High Prevalence of Hepatitis B Virus Drug Resistance Mutations to Lamivudine among People with HIV/HBV Coinfection in Rural and Peri-Urban Communities in Botswana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. HBV Screening
2.3. Nanopore Sequencing
2.4. Sequencing Analysis
2.5. Statistical Analysis
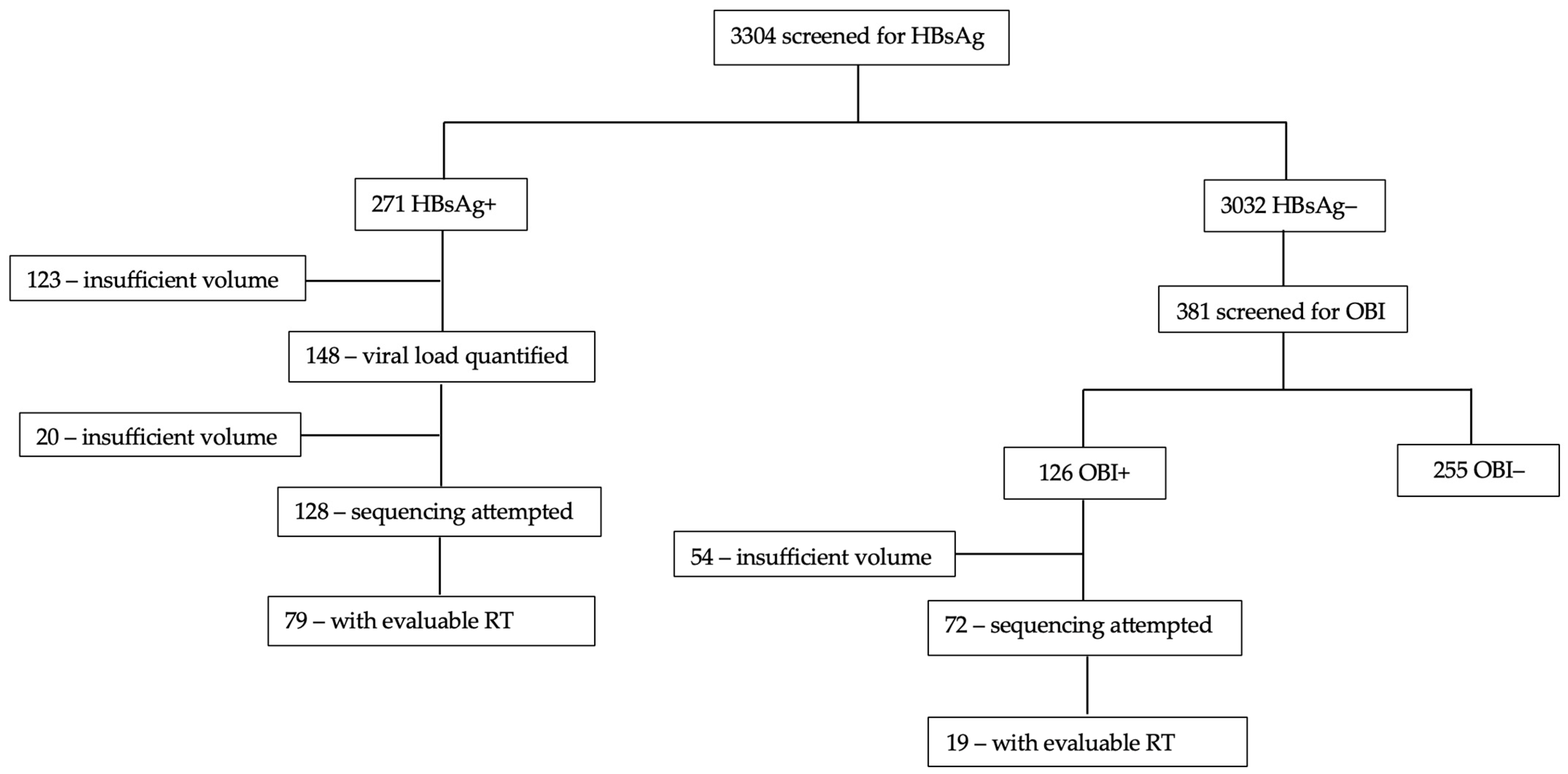
3. Results
3.1. Participant Description
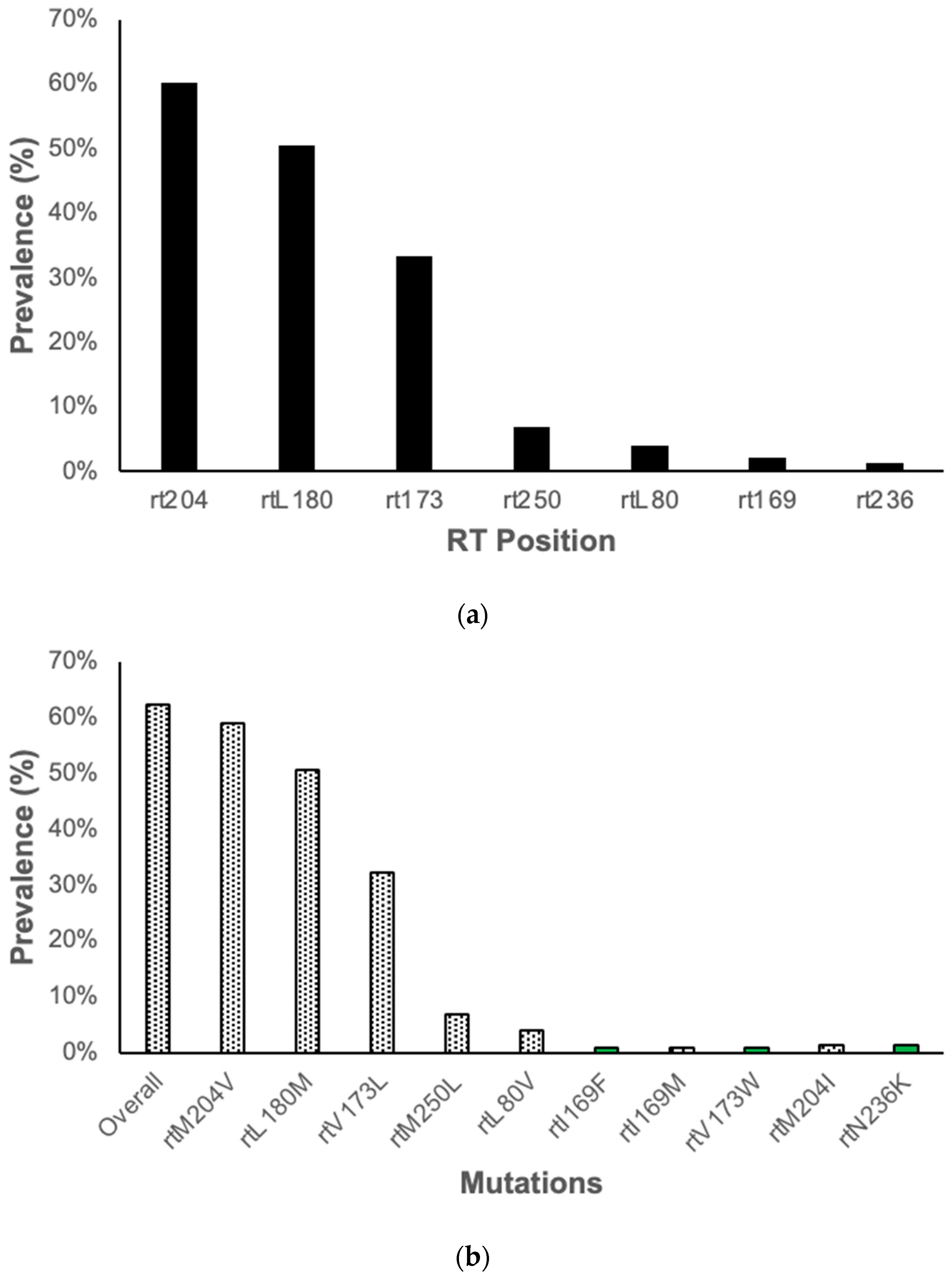
3.2. Resistance Associated Mutations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Interim Guidance for Country Validation of Viral Hepatitis Elimination; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- WHO. Hepatitis B Virus Fact Sheets. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (accessed on 8 April 2024).
- Kramvis, A. Genotypes and genetic variability of hepatitis B virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.; Choga, W.T.; Moyo, S.; Bell, T.G.; Mbangiwa, T.; Phinius, B.B.; Bhebhe, L.; Sebunya, T.K.; Lockman, S.; Marlink, R.; et al. Molecular Characterization of Near Full-Length Genomes of Hepatitis B Virus Isolated from Predominantly HIV Infected Individuals in Botswana. Genes 2018, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- Mbangiwa, T.; Kasvosve, I.; Anderson, M.; Thami, P.K.; Choga, W.T.; Needleman, A.; Phinius, B.B.; Moyo, S.; Leteane, M.; Leidner, J.; et al. Chronic and Occult Hepatitis B Virus Infection in Pregnant Women in Botswana. Genes 2018, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Choga, W.T.; Anderson, M.; Zumbika, E.; Moyo, S.; Mbangiwa, T.; Phinius, B.B.; Melamu, P.; Kayembe, M.K.; Kasvosve, I.; Sebunya, T.K.; et al. Molecular characterization of hepatitis B virus in blood donors in Botswana. Virus Genes 2018, 55, 33–42. [Google Scholar] [CrossRef]
- Spitz, N.; Mello, F.C.A.; Moreira, A.S.; Gusatti, C.S.; Martins, R.M.B.; Gomes, S.A.; Bello, G.; Araujo, N.M. Reconstruction of the spatial and temporal dynamics of hepatitis B virus genotype D in the Americas. PLoS ONE 2019, 14, e0220342. [Google Scholar] [CrossRef] [PubMed]
- Paraskevis, D.; Angelis, K.; Magiorkinis, G.; Kostaki, E.; Ho, S.Y.; Hatzakis, A. Dating the origin of hepatitis B virus reveals higher substitution rate and adaptation on the branch leading to F/H genotypes. Mol. Phylogenet. Evol. 2015, 93, 44–54. [Google Scholar] [CrossRef]
- Zehender, G.; Ebranati, E.; Gabanelli, E.; Sorrentino, C.; Lo Presti, A.; Tanzi, E.; Ciccozzi, M.; Galli, M. Enigmatic origin of hepatitis B virus: An ancient travelling companion or a recent encounter? World J. Gastroenterol. 2014, 20, 7622–7634. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Sarkar, N.; Saha, D.; Guha, S.K.; Saha, B.; Chakrabarti, S.; Chakravarty, R. High incidence of lamivudine-resistance-associated vaccine-escape HBV mutants among HIV-coinfected patients on prolonged antiretroviral therapy. Antivir. Ther. 2015, 20, 545–554. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guidelines for the Prevention, Diagnosis, Care and Treatment for People with Chronic Hepatitis B Infection; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Woo, H.Y.; Park, J.Y.; Bae, S.H.; Kim, C.W.; Jang, J.Y.; Tak, W.Y.; Kim, D.J.; Kim, I.H.; Heo, J.; Ahn, S.H. Entecavir+tenofovir vs. lamivudine/telbivudine+adefovir in chronic hepatitis B patients with prior suboptimal response. Clin. Mol. Hepatol. 2020, 26, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Msomi, N.; Parboosing, R.; Wilkinson, E.; Giandhari, J.; Govender, K.; Chimukangara, B.; Mlisana, K.P. Persistent Hepatitis B Viraemia with Polymerase Mutations among HIV/HBV Co-Infected Patients on HBV-Active ART in KwaZulu-Natal, South Africa. Viruses 2022, 14, 788. [Google Scholar] [CrossRef]
- Mokaya, J.; McNaughton, A.L.; Bester, P.A.; Goedhals, D.; Barnes, E.; Marsden, B.D.; Matthews, P.C. Hepatitis B virus resistance to tenofovir: Fact or fiction? A systematic literature review and structural analysis of drug resistance mechanisms. Wellcome Open Res. 2020, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Amini-Bavil-Olyaee, S.; Herbers, U.; Sheldon, J.; Luedde, T.; Trautwein, C.; Tacke, F. The rtA194T polymerase mutation impacts viral replication and susceptibility to tenofovir in hepatitis B e antigen-positive and hepatitis B e antigen-negative hepatitis B virus strains. Hepatology 2009, 49, 1158–1165. [Google Scholar] [CrossRef] [PubMed]
- Shirvani-Dastgerdi, E.; Winer, B.Y.; Celia-Terrassa, T.; Kang, Y.; Tabernero, D.; Yagmur, E.; Rodriguez-Frias, F.; Gregori, J.; Luedde, T.; Trautwein, C.; et al. Selection of the highly replicative and partially multidrug resistant rtS78T HBV polymerase mutation during TDF-ETV combination therapy. J. Hepatol. 2017, 67, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Mokaya, J.; McNaughton, A.L.; Hadley, M.J.; Beloukas, A.; Geretti, A.M.; Goedhals, D.; Matthews, P.C. A systematic review of hepatitis B virus (HBV) drug and vaccine escape mutations in Africa: A call for urgent action. PLoS Negl. Trop. Dis. 2018, 12, e0006629. [Google Scholar] [CrossRef]
- Mine, M.; Stafford, K.; Laws, R.L.; Marima, M.; Lekone, P.; Ramaabya, D.; Makhaola, K.; Mapondera, P.; Wray-Gordon, F.; Akbakwuru, C.; et al. Botswana achieved the Joint United Nations Programme on HIV/AIDS (UNAIDS) 95-95-95 targets: Results from the Fifth Botswana HIV/AIDS Impact Survey (BAIS V), 2021. In Proceedings of the The 24th International AIDS Conference, Montreal, QC, Canada, 29 July–2 August 2022; p. 231. [Google Scholar]
- Shaver, Z.M.; Anderson, M.; Bhebhe, L.; Baruti, K.; Choga, W.T.; Ngidi, J.; Mbangiwa, T.; Tau, M.; Setlhare, D.R.; Melamu, P.; et al. Decreased hepatitis B virus vaccine response among HIV-positive infants compared with HIV-negative infants in Botswana. AIDS 2022, 36, 755–762. [Google Scholar] [CrossRef]
- Anderson, M.; Gaseitsiwe, S.; Moyo, S.; Thami, K.P.; Mohammed, T.; Setlhare, D.; Sebunya, T.K.; Powell, E.A.; Makhema, J.; Blackard, J.T.; et al. Slow CD4+ T cell Recovery in Human Immunodeficiency Virus/Hepatitis B Virus-Coinfected Patients Initiating Truvada-Based Combination Antiretroviral Therapy in Botswana. Open Forum Infect. Dis. 2016, 3, ofw140. [Google Scholar] [CrossRef] [PubMed]
- Phinius, B.B.; Anderson, M.; Gobe, I.; Mokomane, M.; Choga, W.T.; Mutenga, S.R.; Mpebe, G.; Pretorius-Holme, M.; Musonda, R.; Gaolathe, T.; et al. High Prevalence of Hepatitis B Virus Infection Among People With HIV in Rural and Periurban Communities in Botswana. Open Forum Infect. Dis. 2023, 10, ofac707. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, G.; Pollicino, T.; Cacciola, I.; Squadrito, G. Occult hepatitis B virus infection. J. Hepatol. 2007, 46, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.; Anderson, M.; Gyurova, I.; Ambroggio, L.; Moyo, S.; Sebunya, T.; Makhema, J.; Marlink, R.; Essex, M.; Musonda, R.; et al. High Rates of Occult Hepatitis B Virus Infection in HIV-Positive Individuals Initiating Antiretroviral Therapy in Botswana. Open Forum Infect Dis 2017, 4, ofx195. [Google Scholar] [CrossRef]
- Ministy-of-Health. Handbook of the Botswana 2016 Integrated HIV Clincial Care Guidelines; Ministy-of-Health Botswana: Gaborone, Botswana, 2016. [Google Scholar]
- Makhema, J.; Wirth, K.E.; Pretorius Holme, M.; Gaolathe, T.; Mmalane, M.; Kadima, E.; Chakalisa, U.; Bennett, K.; Leidner, J.; Manyake, K.; et al. Universal Testing, Expanded Treatment, and Incidence of HIV Infection in Botswana. N. Engl. J. Med. 2019, 381, 230–242. [Google Scholar] [CrossRef]
- Choga, W. Complete Hepatitis B Virus Sequencing Using an ONT-Based Next-Generation Sequencing Protocol v1; CERI: Stellenbosch, South Africa, 2023. [Google Scholar]
- Phinius, B.B.; Anderson, M.; Mokomane, M.; Gobe, I.; Choga, W.T.; Ratsoma, T.; Phakedi, B.; Mpebe, G.; Ditshwanelo, D.; Musonda, R.; et al. Atypical Hepatitis B Virus Serology Profile-Hepatitis B Surface Antigen-Positive/Hepatitis B Core Antibody-Negative-In Hepatitis B Virus/HIV Coinfected Individuals in Botswana. Viruses 2023, 15, 1544. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Gulube, Z.; Chirara, M.; Kew, M.; Tanaka, Y.; Mizokami, M.; Kramvis, A. Molecular characterization of hepatitis B virus isolates from Zimbabwean blood donors. J. Med. Virol. 2011, 83, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Hamers, R.L.; Zaaijer, H.L.; Wallis, C.L.; Siwale, M.; Ive, P.; Botes, M.E.; Sigaloff, K.C.E.; Hoepelman, A.I.M.; Stevens, W.S.; Rinke de Wit, T.F.; et al. HIV–HBV Coinfection in Southern Africa and the Effect of Lamivudine- Versus Tenofovir-Containing cART on HBV Outcomes. JAIDS J. Acquir. Immune Defic. Syndr. 2013, 64, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Maepa, M.B.; Ely, A.; Kramvis, A.; Bloom, K.; Naidoo, K.; Simani, O.E.; Maponga, T.G.; Arbuthnot, P. Hepatitis B Virus Research in South Africa. Viruses 2022, 14, 1939. [Google Scholar] [CrossRef]
- Kew, M.C.; Kramvis, A.; Yu, M.C.; Arakawa, K.; Hodkinson, J. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-saharan Africans. J. Med. Virol. 2005, 75, 513–521. [Google Scholar] [CrossRef]
- Tanaka, Y.; Hasegawa, I.; Kato, T.; Orito, E.; Hirashima, N.; Acharya, S.K.; Gish, R.G.; Kramvis, A.; Kew, M.C.; Yoshihara, N.; et al. A case-control study for differences among hepatitis B virus infections of genotypes A (subtypes Aa and Ae) and D. Hepatology 2004, 40, 747–755. [Google Scholar] [CrossRef]
- Svicher, V.; Alteri, C.; Gori, C.; Salpini, R.; Marcuccilli, F.; Bertoli, A.; Longo, R.; Bernassola, M.; Gallinaro, V.; Romano, S.; et al. Lamivudine-resistance mutations can be selected even at very low levels of hepatitis B viraemia. Dig. Liver Dis. 2010, 42, 902–907. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Kou, Z.; Ren, F.; Jin, Y.; Yang, L.; Dong, X.; Yang, M.; Zhao, J.; Liu, H.; et al. Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clin. Microbiol. Infect. 2021, 27, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Ghany, M.G.; Doo, E.C. Antiviral resistance and hepatitis B therapy. Hepatology 2009, 49, S174–S184. [Google Scholar] [CrossRef] [PubMed]
- Shaw, T.; Bartholomeusz, A.; Locarnini, S. HBV drug resistance: Mechanisms, detection and interpretation. J. Hepatol. 2006, 44, 593–606. [Google Scholar] [CrossRef]
- Bivigou-Mboumba, B.; Amougou-Atsama, M.; Zoa-Assoumou, S.; M’Boyis Kamdem, H.; Nzengui-Nzengui, G.F.; Ndojyi-Mbiguino, A.; Njouom, R.; Francois-Souquiere, S. Hepatitis B infection among HIV infected individuals in Gabon: Occult hepatitis B enhances HBV DNA prevalence. PLoS ONE 2018, 13, e0190592. [Google Scholar] [CrossRef] [PubMed]
- Mokaya, J.; Vasylyeva, T.I.; Barnes, E.; Ansari, M.A.; Pybus, O.G.; Matthews, P.C. Global prevalence and phylogeny of hepatitis B virus (HBV) drug and vaccine resistance mutations. J. Viral Hepat. 2021, 28, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Delaney, W.E., IV; Yang, H.; Westland, C.E.; Das, K.; Arnold, E.; Gibbs, C.S.; Miller, M.D.; Xiong, S. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J. Virol. 2003, 77, 11833–11841. [Google Scholar] [CrossRef]
- Geipel, A.; Seiz, P.L.; Niekamp, H.; Neumann-Fraune, M.; Zhang, K.; Kaiser, R.; Protzer, U.; Gerlich, W.H.; Glebe, D. Entecavir allows an unexpectedly high residual replication of HBV mutants resistant to lamivudine. Antivir. Ther. 2015, 20, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Rodriguez, C.V.; Van Rompaey, S.; Eron, J.J.; Thio, C.L.; Crane, H.M.; Overton, E.T.; Saag, M.S.; Martin, J.; Geng, E.; et al. Factors associated with delayed hepatitis B viral suppression on tenofovir among patients coinfected with HBV-HIV in the CNICS cohort. J. Acquir. Immune Defic. Syndr. 2014, 66, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, N.; Cicero, M.; Santana, L.C.; Silveira, C.; do Carmo, E.P.; Abrão, P.R.; Diaz, R.S.; Caseiro, M.M.; Komninakis, S.V. Detection of lamivudine-resistant variants and mutations related to reduced antigenicity of HBsAg in individuals from the cities of Santos and São Paulo, Brazil. Virol. J. 2013, 10, 320. [Google Scholar] [CrossRef] [PubMed]

| No RAMs, n = 36 | RAMs Present, n = 61 | p-Value | |
|---|---|---|---|
| Sex, n (%) | |||
| Female | 24 (64.9) | 42 (68.9) | |
| Male | 13 (35.1) | 19 (31.2) | 0.82 |
| Age, years, median (IQR) | 41 (34–47) | 45 (40–50) | 0.68 |
| Region | |||
| South | 7 (18.9) | 10 (16.4) | |
| Central | 17 (46.0) | 23 (37.7) | |
| North | 13 (35.1) | 28 (45.9) | 0.56 |
| Nadir CD4 T cell count, cells/μL, n (%), n = 19 | |||
| <350 | 7 (70.0) | 3 (33.3) | |
| ≥350 | 3 (30.0) | 6 (66.7) | 0.11 |
| HIV viral load, copies/mL, n (%) | |||
| Suppressed | 30 (81.1) | 56 (91.8) | |
| Unsuppressed | 7 (18.9) | 5 (8.2) | 0.12 |
| HBV infection phase, n (%) | |||
| HBsAg positive | 28 (75.7) | 51 (83.6) | |
| OBI positive | 9 (24.3) | 10 (16.4) | 0.43 |
| HBV viral load, n (%) | |||
| TND | 3 (8.1) | 17 (27.9) | |
| <2000 | 27 (73.0) | 31 (50.8) | |
| >2000 | 7 (18.9) | 13 (21.3) | 0.04 |
| HBeAg status, n (%), n = 76 | |||
| Negative | 24 (88.9) | 39 (79.6) | |
| Positive | 3 (11.1) | 10 (20.4) | 0.30 |
| Anti-HBc IgM, n (%), n = 75 | |||
| Negative | 25 (92.6) | 46 (95.8) | |
| Positive | 2 (7.4) | 2 (4.2) | 0.55 |
| Total anti-HBc, n (%) n = 95 | |||
| Negative | 7 (19.4) | 9 (15.3) | |
| Positive | 29 (80.6) | 61 (85.9) | 0.44 |
| ART status, n (%) | |||
| ART-naive | 3 (8.1) | 2 (3.3) | |
| On ART | 34 (91.9) | 59 (96.7) | 0.29 |
| Current ART regimen, n (%) n = 67 | |||
| 3TC containing, without TDF | 8 (38.1) | 19 (41.3) | |
| TDF containing * | 13 (61.9) | 27 (58.7) | 1.00 |
| Time on ART, years, median (IQR), n = 74 | 5.9 (4.2–10.3) | 7.5 (4.8–10.5) | 0.55 |
| Mutations | Frequency | ART Status | ART Regimen *# |
|---|---|---|---|
| rtV173L/rtL180M/rtM204V | 17 | All on ART | 7 on TDF 5 on 3TC |
| rtL180M/rtM204V | 10 | All on ART | 3 on TDF 5 on 3TC |
| rtM204V | 8 | All on ART | 7 on TDF 1 on 3TC |
| rtL180M | 5 | All on ART | 2 on TDF 2 on 3TC |
| rtV173L/rtL180M | 5 | All on ART | 2 on TDF 2 on 3TC |
| rtL180M/rtM204V/rtM250L | 3 | 1 on ART 2 ART naïve | No data |
| rtV173L/rtM204V | 2 | 1 on ART 1 ART naïve | On 3TC |
| rtV173L | 2 | 1 on ART 1 ART naïve | No data |
| rtI169F/rtV173L/rtL180M | 1 | On ART | On TDF |
| rtV173L/rtL180M/rtM204V/rtM250L | 1 | On ART | On TDF |
| rtL180M/rtM204V/rtN236K | 1 | On ART | On 3TC |
| rtL80V/rtV173L/rtL180M/rtM204I | 1 | On ART | On 3TC |
| rtL80V/rtV173L/rtL180M | 1 | On ART | On 3TC |
| rtL80V/rtV173L/rtL180M/rtM204V | 1 | On ART | On TDF |
| rtV173W/rtL180M | 1 | On ART | On TDF |
| rtI169M/rtL180M | 1 | On ART | On TDF |
| rtM204V/rtM250L | 1 | On ART | On TDF |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phinius, B.B.; Anderson, M.; Gobe, I.; Mokomane, M.; Choga, W.T.; Phakedi, B.; Ratsoma, T.; Mpebe, G.; Makhema, J.; Shapiro, R.; et al. High Prevalence of Hepatitis B Virus Drug Resistance Mutations to Lamivudine among People with HIV/HBV Coinfection in Rural and Peri-Urban Communities in Botswana. Viruses 2024, 16, 592. https://doi.org/10.3390/v16040592
Phinius BB, Anderson M, Gobe I, Mokomane M, Choga WT, Phakedi B, Ratsoma T, Mpebe G, Makhema J, Shapiro R, et al. High Prevalence of Hepatitis B Virus Drug Resistance Mutations to Lamivudine among People with HIV/HBV Coinfection in Rural and Peri-Urban Communities in Botswana. Viruses. 2024; 16(4):592. https://doi.org/10.3390/v16040592
Chicago/Turabian StylePhinius, Bonolo B., Motswedi Anderson, Irene Gobe, Margaret Mokomane, Wonderful T. Choga, Basetsana Phakedi, Tsholofelo Ratsoma, Gorata Mpebe, Joseph Makhema, Roger Shapiro, and et al. 2024. "High Prevalence of Hepatitis B Virus Drug Resistance Mutations to Lamivudine among People with HIV/HBV Coinfection in Rural and Peri-Urban Communities in Botswana" Viruses 16, no. 4: 592. https://doi.org/10.3390/v16040592





