Transcriptomic Analysis of PDCoV-Infected HIEC-6 Cells and Enrichment Pathways PI3K-Akt and P38 MAPK
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Antibodies
2.2. Virus Culture
2.3. Drug Exposure
2.4. One-Step Growth Curve
2.5. Western Blotting
2.6. Transmission Electron Microscopy of PDCoV Virus Particles
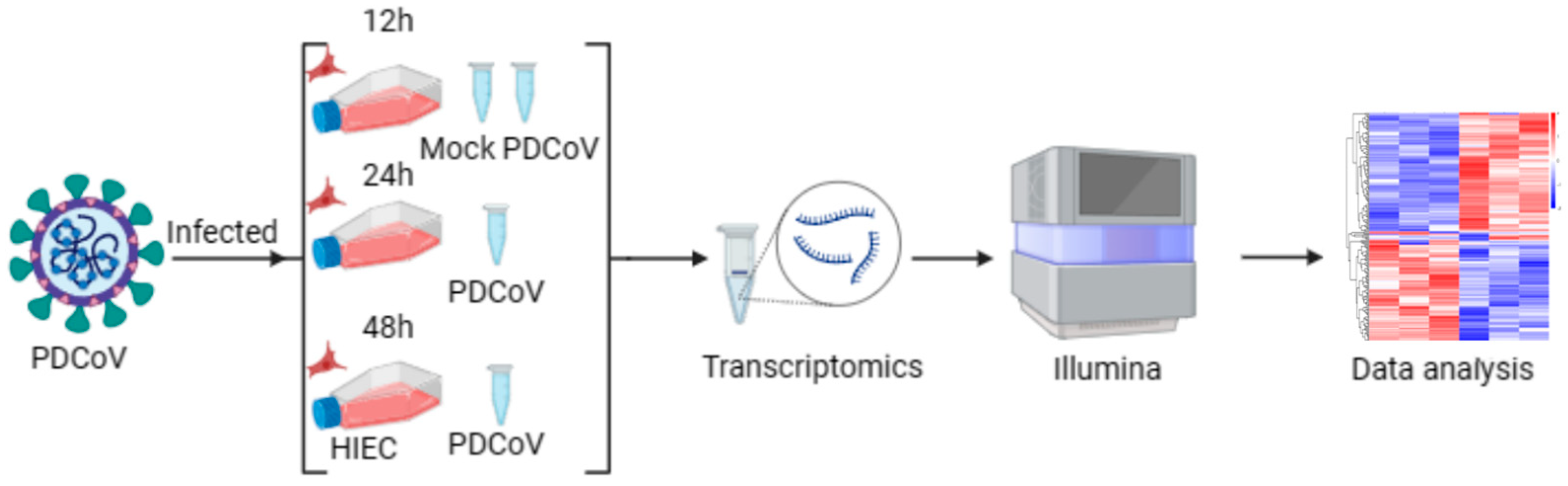
2.7. Sample Preparation and RNA Extraction
2.8. RNA-Seq Analysis
2.9. Differential Gene Analysis
2.10. GO and KEGG Enrichment Analysis
2.11. RT-qPCR Validation
2.12. Statistical Analysis
3. Results
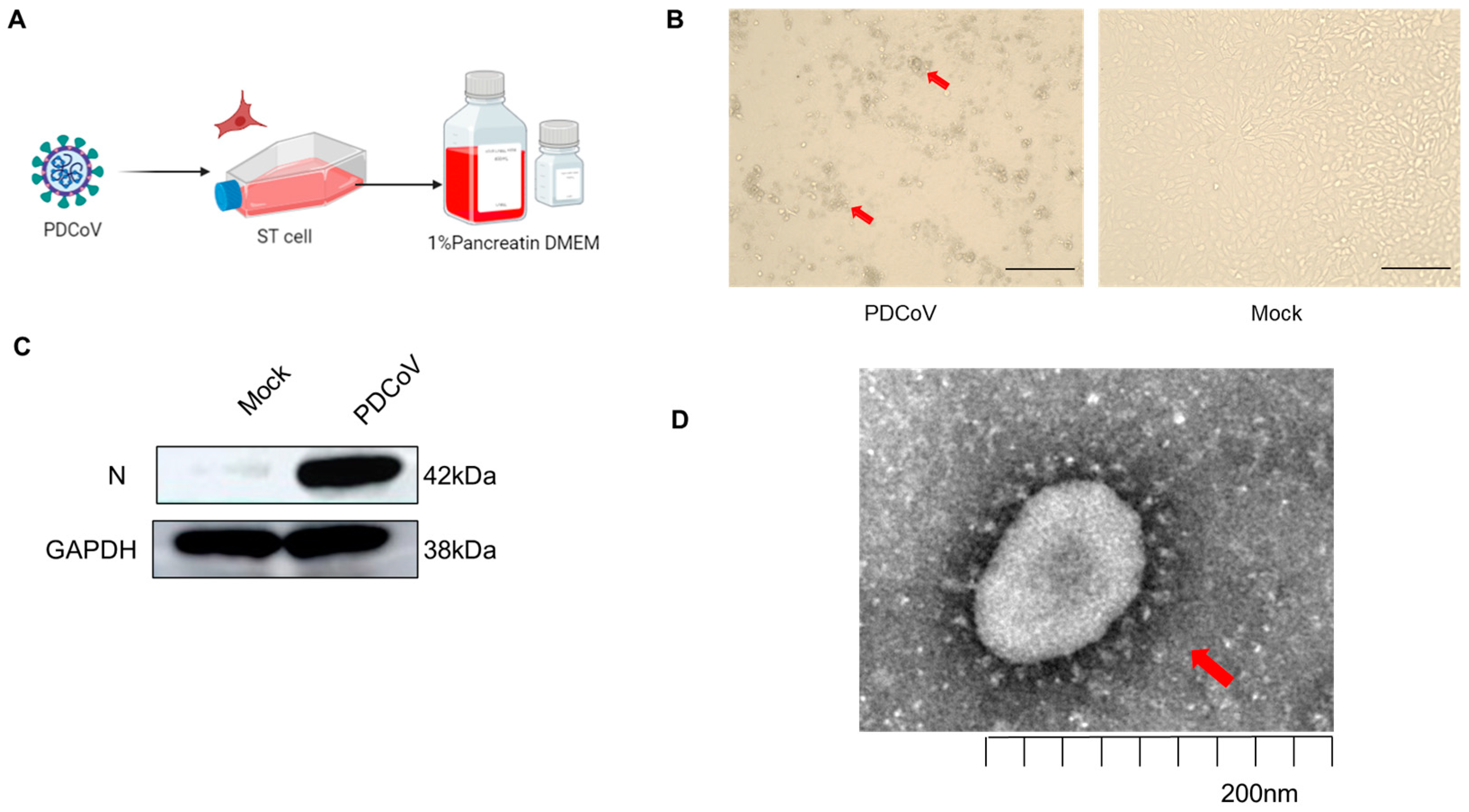
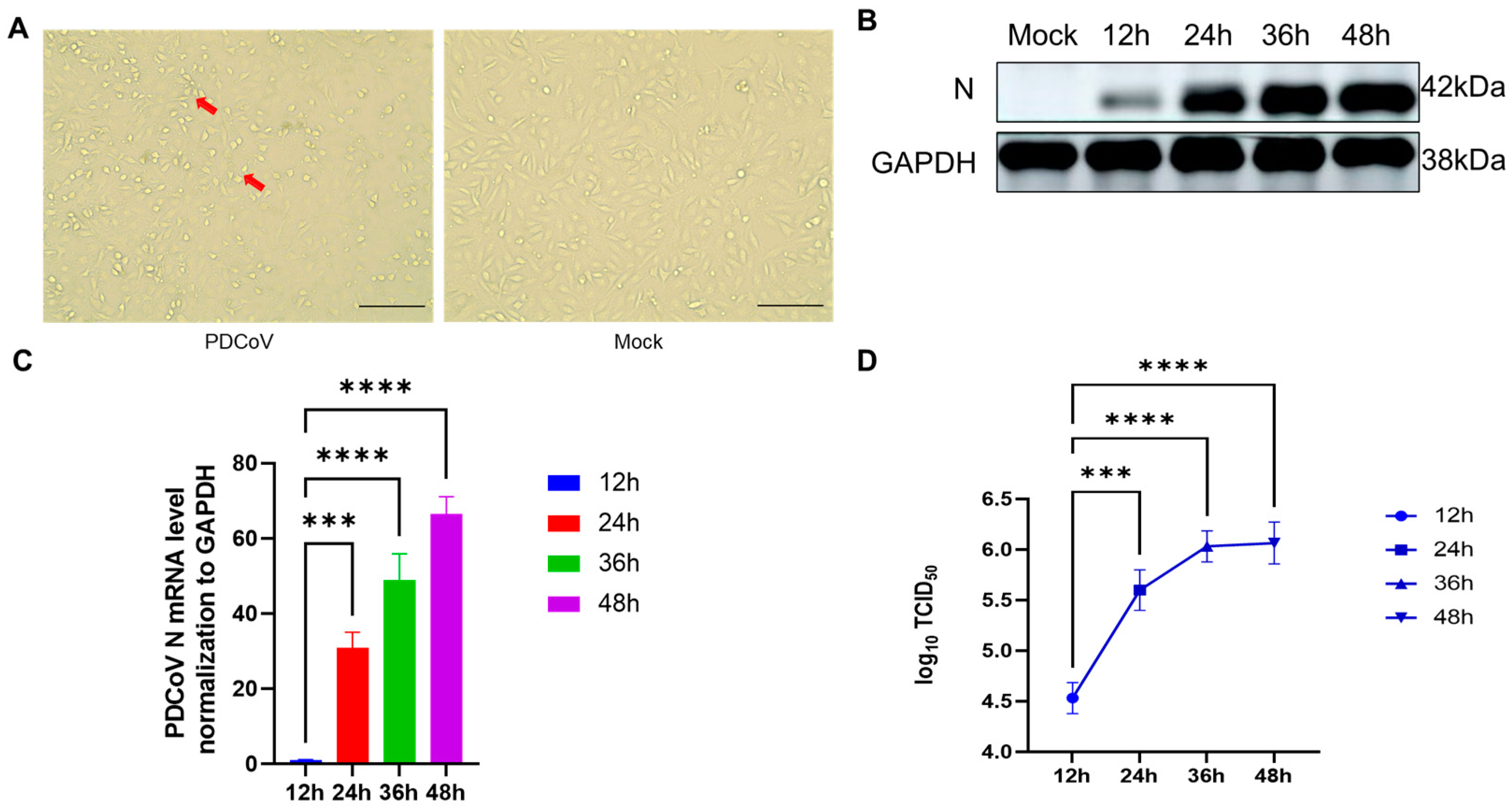
3.1. Validation of PDCoV Virus Infection of Different Cells
3.2. Evaluation of Transcriptome Sequencing Data
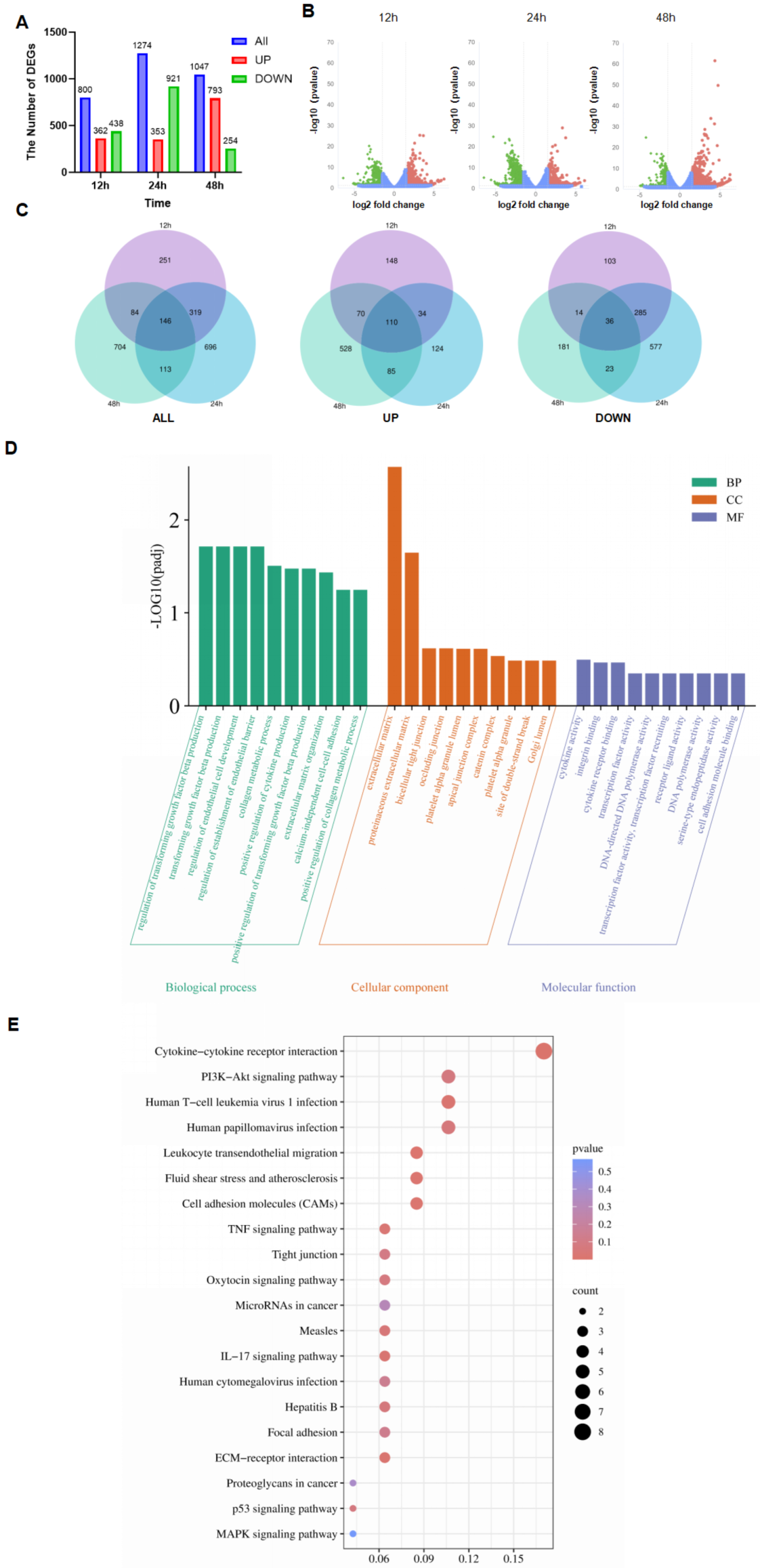
3.3. Transcriptome of PDCoV-Infected HIEC-6
3.4. GO and KEGG Analyses of Shared DEGs
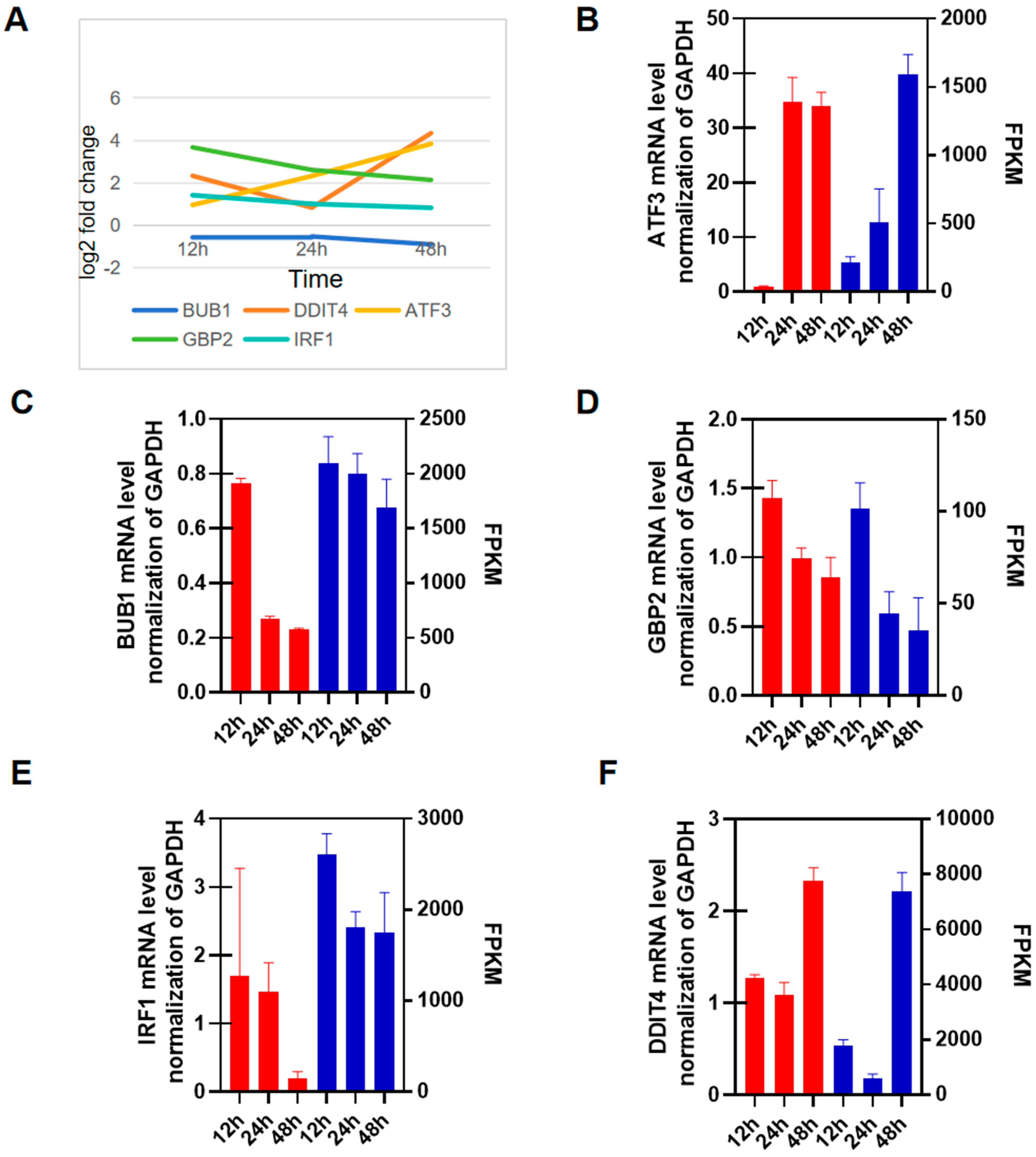
3.5. RT-qPCR Validation
3.6. Comparison Transcriptome Data with the Published Article
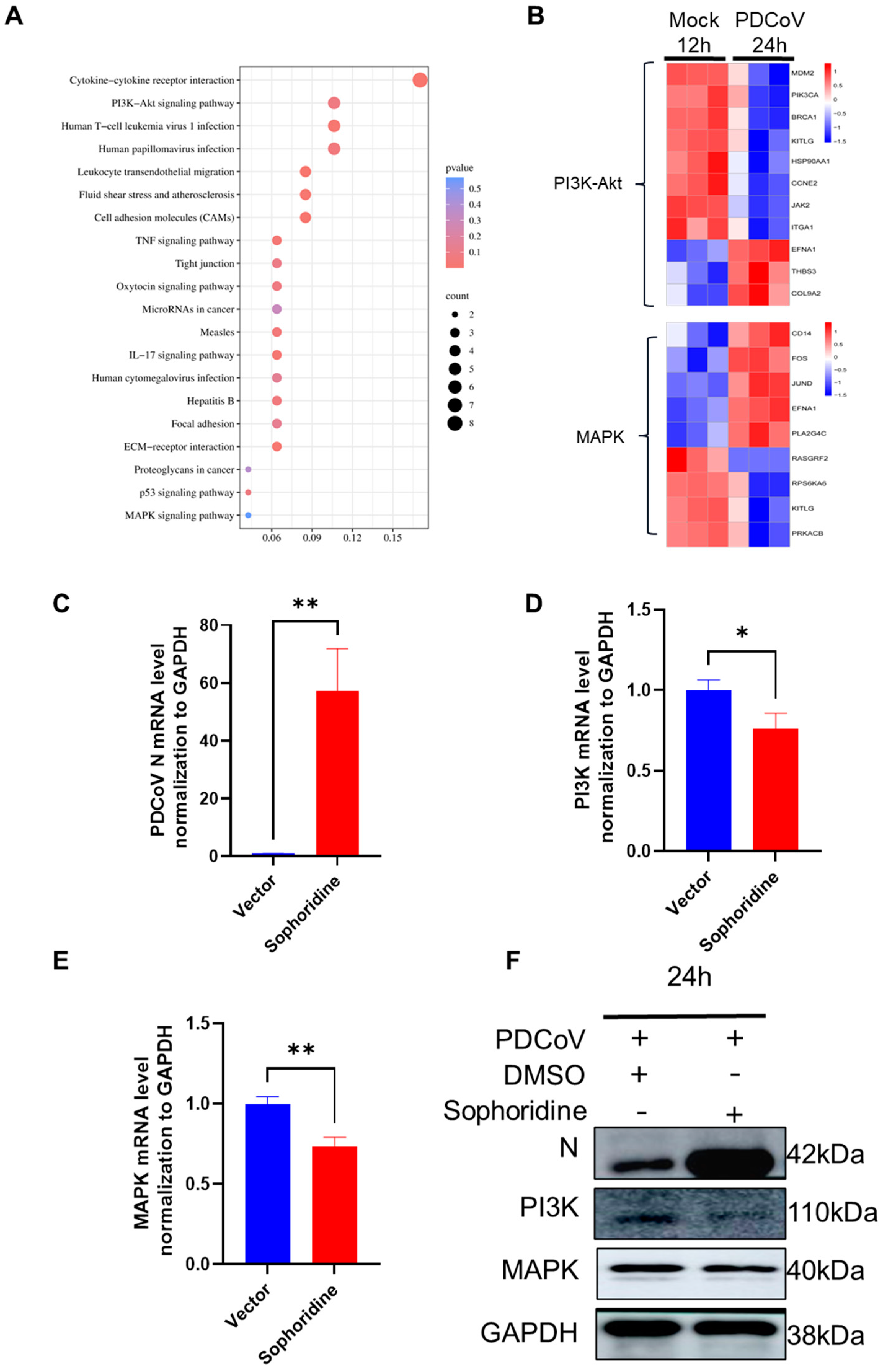
3.7. KEGG Enrichment of DEGs and Heatmap Analysis of Key Pathway DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Byrum, B.; Zhang, Y. Detection and Genetic Characterization of Deltacoronavirus in Pigs, Ohio, USA, 2014. Emerg Infect. Dis. 2014, 20, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, C. Complete Genome Characterization of Korean Porcine Deltacoronavirus Strain Kor/Knu14-04/2014. Genome Announc. 2014, 2, e01191-14. [Google Scholar] [CrossRef] [PubMed]
- Janetanakit, T.; Lumyai, M.; Bunpapong, N.; Boonyapisitsopa, S.; Chaiyawong, S.; Nonthabenjawan, N.; Kesdaengsakonwut, S.; Amonsin, A. Porcine Deltacoronavirus, Thailand, 2015. Emerg. Infect. Dis. 2016, 22, 757–759. [Google Scholar] [CrossRef] [PubMed]
- Lorsirigool, A.; Saeng-Chuto, K.; Temeeyasen, G.; Madapong, A.; Tripipat, T.; Wegner, M.; Tuntituvanont, A.; Intrakamhaeng, M.; Nilubol, D. The First Detection and Full-Length Genome Sequence of Porcine Deltacoronavirus Isolated in Lao Pdr. Arch. Virol. 2016, 161, 2909–2911. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Shibahara, T.; Imai, N.; Yamamoto, T.; Ohashi, S. Genetic Characterization and Pathogenicity of Japanese Porcine Deltacoronavirus. Infect. Genet. Evol. 2018, 61, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Yue, H.; Fang, W.; Huang, Y.W. Complete Genome Sequence of Porcine Deltacoronavirus Strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015, 3, e00945-15. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Hu, H.; Eyerly, B.; Lu, Z.; Chepngeno, J.; Saif, L.J. Pathogenicity of 2 Porcine Deltacoronavirus Strains in Gnotobiotic Pigs. Emerg. Infect. Dis. 2015, 21, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, Y.; Liang, X.; Lou, F.; Oglesbee, M.; Krakowka, S.; Li, J. Origin, Evolution, and Virulence of Porcine Deltacoronaviruses in the United States. mBio 2015, 6, e00064. [Google Scholar] [CrossRef]
- Hu, H.; Jung, K.; Vlasova, A.N.; Chepngeno, J.; Lu, Z.; Wang, Q.; Saif, L.J. Isolation and Characterization of Porcine Deltacoronavirus from Pigs with Diarrhea in the United States. J. Clin. Microbiol. 2015, 53, 1537–1548. [Google Scholar] [CrossRef]
- Li, G.; Chen, Q.; Harmon, K.M.; Yoon, K.J.; Schwartz, K.J.; Hoogland, M.J.; Gauger, P.C.; Main, R.G.; Zhang, J. Full-Length Genome Sequence of Porcine Deltacoronavirus Strain USA/Ia/2014/8734. Genome Announc. 2014, 2, e00278-14. [Google Scholar] [CrossRef]
- Ye, X.; Chen, Y.; Zhu, X.; Guo, J.; Xie, D.; Hou, Z.; Xu, S.; Zhou, J.; Fang, L.; Wang, D.; et al. Cross-Species Transmission of Deltacoronavirus and the Origin of Porcine Deltacoronavirus. Evol. Appl. 2020, 13, 2246–2253. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hulswit, R.J.G.; Kenney, S.P.; Widjaja, I.; Jung, K.; Alhamo, M.A.; van Dieren, B.; van Kuppeveld, F.J.M.; Saif, L.J.; Bosch, B.J. Broad Receptor Engagement of an Emerging Global Coronavirus May Potentiate Its Diverse Cross-Species Transmissibility. Proc. Natl. Acad. Sci. USA 2018, 115, E5135–E5143. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Vasquez-Lee, M.; Saif, L.J. Replicative Capacity of Porcine Deltacoronavirus and Porcine Epidemic Diarrhea Virus in Primary Bovine Mesenchymal Cells. Vet. Microbiol. 2020, 244, 108660. [Google Scholar] [CrossRef]
- Boley, P.A.; Alhamo, M.A.; Lossie, G.; Yadav, K.K.; Vasquez-Lee, M.; Saif, L.J.; Kenney, S.P. Porcine Deltacoronavirus Infection and Transmission in Poultry, United States(1). Emerg. Infect. Dis. 2020, 26, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Zhang, H.; Li, B.; Ding, Q.; Wang, Y.; Gao, W.; Guo, D.; Wei, Z.; Hu, H. Susceptibility of Chickens to Porcine Deltacoronavirus Infection. Viruses 2019, 11, 573. [Google Scholar] [CrossRef]
- Jung, K.; Hu, H.; Saif, L.J. Calves Are Susceptible to Infection with the Newly Emerged Porcine Deltacoronavirus, but Not with the Swine Enteric Alphacoronavirus, Porcine Epidemic Diarrhea Virus. Arch. Virol. 2017, 162, 2357–2362. [Google Scholar] [CrossRef] [PubMed]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent Infections of Porcine Deltacoronavirus among Haitian Children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef]
- Xu, K.; Zhou, Y.; Mu, Y.; Liu, Z.; Hou, S.; Xiong, Y.; Fang, L.; Ge, C.; Wei, Y.; Zhang, X.; et al. CD163 and pAPN Double-Knockout Pigs Are Resistant to PRRSV and TGEV and Exhibit Decreased Susceptibility to PDCoV While Maintaining Normal Production Performance. eLife 2020, 9, e57132. [Google Scholar] [CrossRef]
- Ji, W.; Peng, Q.; Fang, X.; Li, Z.; Li, Y.; Xu, C.; Zhao, S.; Li, J.; Chen, R.; Mo, G.; et al. Structures of a Deltacoronavirus Spike Protein Bound to Porcine and Human Receptors. Nat. Commun. 2022, 13, 1467. [Google Scholar] [CrossRef]
- Juntarachot, N.; Sunpaweravong, S.; Kaewdech, A.; Wongsuwanlert, M.; Ruangsri, P.; Pahumunto, N.; Teanpaisan, R. Characterization of Adhesion, Anti-Adhesion, Co-Aggregation, and Hydrophobicity of Helicobacter Pylori and Probiotic Strains. J. Taibah Univ. Med. Sci. 2023, 18, 1048–1054. [Google Scholar] [CrossRef]
- Cruz-Pulido, D.; Boley, P.A.; Ouma, W.Z.; Alhamo, M.A.; Saif, L.J.; Kenney, S.P. Comparative Transcriptome Profiling of Human and Pig Intestinal Epithelial Cells after Porcine Deltacoronavirus Infection. Viruses 2021, 13, 292. [Google Scholar] [CrossRef] [PubMed]
- Alhamo, M.A.; Boley, P.A.; Liu, M.; Niu, X.; Yadav, K.K.; Lee, C.; Saif, L.J.; Wang, Q.; Kenney, S.P. Characterization of the Cross-Species Transmission Potential for Porcine Deltacoronaviruses Expressing Sparrow Coronavirus Spike Protein in Commercial Poultry. Viruses 2022, 14, 1225. [Google Scholar] [CrossRef] [PubMed]
- Zheng, A.; Shi, Y.; Shen, Z.; Wang, G.; Shi, J.; Xiong, Q.; Fang, L.; Xiao, S.; Fu, Z.F.; Peng, G. Insight into the Evolution of Nidovirus Endoribonuclease Based on the Finding that Nsp15 from Porcine Deltacoronavirus Functions as a Dimer. J. Biol. Chem. 2018, 293, 12054–12067. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Lin, C.; Du, L.; Liu, J.; Gu, J.; Zhou, J. Porcine Deltacoronavirus Induces Mitochondrial Apoptosis in St Cells. Sheng Wu Gong Cheng Xue Bao 2019, 35, 1050–1058. [Google Scholar] [PubMed]
- Jin, X.H.; Zhang, Y.F.; Yuan, Y.X.; Han, L.; Zhang, G.P.; Hu, H. Isolation, Characterization and Transcriptome Analysis of Porcine Deltacoronavirus Strain Hnzk-02 from Henan Province, China. Mol. Immunol. 2021, 134, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Li, F.; Li, X.; Wang, L.; Zhang, L.; Lu, C.; Zheng, L.; Yan, M. Transcriptome Analysis of Pk-15 Cells in Innate Immune Response to Porcine Deltacoronavirus Infection. PLoS ONE 2019, 14, e0223177. [Google Scholar] [CrossRef]
- Tang, Q.; Luan, F.; Yuan, A.; Sun, J.; Rao, Z.; Wang, B.; Liu, Y.; Zeng, N. Sophoridine Suppresses Herpes Simplex Virus Type 1 Infection by Blocking the Activation of Cellular PI3K/Akt and p38 MAPK Pathways. Front. Microbiol. 2022, 13, 872505. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, L.; Liu, W.; Li, D.; Zeng, J.; Tang, Q.; Zhang, Y.; Luan, F.; Zeng, N. Cepharanthine Suppresses Herpes Simplex Virus Type 1 Replication Through the Downregulation of the PI3K/Akt and p38 MAPK Signaling Pathways. Front. Microbiol. 2021, 12, 795756. [Google Scholar] [CrossRef]
- Lu, Q.; Li, L.; Huang, A.; Cui, L.; Zhang, Y.; Liu, Q.; Wang, X.; Wang, Y.; Liu, Z.; Yuan, Z.; et al. Molecular Characterization and Biological Function of a Novel LncRNA CRNG in Swine. Front. Pharmacol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Potla, L.; Boghaert, E.R.; Armellino, D.; Frost, P.; Damle, N.K. Reduced Expression of Ephrina1 (Efna1) Inhibits Three-Dimensional Growth of Ht29 Colon Carcinoma Cells. Cancer Lett. 2002, 175, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Xiao, T.; Hu, Y.; Yang, Y.; Gu, X.; Jin, G.; Cao, H.; Zhou, H.; Yang, C. Nintedanib Alleviates Experimental Colitis by Inhibiting CEBPB/PCK1 and CEBPB/EFNA1 Pathways. Front. Pharmacol. 2022, 13, 904420. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence |
|---|---|
| PDCoV-N-F | CTATGAGCCACCCACCAA |
| PDCoV-N-R | TCCCACTCCCAATCCTGT |
| BUB1-F | GTTGCATCAGGTGGTGGAGA |
| BUB1-R | GAGGTGCCTCTCTTGGGTTC |
| DDIT4-F | GCTTACCTGGATGGGGTGTC |
| DDIT4-R | GCATCAGGTTGGCACACAAG |
| ATF3-F | CTGTCTCGAGACCATGATGCTTCAACATCCA |
| ATF3-R | CTGTCCCGGGTTAGCTCTGCAATGTTCC |
| GBP2-F | ATGAACAAGCTGGCTGGGAA |
| GBP2-R | TCTTGGGATGAGGCACACAC |
| IRF1-F | ACCCTGGCTAGAGATGCAGA |
| IRF1-R | TGCTTTGTATCGGCCTGTGT |
| GAPDH-F | CTACATGGTTTACATGTTCC |
| GAPDH-R | GGATCTCGCTCCTGGAAGAT |
| qPI3K-F | CTGCAGTTCAACAGCCACAC |
| qPI3K-R | ACAGGTCAATGGCTGCATCA |
| qMAPK-F | AGGCTGTTCCCAAATGCTGA |
| qMAPK-R | CAGATATGGGTGGGCCAGAG |
| Sample | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Q30 | GCpct | Total Map | Multimap | Unique Map |
|---|---|---|---|---|---|---|---|---|---|
| Mock-1 | 45142592 | 6.77 G | 44517704 | 6.68 G | 90.83 | 52.52 | 42706945 | 1024389 | 41682556 |
| Mock-2 | 46280682 | 6.94 G | 45076696 | 6.76 G | 91.35 | 52.98 | 43027266 | 1136101 | 41891165 |
| Mock-3 | 44625196 | 6.69 G | 43355404 | 6.5 G | 91.32 | 52 | 41228385 | 1115310 | 40113075 |
| PDCoV-12h-1 | 44691544 | 6.7 G | 43856336 | 6.58 G | 90.88 | 53.47 | 41847210 | 1018571 | 40828639 |
| PDCoV-12h-2 | 45824392 | 6.87 G | 44782282 | 6.72 G | 90.4 | 55.32 | 42330576 | 1111281 | 41219295 |
| PDCoV-12h-3 | 45255846 | 6.79 G | 44379500 | 6.66 G | 90.68 | 53.71 | 42067900 | 1167913 | 40899987 |
| PDCoV-24h-1 | 42819744 | 6.42 G | 41660400 | 6.25 G | 91.71 | 55.48 | 39508414 | 1080319 | 38428095 |
| PDCoV-24h-2 | 44243262 | 6.64 G | 42104606 | 6.32 G | 91.31 | 53.55 | 39879439 | 1055958 | 38823481 |
| PDCoV-24h-3 | 45048018 | 6.76 G | 43430312 | 6.51 G | 91.04 | 54.52 | 40885945 | 1143274 | 39742671 |
| PDCoV-48h-1 | 46048916 | 6.91 G | 43818106 | 6.57 G | 90.29 | 54.21 | 41277870 | 1128930 | 40148940 |
| PDCoV-24h-2 | 45317606 | 6.8 G | 42285954 | 6.34 G | 90.57 | 53.06 | 39831492 | 1093925 | 38737567 |
| PDCoV-24h-3 | 44781216 | 6.72 G | 42845208 | 6.43 G | 90.56 | 50.91 | 40670083 | 1013339 | 39656744 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Zhang, G.; Li, L.; Wang, M.; Chen, J.; Hao, P.; Gao, Z.; Hao, J.; Li, C.; Jin, N. Transcriptomic Analysis of PDCoV-Infected HIEC-6 Cells and Enrichment Pathways PI3K-Akt and P38 MAPK. Viruses 2024, 16, 579. https://doi.org/10.3390/v16040579
Jiang Y, Zhang G, Li L, Wang M, Chen J, Hao P, Gao Z, Hao J, Li C, Jin N. Transcriptomic Analysis of PDCoV-Infected HIEC-6 Cells and Enrichment Pathways PI3K-Akt and P38 MAPK. Viruses. 2024; 16(4):579. https://doi.org/10.3390/v16040579
Chicago/Turabian StyleJiang, Yuhang, Guoqing Zhang, Letian Li, Maopeng Wang, Jing Chen, Pengfei Hao, Zihan Gao, Jiayi Hao, Chang Li, and Ningyi Jin. 2024. "Transcriptomic Analysis of PDCoV-Infected HIEC-6 Cells and Enrichment Pathways PI3K-Akt and P38 MAPK" Viruses 16, no. 4: 579. https://doi.org/10.3390/v16040579
APA StyleJiang, Y., Zhang, G., Li, L., Wang, M., Chen, J., Hao, P., Gao, Z., Hao, J., Li, C., & Jin, N. (2024). Transcriptomic Analysis of PDCoV-Infected HIEC-6 Cells and Enrichment Pathways PI3K-Akt and P38 MAPK. Viruses, 16(4), 579. https://doi.org/10.3390/v16040579








