever virus neutralizing antibody response in sheep. Vector-Borne Zoonotic Dis. 2014, 14, 746–756. [Google Scholar] [CrossRef]
- Chrun, T.; Lacôte, S.; Urien, C.; Richard, C.A.; Tenbusch, M.; Aubrey, N.; Pulido, C.; Lakhdar, L.; Marianneau, P.; Schwartz-Cornil, I. A DNA Vaccine Encoding the Gn Ectodomain of Rift Valley Fever Virus Protects Mice via a Humoral Response Decreased by DEC205 Targeting. Front. Immunol. 2019, 10, 860. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lai, C.J.; Cha, I.; Kang, S.; Yang, W.S.; Choi, Y.; Jung, J.U. SFTSV Gn-Head mRNA vaccine confers efficient protection against lethal viral challenge. J. Med. Virol. 2023, 95, e29203. [Google Scholar] [CrossRef] [PubMed]
- Vapalahti, O.; Kallio-Kokko, H.; Närvänen, A.; Julkunen, I.; Lundkvist, A.; Plyusnin, A.; Lehväslaiho, H.; Brummer-Korvenkontio, M.; Vaheri, A.; Lankinen, H. Human B-cell epitopes of Puumala virus nucleocapsid protein, the major antigen in early serological response. J. Med. Virol. 1995, 46, 293–303. [Google Scholar] [CrossRef]
- Engdahl, T.B.; Crowe, J.E., Jr. Humoral Immunity to Hantavirus Infection. mSphere 2020, 5, e00482-20. [Google Scholar] [CrossRef]
- Hepojoki, J.; Strandin, T.; Vaheri, A.; Lankinen, H. Interactions and oligomerization of hantavirus glycoproteins. J. Virol. 2010, 84, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Battisti, A.J.; Chu, Y.K.; Chipman, P.R.; Kaufmann, B.; Jonsson, C.B.; Rossmann, M.G. Structural studies of Hantaan virus. J. Virol. 2011, 85, 835–841. [Google Scholar] [CrossRef]
- Li, S.; Rissanen, I.; Zeltina, A.; Hepojoki, J.; Raghwani, J.; Harlos, K.; Pybus, O.G.; Huiskonen, J.T.; Bowden, T.A. A Molecular-Level Account of the Antigenic Hantaviral Surface. Cell Rep. 2016, 15, 959–967. [Google Scholar] [CrossRef]
- Engdahl, T.B.; Binshtein, E.; Brocato, R.L.; Kuzmina, N.A.; Principe, L.M.; Kwilas, S.A.; Kim, R.K.; Chapman, N.S.; Porter, M.S.; Guardado-Calvo, P.; et al. Antigenic mapping and functional characterization of human New World hantavirus neutralizing antibodies. Elife 2023, 12, e81743. [Google Scholar] [CrossRef]
- Stass, R.; Engdahl, T.B.; Chapman, N.S.; Wolters, R.M.; Handal, L.S.; Diaz, S.M.; Crowe, J.E., Jr.; Bowden, T.A. Mechanistic basis for potent neutralization of Sin Nombre hantavirus by a human monoclonal antibody. Nat. Microbiol. 2023, 8, 1293–1303. [Google Scholar] [CrossRef]
- Levanov, L.; Iheozor-Ejiofor, R.P.; Lundkvist, Å.; Vapalahti, O.; Plyusnin, A. Defining of MAbs-neutralizing sites on the surface glycoproteins Gn and Gc of a hantavirus using vesicular stomatitis virus pseudotypes and site-directed mutagenesis. J. Gen. Virol. 2019, 100, 145–155. [Google Scholar] [CrossRef]
- Plyusnin, A.; Kedari, A.; Rissanen, I.; Iheozor-Ejiofor, R.P.; Lundkvist, Å.; Vapalahti, O.; Levanov, L. Validation of an antigenic site targeted by monoclonal antibodies against Puumala virus. J. Gen. Virol. 2023, 104, 001901. [Google Scholar] [CrossRef]
- Mittler, E.; Serris, A.; Esterman, E.S.; Florez, C.; Polanco, L.C.; O’Brien, C.M.; Slough, M.M.; Tynell, J.; Gröning, R.; Sun, Y.; et al. Structural and mechanistic basis of neutralization by a pan-hantavirus protective antibody. Sci. Transl. Med. 2023, 15, eadg1855. [Google Scholar] [CrossRef] [PubMed]
- Mittler, E.; Wec, A.Z.; Tynell, J.; Guardado-Calvo, P.; Wigren-Byström, J.; Polanco, L.C.; O’Brien, C.M.; Slough, M.M.; Abelson, D.M.; Serris, A.; et al. Human antibody recognizing a quaternary epitope in the Puumala virus glycoprotein provides broad protection against orthohantaviruses. Sci. Transl. Med. 2022, 14, eabl5399. [Google Scholar] [CrossRef]
- Rissanen, I.; Krumm, S.A.; Stass, R.; Whitaker, A.; Voss, J.E.; Bruce, E.A.; Rothenberger, S.; Kunz, S.; Burton, D.R.; Huiskonen, J.T.; et al. Structural Basis for a Neutralizing Antibody Response Elicited by a Recombinant Hantaan Virus Gn Immunogen. mBio 2021, 12, e0253120. [Google Scholar] [CrossRef]
- Lundkvist, A.; Kallio-Kokko, H.; Sjölander, K.B.; Lankinen, H.; Niklasson, B.; Vaheri, A.; Vapalahti, O. Characterization of Puumala virus nucleocapsid protein: Identification of B-cell epitopes and domains involved in protective immunity. Virology 1996, 216, 397–406. [Google Scholar] [CrossRef]
- Kalaiselvan, S.; Sankar, S.; Ramamurthy, M.; Ghosh, A.R.; Nandagopal, B.; Sridharan, G. Prediction of Pan-Specific B-Cell Epitopes from Nucleocapsid Protein of Hantaviruses Causing Hantavirus Cardiopulmonary Syndrome. J. Cell Biochem. 2017, 118, 2320–2324. [Google Scholar] [CrossRef] [PubMed]
- Duehr, J.; McMahon, M.; Williamson, B.; Amanat, F.; Durbin, A.; Hawman, D.W.; Noack, D.; Uhl, S.; Tan, G.S.; Feldmann, H.; et al. Neutralizing Monoclonal Antibodies against the Gn and the Gc of the Andes Virus Glycoprotein Spike Complex Protect from Virus Challenge in a Preclinical Hamster Model. mBio 2020, 11, e00028-20. [Google Scholar] [CrossRef]
- Garrido, J.L.; Prescott, J.; Calvo, M.; Bravo, F.; Alvarez, R.; Salas, A.; Riquelme, R.; Rioseco, M.L.; Williamson, B.N.; Haddock, E.; et al. Two recombinant human monoclonal antibodies that protect against lethal Andes hantavirus infection in vivo. Sci. Transl. Med. 2018, 10, eaat6420. [Google Scholar] [CrossRef] [PubMed]
- Schmaljohn, C.S.; Chu, Y.K.; Schmaljohn, A.L.; Dalrymple, J.M. Antigenic subunits of Hantaan virus expressed by baculovirus and vaccinia virus recombinants. J. Virol. 1990, 64, 3162–3170. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Chu, Y.K.; Schmaljohn, C. Bacterial expression of neutralizing mouse monoclonal antibody Fab fragments to Hantaan virus. Virology 1996, 217, 262–271. [Google Scholar] [CrossRef]
- Arikawa, J.; Yao, J.S.; Yoshimatsu, K.; Takashima, I.; Hashimoto, N. Protective role of antigenic sites on the envelope protein of Hantaan virus defined by monoclonal antibodies. Arch. Virol. 1992, 126, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wei, L.; Wang, L.; Wang, H.; Jiang, S. The in vitro and in vivo protective activity of monoclonal antibodies directed against Hantaan virus: Potential application for immunotherapy and passive immunization. Biochem. Biophys. Res. Commun. 2002, 298, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Vial, P.A.; Valdivieso, F.; Calvo, M.; Rioseco, M.L.; Riquelme, R.; Araneda, A.; Tomicic, V.; Graf, J.; Paredes, L.; Florenzano, M.; et al. A non-randomized multicentre trial of human immune plasma for treatment of hantavirus cardiopulmonary syndrome caused by Andes virus. Antivir. Ther. 2015, 20, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Engdahl, T.B.; Kuzmina, N.A.; Ronk, A.J.; Mire, C.E.; Hyde, M.A.; Kose, N.; Josleyn, M.D.; Sutton, R.E.; Mehta, A.; Wolters, R.M.; et al. Broad and potently neutralizing monoclonal antibodies isolated from human survivors of New World hantavirus infection. Cell Rep. 2021, 35, 109086. [Google Scholar] [CrossRef] [PubMed]
- Hörling, J.; Lundkvist, A.; Huggins, J.W.; Niklasson, B. Antibodies to Puumala virus in humans determined by neutralization test. J. Virol. Methods 1992, 39, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Iheozor-Ejiofor, R.; Vapalahti, K.; Sironen, T.; Levanov, L.; Hepojoki, J.; Lundkvist, Å.; Mäkelä, S.; Vaheri, A.; Mustonen, J.; Plyusnin, A.; et al. Neutralizing Antibody Titers in Hospitalized Patients with Acute Puumala Orthohantavirus Infection Do Not Associate with Disease Severity. Viruses 2022, 14, 901. [Google Scholar] [CrossRef]
- Valdivieso, F.; Vial, P.; Ferres, M.; Ye, C.; Goade, D.; Cuiza, A.; Hjelle, B. Neutralizing antibodies in survivors of Sin Nombre and Andes hantavirus infection. Emerg. Infect. Dis. 2006, 12, 166–168. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, M.; Nofchissey, R.; Goade, D.; Koster, F.; Hjelle, B. Humoral Immune Responses in the Hantavirus Cardiopulmonary Syndrome. J. Infect. Dis. 2000, 182, 43–48. [Google Scholar] [CrossRef]
- Pettersson, L.; Thunberg, T.; Rocklöv, J.; Klingström, J.; Evander, M.; Ahlm, C. Viral load and humoral immune response in association with disease severity in Puumala hantavirus-infected patients—Implications for treatment. Clin. Microbiol. Infect. 2014, 20, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Tuiskunen Bäck, A.; Rasmuson, J.; Thunberg, T.; Rankin, G.; Wigren Byström, J.; Andersson, C.; Sjödin, A.; Forsell, M.; Ahlm, C. Clinical and genomic characterisation of a fatal Puumala orthohantavirus case with low levels of neutralising antibodies. Infect. Dis. 2022, 54, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ma, H.; Shu, J.; Zhang, Q.; Han, M.; Liu, Z.; Jin, X.; Zhang, F.; Wu, X. Vaccines and Therapeutics against Hantaviruses. Front. Microbiol. 2019, 10, 2989. [Google Scholar] [CrossRef] [PubMed]
- Bertolotti-Ciarlet, A.; Smith, J.; Strecker, K.; Paragas, J.; Altamura, L.A.; McFalls, J.M.; Frias-Stäheli, N.; García-Sastre, A.; Schmaljohn, C.S.; Doms, R.W. Cellular localization and antigenic characterization of crimean-congo hemorrhagic fever virus glycoproteins. J. Virol. 2005, 79, 6152–6161. [Google Scholar] [CrossRef]
- Zivcec, M.; Guerrero, L.I.W.; Albariño, C.G.; Bergeron, É.; Nichol, S.T.; Spiropoulou, C.F. Identification of broadly neutralizing monoclonal antibodies against Crimean-Congo hemorrhagic fever virus. Antivir. Res. 2017, 146, 112–120. [Google Scholar] [CrossRef]
- Li, N.; Rao, G.; Li, Z.; Yin, J.; Chong, T.; Tian, K.; Fu, Y.; Cao, S. Cryo-EM structure of glycoprotein C from Crimean-Congo hemorrhagic fever virus. Virol. Sin. 2022, 37, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Simayi, A.; Wang, M.; Moming, A.; Xu, W.; Wang, C.; Li, Y.; Ding, J.; Deng, F.; Zhang, Y.; et al. Fine mapping epitope on glycoprotein Gc from Crimean-Congo hemorrhagic fever virus. Comp. Immunol. Microbiol. Infect. Dis. 2019, 67, 101371. [Google Scholar] [CrossRef]
- Durie, I.A.; Tehrani, Z.R.; Karaaslan, E.; Sorvillo, T.E.; McGuire, J.; Golden, J.W.; Welch, S.R.; Kainulainen, M.H.; Harmon, J.R.; Mousa, J.J.; et al. Structural characterization of protective non-neutralizing antibodies targeting Crimean-Congo hemorrhagic fever virus. Nat. Commun. 2022, 13, 7298. [Google Scholar] [CrossRef]
- Lasecka, L.; Bin-Tarif, A.; Bridgen, A.; Juleff, N.; Waters, R.A.; Baron, M.D. Antibodies to the core proteins of Nairobi sheep disease virus/Ganjam virus reveal details of the distribution of the proteins in infected cells and tissues. PLoS ONE 2015, 10, e0124966. [Google Scholar] [CrossRef] [PubMed]
- Lombe, B.P.; Saito, T.; Miyamoto, H.; Mori-Kajihara, A.; Kajihara, M.; Saijo, M.; Masumu, J.; Hattori, T.; Igarashi, M.; Takada, A. Mapping of Antibody Epitopes on the Crimean-Congo Hemorrhagic Fever Virus Nucleoprotein. Viruses 2022, 14, 544. [Google Scholar] [CrossRef] [PubMed]
- Burt, F.J.; Samudzi, R.R.; Randall, C.; Pieters, D.; Vermeulen, J.; Knox, C.M. Human defined antigenic region on the nucleoprotein of Crimean-Congo hemorrhagic fever virus identified using truncated proteins and a bioinformatics approach. J. Virol. Methods 2013, 193, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Golden, J.W.; Shoemaker, C.J.; Lindquist, M.E.; Zeng, X.; Daye, S.P.; Williams, J.A.; Liu, J.; Coffin, K.M.; Olschner, S.; Flusin, O.; et al. GP38-targeting monoclonal antibodies protect adult mice against lethal Crimean-Congo hemorrhagic fever virus infection. Sci. Adv. 2019, 5, eaaw9535. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Moyer, C.L.; Abelson, D.M.; Deer, D.J.; El Omari, K.; Duman, R.; Lobel, L.; Lutwama, J.J.; Dye, J.M.; Wagner, A.; et al. Structure and Characterization of Crimean-Congo Hemorrhagic Fever Virus GP38. J. Virol. 2020, 94, e02005-19. [Google Scholar] [CrossRef] [PubMed]
- Fels, J.M.; Maurer, D.P.; Herbert, A.S.; Wirchnianski, A.S.; Vergnolle, O.; Cross, R.W.; Abelson, D.M.; Moyer, C.L.; Mishra, A.K.; Aguilan, J.T.; et al. Protective neutralizing antibodies from human survivors of Crimean-Congo hemorrhagic fever. Cell 2021, 184, 3486–3501.e3421. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Hellert, J.; Freitas, N.; Guardado-Calvo, P.; Haouz, A.; Fels, J.M.; Maurer, D.P.; Abelson, D.M.; Bornholdt, Z.A.; Walker, L.M.; et al. Structural basis of synergistic neutralization of Crimean-Congo hemorrhagic fever virus by human antibodies. Science 2022, 375, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, A.J.; Swanepoel, R.; Leman, P.A. Antibody Response in Crimean-Congo Hemorrhagic Fever. Rev. Infect. Dis. 1989, 11 (Suppl. S4), S801–S806. [Google Scholar] [CrossRef] [PubMed]
- Ly, H. Differential Immune Responses to New World and Old World Mammalian Arenaviruses. Int. J. Mol. Sci. 2017, 18, 1040. [Google Scholar] [CrossRef] [PubMed]
- Borenstein-Katz, A.; Shulman, A.; Hamawi, H.; Leitner, O.; Diskin, R. Differential Antibody-Based Immune Response against Isolated GP1 Receptor-Binding Domains from Lassa and Junín Viruses. J. Virol. 2019, 93, e00090-19. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, Z.; Zhang, L.; Wang, S.; Xiao, G. Structure-function relationship of the mammarenavirus envelope glycoprotein. Virol. Sin. 2016, 31, 380–394. [Google Scholar] [CrossRef]
- Sommerstein, R.; Flatz, L.; Remy, M.M.; Malinge, P.; Magistrelli, G.; Fischer, N.; Sahin, M.; Bergthaler, A.; Igonet, S.; Ter Meulen, J.; et al. Arenavirus Glycan Shield Promotes Neutralizing Antibody Evasion and Protracted Infection. PLoS Pathog. 2015, 11, e1005276. [Google Scholar] [CrossRef]
- Cross, R.W.; Hastie, K.M.; Mire, C.E.; Robinson, J.E.; Geisbert, T.W.; Branco, L.M.; Ollmann Saphire, E.; Garry, R.F. Antibody therapy for Lassa fever. Curr. Opin. Virol. 2019, 37, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Buck, T.K.; Enriquez, A.S.; Schendel, S.L.; Zandonatti, M.A.; Harkins, S.S.; Li, H.; Moon-Walker, A.; Robinson, J.E.; Branco, L.M.; Garry, R.F.; et al. Neutralizing Antibodies against Lassa Virus Lineage I. mBio 2022, 13, e01278-22. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.E.; Hastie, K.M.; Cross, R.W.; Yenni, R.E.; Elliott, D.H.; Rouelle, J.A.; Kannadka, C.B.; Smira, A.A.; Garry, C.E.; Bradley, B.T.; et al. Most neutralizing human monoclonal antibodies target novel epitopes requiring both Lassa virus glycoprotein subunits. Nat. Commun. 2016, 7, 11544. [Google Scholar] [CrossRef] [PubMed]
- Perrett, H.R.; Brouwer, P.J.M.; Hurtado, J.; Newby, M.L.; Liu, L.; Müller-Kräuter, H.; Müller Aguirre, S.; Burger, J.A.; Bouhuijs, J.H.; Gibson, G.; et al. Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies. Cell Rep. 2023, 42, 112524. [Google Scholar] [CrossRef] [PubMed]
- Enriquez, A.S.; Buck, T.K.; Li, H.; Norris, M.J.; Moon-Walker, A.; Zandonatti, M.A.; Harkins, S.S.; Robinson, J.E.; Branco, L.M.; Garry, R.F.; et al. Delineating the mechanism of anti-Lassa virus GPC-A neutralizing antibodies. Cell Rep. 2022, 39, 110841. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Zandonatti, M.A.; Kleinfelter, L.M.; Heinrich, M.L.; Rowland, M.M.; Chandran, K.; Branco, L.M.; Robinson, J.E.; Garry, R.F.; Saphire, E.O. Structural basis for antibody-mediated neutralization of Lassa virus. Science 2017, 356, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Cross, R.W.; Harkins, S.S.; Zandonatti, M.A.; Koval, A.P.; Heinrich, M.L.; Rowland, M.M.; Robinson, J.E.; Geisbert, T.W.; Garry, R.F.; et al. Convergent Structures Illuminate Features for Germline Antibody Binding and Pan-Lassa Virus Neutralization. Cell 2019, 178, 1004–1015.e1014. [Google Scholar] [CrossRef]
- Mahmutovic, S.; Clark, L.; Levis, S.C.; Briggiler, A.M.; Enria, D.A.; Harrison, S.C.; Abraham, J. Molecular Basis for Antibody-Mediated Neutralization of New World Hemorrhagic Fever Mammarenaviruses. Cell Host Microbe 2015, 18, 705–713. [Google Scholar] [CrossRef]
- Pan, X.; Wu, Y.; Wang, W.; Zhang, L.; Xiao, G. Novel neutralizing monoclonal antibodies against Junin virus. Antivir. Res. 2018, 156, 21–28. [Google Scholar] [CrossRef]
- York, J.; Berry, J.D.; Ströher, U.; Li, Q.; Feldmann, H.; Lu, M.; Trahey, M.; Nunberg, J.H. An antibody directed against the fusion peptide of Junin virus envelope glycoprotein GPC inhibits pH-induced membrane fusion. J. Virol. 2010, 84, 6119–6129. [Google Scholar] [CrossRef]
- Amanat, F.; Duehr, J.; Huang, C.; Paessler, S.; Tan, G.S.; Krammer, F. Monoclonal Antibodies with Neutralizing Activity and Fc-Effector Functions against the Machupo Virus Glycoprotein. J. Virol. 2020, 94, e01741-19. [Google Scholar] [CrossRef] [PubMed]
- Oestereich, L.; Müller-Kräuter, H.; Pallasch, E.; Strecker, T. Passive Transfer of Animal-Derived Polyclonal Hyperimmune Antibodies Provides Protection of Mice from Lethal Lassa Virus Infection. Viruses 2023, 15, 1436. [Google Scholar] [CrossRef] [PubMed]
- Abreu-Mota, T.; Hagen, K.R.; Cooper, K.; Jahrling, P.B.; Tan, G.; Wirblich, C.; Johnson, R.F.; Schnell, M.J. Non-neutralizing antibodies elicited by recombinant Lassa-Rabies vaccine are critical for protection against Lassa fever. Nat. Commun. 2018, 9, 4223. [Google Scholar] [CrossRef] [PubMed]
- Ronk, A.J.; Lloyd, N.M.; Zhang, M.; Atyeo, C.; Perrett, H.R.; Mire, C.E.; Hastie, K.M.; Sanders, R.W.; Brouwer, P.J.M.; Saphire, E.O.; et al. A Lassa virus mRNA vaccine confers protection but does not require neutralizing antibody in a guinea pig model of infection. Nat. Commun. 2023, 14, 5603. [Google Scholar] [CrossRef]
- Battegay, M.; Moskophidis, D.; Waldner, H.; Bründler, M.A.; Fung-Leung, W.P.; Mak, T.W.; Hengartner, H.; Zinkernagel, R.M. Impairment and delay of neutralizing antiviral antibody responses by virus-specific cytotoxic T cells. J. Immunol. 1993, 151, 5408–5415. [Google Scholar] [CrossRef]
- Eschli, B.; Zellweger, R.M.; Wepf, A.; Lang, K.S.; Quirin, K.; Weber, J.; Zinkernagel, R.M.; Hengartner, H. Early antibodies specific for the neutralizing epitope on the receptor binding subunit of the lymphocytic choriomeningitis virus glycoprotein fail to neutralize the virus. J. Virol. 2007, 81, 11650–11657. [Google Scholar] [CrossRef] [PubMed]
- Bergthaler, A.; Flatz, L.; Verschoor, A.; Hegazy, A.N.; Holdener, M.; Fink, K.; Eschli, B.; Merkler, D.; Sommerstein, R.; Horvath, E.; et al. Impaired antibody response causes persistence of prototypic T cell-contained virus. PLoS Biol. 2009, 7, e1000080. [Google Scholar] [CrossRef]
- Buchmeier, M. Arenaviruses: Protein structure and function. Curr. Top. Microbiol. Immunol. 2002, 262, 159–173. [Google Scholar]
- McCormick, J.B.; King, I.J.; Webb, P.A.; Scribner, C.L.; Craven, R.B.; Johnson, K.M.; Elliott, L.H.; Belmont-Williams, R. Lassa Fever. N. Engl. J. Med. 1986, 314, 20–26. [Google Scholar] [CrossRef]
- Fisher-Hoch, S.P.; McCormick, J.B.; Auperin, D.; Brown, B.G.; Castor, M.; Perez, G.; Ruo, S.; Conaty, A.; Brammer, L.; Bauer, S. Protection of rhesus monkeys from fatal Lassa fever by vaccination with a recombinant vaccinia virus containing the Lassa virus glycoprotein gene. Proc. Natl. Acad. Sci. USA 1989, 86, 317–321. [Google Scholar] [CrossRef]
- Jahrling, P.B.; Peters, C.J. Passive antibody therapy of Lassa fever in cynomolgus monkeys: Importance of neutralizing antibody and Lassa virus strain. Infect. Immun. 1984, 44, 528–533. [Google Scholar] [CrossRef]
- Branco, L.M.; Grove, J.N.; Boisen, M.L.; Shaffer, J.G.; Goba, A.; Fullah, M.; Momoh, M.; Grant, D.S.; Garry, R.F. Emerging trends in Lassa fever: Redefining the role of immunoglobulin M and inflammation in diagnosing acute infection. Virol. J. 2011, 8, 1–15. [Google Scholar] [CrossRef]
- Mire, C.E.; Cross, R.W.; Geisbert, J.B.; Borisevich, V.; Agans, K.N.; Deer, D.J.; Heinrich, M.L.; Rowland, M.M.; Goba, A.; Momoh, M.; et al. Human-monoclonal-antibody therapy protects nonhuman primates against advanced Lassa fever. Nat. Med. 2017, 23, 1146–1149. [Google Scholar] [CrossRef]
- Grant, A.; Seregin, A.; Huang, C.; Kolokoltsova, O.; Brasier, A.; Peters, C.; Paessler, S. Junín Virus Pathogenesis and Virus Replication. Viruses 2012, 4, 2317–2339. [Google Scholar] [CrossRef]
- Enria, D.A.; Briggiler, A.M.; Sánchez, Z. Treatment of Argentine hemorrhagic fever. Antivir. Res. 2008, 78, 132–139. [Google Scholar] [CrossRef]
- Enria, D.; Fernandez, N.; Briggiler, A.; Levis, S.; Maiztegui, J. Importance of dose of neutralising antibodies in treatment of Argentine haemorrhagic fever with immune plasma. Lancet 1984, 324, 255–256. [Google Scholar] [CrossRef]
- Maiztegui, J.I.; McKee, K.T., Jr.; Oro, J.G.B.; Harrison, L.H.; Gibbs, P.H.; Feuillade, M.R.; Enria, D.A.; Briggiler, A.M.; Levis, S.C.; Ambrosio, A.M. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. J. Infect. Dis. 1998, 177, 277–283. [Google Scholar] [CrossRef]
- Zeitlin, L.; Cross, R.W.; Geisbert, J.B.; Borisevich, V.; Agans, K.N.; Prasad, A.N.; Enterlein, S.; Aman, M.J.; Bornholdt, Z.A.; Brennan, M.B.; et al. Therapy for Argentine hemorrhagic fever in nonhuman primates with a humanized monoclonal antibody. Proc. Natl. Acad. Sci. USA 2021, 118, e2023332118. [Google Scholar] [CrossRef]
- Saito, T.; Reyna, R.A.; Taniguchi, S.; Littlefield, K.; Paessler, S.; Maruyama, J. Vaccine Candidates against Arenavirus Infections. Vaccines 2023, 11, 635. [Google Scholar] [CrossRef]
- CEPI Awards Funding Agreement Worth Up to US$9.5 Million to Colorado State University to Develop a Human Vaccine against Rift Valley Fever. Available online: https://cepi.net/cepi-awards-funding-agreement-worth-us95-million-colorado-state-university-develop-human-vaccine (accessed on 31 January 2024).
- Stedman, A.; Wright, D.; Wichgers Schreur, P.J.; Clark, M.H.A.; Hill, A.V.S.; Gilbert, S.C.; Francis, M.J.; van Keulen, L.; Kortekaas, J.; Charleston, B.; et al. Safety and efficacy of ChAdOx1 RVF vaccine against Rift Valley fever in pregnant sheep and goats. NPJ Vaccines 2019, 4, 44. [Google Scholar] [CrossRef]
- Safety and Immunogenicity of a Candidate RVFV Vaccine (RVF001). Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04754776 (accessed on 31 January 2024).
- Song, J.Y.; Jeong, H.W.; Yun, J.W.; Lee, J.; Woo, H.J.; Bae, J.Y.; Park, M.S.; Choi, W.S.; Park, D.W.; Noh, J.Y.; et al. Immunogenicity and safety of a modified three-dose priming and booster schedule for the Hantaan virus vaccine (Hantavax): A multi-center phase III clinical trial in healthy adults. Vaccine 2020, 38, 8016–8023. [Google Scholar] [CrossRef]
- Song, J.Y.; Woo, H.J.; Cheong, H.J.; Noh, J.Y.; Baek, L.J.; Kim, W.J. Long-term immunogenicity and safety of inactivated Hantaan virus vaccine (Hantavax™) in healthy adults. Vaccine 2016, 34, 1289–1295. [Google Scholar] [CrossRef]
- Zeitlin, L.; Geisbert, J.B.; Deer, D.J.; Fenton, K.A.; Bohorov, O.; Bohorova, N.; Goodman, C.; Kim, D.; Hiatt, A.; Pauly, M.H.; et al. Monoclonal antibody therapy for Junin virus infection. Proc. Natl. Acad. Sci. USA 2016, 113, 4458–4463. [Google Scholar] [CrossRef]
- Chapman, N.S.; Hulswit, R.J.G.; Westover, J.L.B.; Stass, R.; Paesen, G.C.; Binshtein, E.; Reidy, J.X.; Engdahl, T.B.; Handal, L.S.; Flores, A.; et al. Multifunctional human monoclonal antibody combination mediates protection against Rift Valley fever virus at low doses. Nat. Commun. 2023, 14, 5650. [Google Scholar] [CrossRef]
- Hooper, J.W.; Brocato, R.L.; Kwilas, S.A.; Hammerbeck, C.D.; Josleyn, M.D.; Royals, M.; Ballantyne, J.; Wu, H.; Jiao, J.A.; Matsushita, H.; et al. DNA vaccine-derived human IgG produced in transchromosomal bovines protect in lethal models of hantavirus pulmonary syndrome. Sci. Transl. Med. 2014, 6, 264ra162. [Google Scholar] [CrossRef]
- Bryden, S.R.; Dunlop, J.I.; Clarke, A.T.; Fares, M.; Pingen, M.; Wu, Y.; Willett, B.J.; Patel, A.H.; Gao, G.F.; Kohl, A.; et al. Exploration of immunological responses underpinning severe fever with thrombocytopenia syndrome virus infection reveals IL-6 as a therapeutic target in an immunocompromised mouse model. PNAS Nexus 2022, 1, pgac024. [Google Scholar] [CrossRef]
- Lu, Y.; Friedman, R.; Kushner, N.; Doling, A.; Thomas, L.; Touzjian, N.; Starnbach, M.; Lieberman, J. Genetically modified anthrax lethal toxin safely delivers whole HIV protein antigens into the cytosol to induce T cell immunity. Proc. Natl. Acad. Sci. USA 2000, 97, 8027–8032. [Google Scholar] [CrossRef]
- Doling, A.M.; Ballard, J.D.; Shen, H.; Krishna, K.M.; Ahmed, R.; Collier, R.J.; Starnbach, M.N. Cytotoxic T-lymphocyte epitopes fused to anthrax toxin induce protective antiviral immunity. Infect. Immun. 1999, 67, 3290–3296. [Google Scholar] [CrossRef]
- Friebe, S.; van der Goot, F.G.; Bürgi, J. The Ins and Outs of Anthrax Toxin. Toxins 2016, 8, 69. [Google Scholar] [CrossRef]
- Akanmu, S.; Herrera, B.B.; Chaplin, B.; Ogunsola, S.; Osibogun, A.; Onawoga, F.; John-Olabode, S.; Akase, I.E.; Nwosu, A.; Hamel, D.J.; et al. High SARS-CoV-2 seroprevalence in Lagos, Nigeria with robust antibody and cellular immune responses. J. Clin. Virol. Plus 2023, 3, 100156. [Google Scholar] [CrossRef]
- Herrera, B.B.; Hamel, D.J.; Oshun, P.; Akinsola, R.; Akanmu, A.S.; Chang, C.A.; Eromon, P.; Folarin, O.; Adeyemi, K.T.; Happi, C.T.; et al. A modified anthrax toxin-based enzyme-linked immunospot assay reveals robust T cell responses in symptomatic and asymptomatic Ebola virus exposed individuals. PLoS Negl. Trop. Dis. 2018, 12, e0006530. [Google Scholar] [CrossRef]
- Herrera, B.B.; Tsai, W.Y.; Chang, C.A.; Hamel, D.J.; Wang, W.K.; Lu, Y.; Mboup, S.; Kanki, P.J. Sustained Specific and Cross-Reactive T Cell Responses to Zika and Dengue Virus NS3 in West Africa. J. Virol. 2018, 92, e01992-17. [Google Scholar] [CrossRef]
- Yadav, P.D.; Chaubal, G.Y.; Shete, A.M.; Mourya, D.T. A mini-review of Bunyaviruses recorded in India. Indian. J. Med. Res. 2017, 145, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Gaillet, M.; Pichard, C.; Restrepo, J.; Lavergne, A.; Perez, L.; Enfissi, A.; Abboud, P.; Lambert, Y.; Ma, L.; Monot, M.; et al. Outbreak of Oropouche Virus in French Guiana. Emerg. Infect. Dis. 2021, 27, 2711–2714. [Google Scholar] [CrossRef]
- Yoshimatsu, K.; Arikawa, J. Bunyavirus and its ecology. Uirusu 2012, 62, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.; Kortekaas, J.; Bowden, T.A.; Warimwe, G.M. Rift Valley fever: Biology and epidemiology. J. Gen. Virol. 2019, 100, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Elgh, F.; Linderholm, M.; Wadell, G.; Tärnvik, A.; Juto, P. Development of humoral cross-reactivity to the nucleocapsid protein of heterologous hantaviruses in nephropathia epidemica. FEMS Immunol. Med. Microbiol. 1998, 22, 309–315. [Google Scholar] [CrossRef]
- Avižinienė, A.; Kučinskaitė-Kodzė, I.; Petraitytė-Burneikienė, R.; Žvirblienė, A.; Mertens, M.L.; Schmidt, S.; Schlegel, M.; Lattwein, E.; Koellner, B.; Ulrich, R.G. Characterization of a Panel of Cross-Reactive Hantavirus Nucleocapsid Protein-Specific Monoclonal Antibodies. Viruses 2023, 15, 532. [Google Scholar] [CrossRef]
- Kalkan-Yazıcı, M.; Karaaslan, E.; Çetin, N.S.; Hasanoğlu, S.; Güney, F.; Zeybek, Ü.; Doymaz, M.Z. Cross-Reactive anti-Nucleocapsid Protein Immunity against Crimean-Congo Hemorrhagic Fever Virus and Hazara Virus in Multiple Species. J. Virol. 2021, 95, e02156-20. [Google Scholar] [CrossRef]
- Sanchez, A.; Pifat, D.Y.; Kenyon, R.H.; Peters, C.J.; McCormick, J.B.; Kiley, M.P. Junin Virus Monoclonal Antibodies: Characterization and Cross-reactivity with Other Arenaviruses. J. Gen. Virol. 1989, 70, 1125–1132. [Google Scholar] [CrossRef]
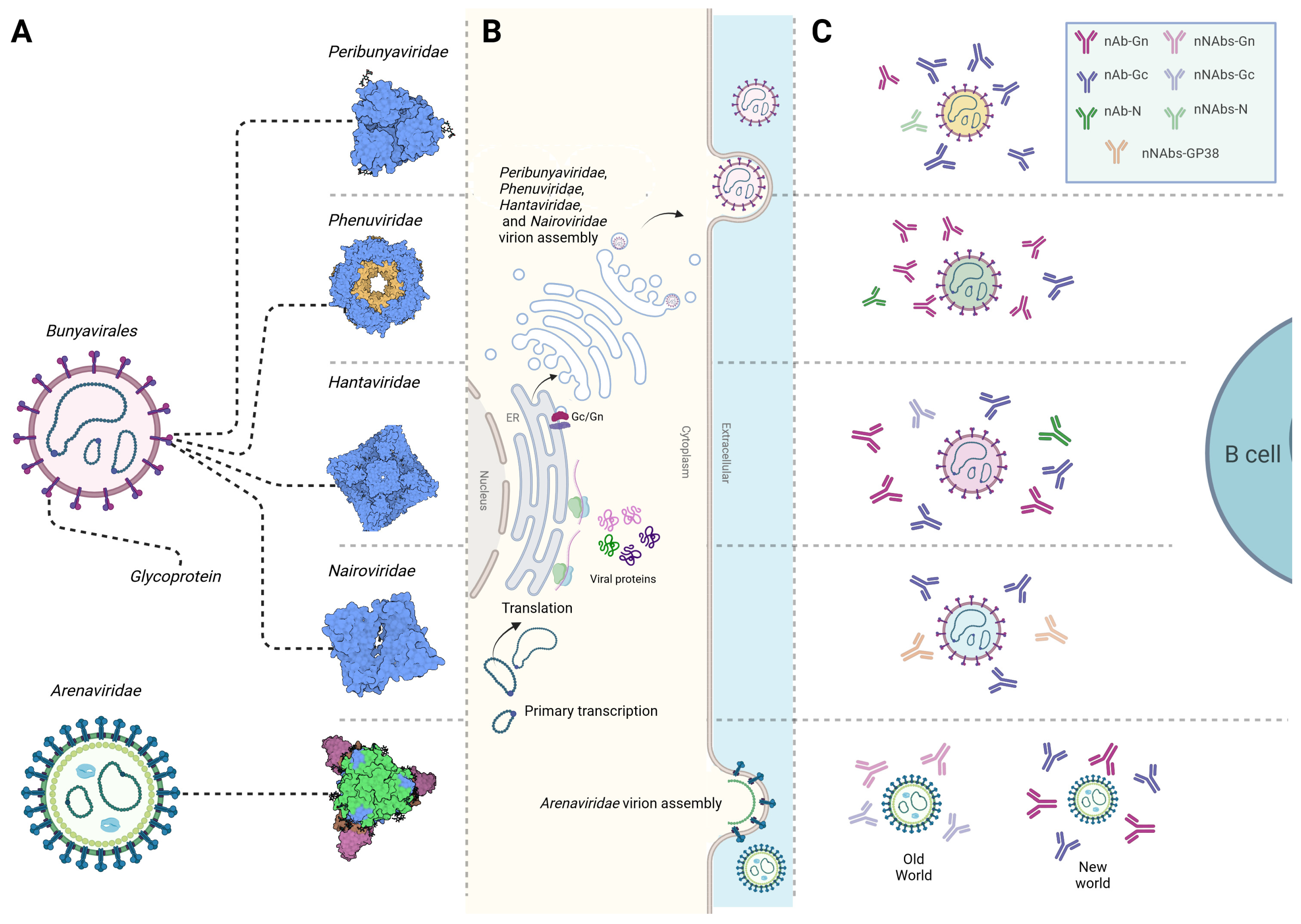
| Virus | T Cell Type | Epitope | Host | Approach |
|---|---|---|---|---|
| Peribunyaviridae | ||||
| OROV | CD8+ | Glycoproteins TSSWGCEEY (1043–1051) CSMCGLIHY (48–56) LAIDTGCLY (4–12) | Humans | Immunoinformatics [30] |
| BUNV | CD8+/CD4+ | N protein KRSEWEVTL (55–63) AIGIYKVQRKEMEPK (161–75) | Humans | Immunoinformatics [32] |
| Glycoproteins YQPTELTRS (716–724) YKAHDKEET (782–790) ILGTGTPKF (1172–1180) | Immunoinformatics [33] | |||
| JCV | CD8+ | N protein AAKAKAALA (26–34) AALARKPER (152–161) ADHGESVSL (175–183) ADHGESVSLS (157–165) YPLTIGIYRV (108–117) | Humans | Immunoinformatics [31] |
| CD4+ | N protein AALARKPER (146–154) ADHGESVSL (160–169) DVEQLKWGR (119–127) EIYLSFFPG (183–191) FLIKFGVKL (141–149) | |||
| SBV | CD8+ | N protein Glycoprotein Gc (678–947) | IFNAR−/− mice | Ubiquitinated and non-ubiquitinated cDNA immunization [34] |
| LACV | CD4+ | Glycoprotein N protein | IFNAR−/− mice | DNA vaccination [35] |
| SBV | CD8+ | N protein | IFNAR−/− mice | Bacterially expressed (SBV-N) [36] |
| Phenuiviridae | ||||
| TOSV | CD4+ | N protein VKMMIVLNL (58–66) Glycoprotein VMILGLLSS (824–832) | Humans | Immunoinformatics [37] |
| SFTSV | CD8+/CD4+ | Panel of peptides 8 peptides within RdRp 8 peptides within glycoprotein | Humans | Immunoinformatics [38] |
| RVFV | CD4+ | Glycoprotein LPALAVFALAPVVFA (139–153) PALAVFALAPVVFAE (140–154) GIAMTVLPALAVFAL (133–147) GSWNFFDWFSGLMSW (1138–1152) FFLLIYLGRTGLSKM (1174–1188) N protein HMMHPSFAGMVDPSL (143–158) | Humans | Immunoinformatics [39] |
| CD8+ | Glycoprotein AVFALAPVV (143–151) LAVFALAPV (142–150) FALAPVVFA (145–153) VFALAPVVF (144–152) IAMTVLPAL (134–142) FFDWFSGLM (1142–1150) FLLIYLGRT (1142–1150) N protein MMHPSFAGM (144–152) | |||
| CD8+/CD4+ | Panel of peptides 14 peptides within N 13 peptides within Gn 16 peptides within Gc | Humans | Ex vivo stimulation assays/immunoinformatics [40] | |
| CD8+ | N protein VLSEWLPVT (121–129) ILDAHSLYL (165–173) | Humans | Ex vivo assays using N-transduced dendritic cells primed with CD8 T cells from HLA-A2 donors [41] | |
| CD8+ | N protein NAAVNSNFI (201–210) | Mice C57BL/6 | Ex vivo stimulation assay of vaccinated mice [42] | |
| CD4+ | N protein VREFAYQGFDARRVI (25–40) AYQGFDARRVIELLK (29–44) | |||
| Hantaviridae | ||||
| Orthohantaviruses (multiple) | CD8+/CD4+ | A panel of cross-reactive epitopes between multiple orthohantaviruses 6 peptides within glycoprotein 2 peptides within nucleocapsid 2 peptides within RdRp 1 peptide within NS protein | Humans | Immunoinformatics [43] |
| HTNV | CD8+ | N protein NAHEGQLVI (12–20) ISNQEPLKL (421–429) | Humans | Ex vivo stimulation [44] |
| N protein TSFVVPILLKALYML (127–141) YMLTTRGRQTTKDNK (139–153) IEPCKLLPDTAAVSL (241–255) LRKKSSFYQSYLRRT (355–369) | Humans | Ex vivo stimulation [45] | ||
| N protein RYRTAVCGL (197–205) KLLPDTAAV (245–253) GPATNRDYL (258–266) | Humans | Ex vivo stimulation [46] | ||
| HTNV/SNV | CD8+ | ILQDMRNTI (HTNV, aa 334–342; SNV, aa 333–341) | Humans | In silico prediction of conserved epitopes, validation of peptides ex vivo using patients’ PBMCs [47] |
| CD8+/CD4+ | N protein ERIDDFLAA (234–242) LPIILKALY (131–139) GIQLDQKIII (372–380) | Humans | Ex vivo stimulation assays [48] | |
| HTNV | Glycoprotein LIWTGMIDL (358–366) | Humans | In silico prediction and evaluation of efficacy in transgenic mice [49] | |
| ANDV | CD8+ | Glycoprotein Gn SLFSLMPDVAHSLAV (461–475) | Humans | Ex vivo stimulation [50] |
| PUUV | CD8+ | Glycoprotein Gn HWMDATFNL (731–739) | Humans | Ex vivo stimulation [51] |
| Nairoviridae | ||||
| CCHFV | CD8+/CD4+ | Panel of peptides 3 peptides within Gc 2 peptides within Gn | Humans | Immunoinformatics [52] |
| A panel of peptides 5 peptides within N protein 4 peptides within Glycoprotein | Humans | Immunoinformatics [53] | ||
| RdRp DCSSTPPDR (197–202) | Humans | Immunoinformatic [54] | ||
| CD8+ | A panel of peptides 4 peptides within NSm 5 peptides within GP38 | Humans | Ex vivo stimulation [55] | |
| N protein Gc | Humans | Ex vivo stimulation [56] | ||
| Gc NSm | C57BL/6 mice | Ex vivo stimulation [57] | ||
| Arenaviridae | ||||
| LASV | CD8+ | Glycoprotein MRMAWGGSY (192–200) N protein ALTDLGLIY (201–210) | Humans | Immunoinformatics [58] |
| CD4+ | A panel of peptides 4 peptides within glycoprotein 4 peptides within N protein | Humans | Immunoinformatics [58] | |
| LASV | CD8+ | A panel of peptides 12 epitopes within glycoprotein and/or N | Humans | Ex vivo stimulation [59] |
| N (1–91) N (411–491) GP2 (92–172) | Humans | Ex vivo stimulation [60] | ||
| LASV | CD4+ | 4 highly conserved peptides between Old and New World arenaviruses within GP2 (289–301) | Humans | Ex vivo stimulation [61] |
| 6 peptides within N protein | Humans | Ex vivo stimulation [62] | ||
| LASV/cross-reacts with LCMV | CD4+ | Glycoprotein IEQQADNMITEMLQK (403–417) | C3H/HeJ mice | Ex vivo stimulation [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alatrash, R.; Herrera, B.B. The Adaptive Immune Response against Bunyavirales. Viruses 2024, 16, 483. https://doi.org/10.3390/v16030483
Alatrash R, Herrera BB. The Adaptive Immune Response against Bunyavirales. Viruses. 2024; 16(3):483. https://doi.org/10.3390/v16030483
Chicago/Turabian StyleAlatrash, Reem, and Bobby Brooke Herrera. 2024. "The Adaptive Immune Response against Bunyavirales" Viruses 16, no. 3: 483. https://doi.org/10.3390/v16030483





