Development of a Mouse-Adapted Reporter SARS-CoV-2 as a Tool for Two-Photon In Vivo Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Viruses
2.3. Mouse Adaptation of SARS-CoV-2
2.4. Generation of Mouse-Adapted Reporter SARS-CoV-2
2.5. Experimental Infection of Mice
2.6. Pathologic Examination
2.7. In Vivo Imaging of Mouse Lung
2.8. Virus Titration
3. Results
3.1. Characterization of a Mouse-Adapted Strain of SARS-CoV-2 in BALB/c Mice
3.2. Characterization of a Mouse-Adapted Strain of SARS-CoV-2 in C57BL/6J Mice
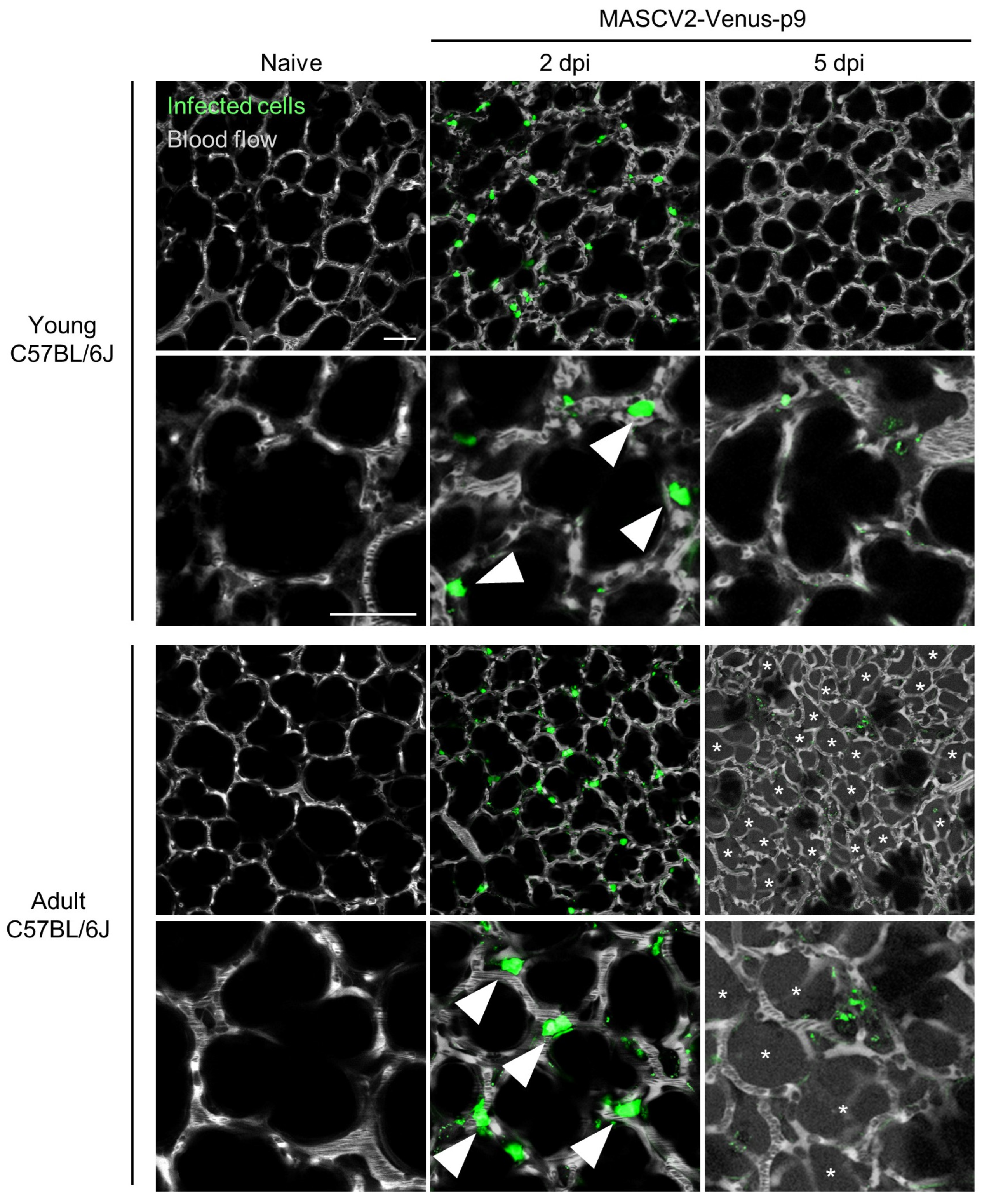
3.3. Characterization of a Mouse-Adapted Reporter SARS-CoV-2 in C57BL/6J Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Blish, C.A.; Sallusto, F.; Iwasaki, A. The immunology and immunopathology of COVID-19. Science 2022, 375, 1122–1127. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R.; Mogensen, T.H. Innate immunological pathways in COVID-19 pathogenesis. Sci. Immunol. 2022, 7, eabm5505. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Iida, S.; Iwatsuki-Horimoto, K.; Maemura, T.; Kiso, M.; Scheaffer, S.M.; Darling, T.L.; Joshi, A.; Loeber, S.; Singh, G.; et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 2022, 603, 687–692. [Google Scholar] [CrossRef]
- Ullah, I.; Prévost, J.; Ladinsky, M.S.; Stone, H.; Lu, M.; Anand, S.P.; Beaudoin-Bussières, G.; Symmes, K.; Benlarbi, M.; Ding, S.; et al. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity 2021, 54, 2143–2158.e15. [Google Scholar] [CrossRef]
- Pittet, M.J.; Weissleder, R. Intravital imaging. Cell 2011, 147, 983–991. [Google Scholar] [CrossRef]
- Choi, M.; Kwok, S.J.; Yun, S.H. In vivo fluorescence microscopy: Lessons from observing cell behavior in their native environment. Physiology 2015, 30, 40–49. [Google Scholar] [CrossRef]
- Ueki, H.; Wang, I.H.; Zhao, D.; Gunzer, M.; Kawaoka, Y. Multicolor two-photon imaging of in vivo cellular pathophysiology upon influenza virus infection using the two-photon IMPRESS. Nat. Protoc. 2020, 15, 1041–1065. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, N.; Brown, S. The mouse ascending: Perspectives for human-disease models. Nat. Cell Biol. 2007, 9, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- McCray, P.B., Jr.; Pewe, L.; Wohlford-Lenane, C.; Hickey, M.; Manzel, L.; Shi, L.; Netland, J.; Jia, H.P.; Halabi, C.; Sigmund, C.D.; et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007, 81, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Winkler, E.S.; Bailey, A.L.; Kafai, N.M.; Nair, S.; McCune, B.T.; Yu, J.; Fox, J.M.; Chen, R.E.; Earnest, J.T.; Keeler, S.P.; et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020, 21, 1327–1335. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.; Yuen, K.Y. Animal models in SARS-CoV-2 research. Nat. Methods 2022, 19, 392–394. [Google Scholar] [CrossRef] [PubMed]
- Leist, S.R.; Schäfer, A.; Martinez, D.R. Cell and animal models of SARS-CoV-2 pathogenesis and immunity. Dis. Model Mech. 2020, 13, dmm046581. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.-M.; Chin, A.W.H.; Fung, K.; Choy, K.-T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.A.P.M.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef]
- Lakdawala, S.S.; Menachery, V.D. The search for a COVID-19 animal model. Science 2020, 368, 942–943. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Iwatsuki-Horimoto, K.; Hatta, M.; Loeber, S.; Halfmann, P.J.; Nakajima, N.; Watanabe, T.; Ujie, M.; Takahashi, K.; Ito, M.; et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA 2020, 117, 16587–16595. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Justice, M.J.; Dhillon, P. Using the mouse to model human disease: Increasing validity and reproducibility. Dis. Model Mech. 2016, 9, 101–103. [Google Scholar] [CrossRef] [PubMed]
- Leist, S.R.; Dinnon, K.H., 3rd; Schäfer, A.; Tse, L.V.; Okuda, K.; Hou, Y.J.; West, A.; Edwards, C.E.; Sanders, W.; Fritch, E.J.; et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 2020, 183, 1070–1085.e1012. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Li, E.; Wang, T.; Li, Y.; Liu, J.; Wang, W.; Qin, T.; Su, R.; Pei, H.; Wang, S.; et al. Characterization of Two Heterogeneous Lethal Mouse-Adapted SARS-CoV-2 Variants Recapitulating Representative Aspects of Human COVID-19. Front. Immunol. 2022, 13, 821664. [Google Scholar] [CrossRef] [PubMed]
- Thieulent, C.J.; Dittmar, W.; Balasuriya, U.B.R.; Crossland, N.A.; Wen, X.; Richt, J.A.; Carossino, M. Mouse-Adapted SARS-CoV-2 MA10 Strain Displays Differential Pulmonary Tropism and Accelerated Viral Replication, Neurodissemination, and Pulmonary Host Responses in K18-hACE2 Mice. mSphere 2023, 8, e00558-22. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- DuBridge, R.B.; Tang, P.; Hsia, H.C.; Leong, P.-M.; Miller, J.H.; Calos, M.P. Analysis of Mutation in Human Cells by Using an Epstein-Barr Virus Shuttle System. Mol. Cell. Biol. 1987, 7, 379–387. [Google Scholar] [CrossRef]
- Imai, M.; Halfmann, P.J.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Chiba, S.; Watanabe, T.; Nakajima, N.; Ito, M.; Kuroda, M.; Kiso, M.; et al. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. Proc. Natl. Acad. Sci. USA 2021, 118, e2106535118. [Google Scholar] [CrossRef]
- Fukuyama, S.; Katsura, H.; Zhao, D.; Ozawa, M.; Ando, T.; Shoemaker, J.E.; Ishikawa, I.; Yamada, S.; Neumann, G.; Watanabe, S.; et al. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nat. Commun. 2015, 6, 6600. [Google Scholar] [CrossRef]
- Furusawa, Y.; Kiso, M.; Iida, S.; Uraki, R.; Hirata, Y.; Imai, M.; Suzuki, T.; Yamayoshi, S.; Kawaoka, Y. In SARS-CoV-2 delta variants, Spike-P681R and D950N promote membrane fusion, Spike-P681R enhances spike cleavage, but neither substitution affects pathogenicity in hamsters. EBioMedicine 2023, 91, 104561. [Google Scholar] [CrossRef]
- Ye, C.; Chiem, K.; Park, J.G.; Silvas, J.A.; Morales Vasquez, D.; Sourimant, J.; Lin, M.J.; Greninger, A.L.; Plemper, R.K.; Torrelles, J.B.; et al. Analysis of SARS-CoV-2 infection dynamic in vivo using reporter-expressing viruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2111593118. [Google Scholar] [CrossRef]
- Ueki, H.; Wang, I.H.; Fukuyama, S.; Katsura, H.; da Silva Lopes, T.J.; Neumann, G.; Kawaoka, Y. In vivo imaging of the pathophysiological changes and neutrophil dynamics in influenza virus-infected mouse lungs. Proc. Natl. Acad. Sci. USA 2018, 115, E6622–E6629. [Google Scholar] [CrossRef]
- Shou, S.; Liu, M.; Yang, Y.; Kang, N.; Song, Y.; Tan, D.; Liu, N.; Wang, F.; Liu, J.; Xie, Y. Animal Models for COVID-19: Hamsters, Mouse, Ferret, Mink, Tree Shrew, and Non-human Primates. Front. Microbiol. 2021, 12, 626553. [Google Scholar] [CrossRef]
- Huang, K.; Zhang, Y.; Hui, X.; Zhao, Y.; Gong, W.; Wang, T.; Zhang, S.; Yang, Y.; Deng, F.; Zhang, Q.; et al. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine 2021, 67, 103381. [Google Scholar] [CrossRef]
- Zúñiga, S.; Sola, I.; Alonso, S.; Enjuanes, L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004, 78, 980–994. [Google Scholar] [CrossRef]
- Gu, H.; Chen, Q.; Yang, G.; He, L.; Fan, H.; Deng, Y.Q.; Wang, Y.; Teng, Y.; Zhao, Z.; Cui, Y.; et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020, 369, 1603–1607. [Google Scholar] [CrossRef]
- Sun, S.; Gu, H.; Cao, L.; Chen, Q.; Ye, Q.; Yang, G.; Li, R.-T.; Fan, H.; Deng, Y.-Q.; Song, X.; et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat. Commun. 2021, 12, 5654. [Google Scholar] [CrossRef]
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216. [Google Scholar] [CrossRef]



| Viral Strain | Production Process |
|---|---|
| MASCV2-p9 | MASCV2-p9 was derived from TY7-501 by sequential lung-to-lung passages in BALB/c mice (9 passages). |
| MASCV2-p25 | MASCV2-p25 was derived from TY7-501 by sequential lung-to-lung passages in BALB/c mice (10 passages) followed by sequential lung-to-lung passages in C57BL/6J mice (15 passages). |
| MASCV2-Venus | MASCV2-Venus was generated by inserting the fluorescent gene Venus into the genome of MASCV2-p25 by use of reverse genetics. |
| MASCV2-Venus-p9 | MASCV2-Venus-p9 was derived from MASCV2-Venus by sequential lung-to-lung passages in C57BL/6J mice (9 passages). |
| MASCV2-p9 | Amino acid substitution | - | - | - | NSP4 A307V | NSP5 F294L | - | - | S Q493R | - | - | - | ORF7a T14I | - |
| Gene mutation | - | - | - | c9474t | t10934c | - | - | a23040g | - | - | - | c27434t | - | |
| MASCV2-p25 | Amino acid substitution | - | - | NSP4 T295I | - | NSP5 F294L | NSP9 R39K | - | S Q493K | - | - | M T7I | - | - |
| Gene mutation | t2365c | - | c9438t | - | t10934c | g12801a | - | c23039a | t24706c | - | c26542t | - | - | |
| MASCV2-Venus | Amino acid substitution | - | - | NSP4 T295I | - | NSP5 F294L | NSP9 R39K | - | S Q493K | - | - | M T7I | - | - |
| Gene mutation | - | - | c9438t | - | t10934c | g12801a | - | c23039a | - | - | c26542t | - | g28262a | |
| MASCV2-Venus-p9 | Amino acid substitution | - | NSP4 V233l | NSP4 T295I | - | NSP5 F294L | NSP9 R39K | NSP13 L83F | S Q493K | - | - | M T7I | - | - |
| Gene mutation | - | g9251a | c9438t | - | t10934c | g12801a | g16476t | c23039a | - | c24904t | c26542t | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueki, H.; Kiso, M.; Furusawa, Y.; Iida, S.; Yamayoshi, S.; Nakajima, N.; Imai, M.; Suzuki, T.; Kawaoka, Y. Development of a Mouse-Adapted Reporter SARS-CoV-2 as a Tool for Two-Photon In Vivo Imaging. Viruses 2024, 16, 537. https://doi.org/10.3390/v16040537
Ueki H, Kiso M, Furusawa Y, Iida S, Yamayoshi S, Nakajima N, Imai M, Suzuki T, Kawaoka Y. Development of a Mouse-Adapted Reporter SARS-CoV-2 as a Tool for Two-Photon In Vivo Imaging. Viruses. 2024; 16(4):537. https://doi.org/10.3390/v16040537
Chicago/Turabian StyleUeki, Hiroshi, Maki Kiso, Yuri Furusawa, Shun Iida, Seiya Yamayoshi, Noriko Nakajima, Masaki Imai, Tadaki Suzuki, and Yoshihiro Kawaoka. 2024. "Development of a Mouse-Adapted Reporter SARS-CoV-2 as a Tool for Two-Photon In Vivo Imaging" Viruses 16, no. 4: 537. https://doi.org/10.3390/v16040537





