Molecular and Serological Detection of Bovine Coronaviruses in Marmots (Marmota marmota) in the Alpine Region
Abstract
:1. Introduction
2. Materials and Methods
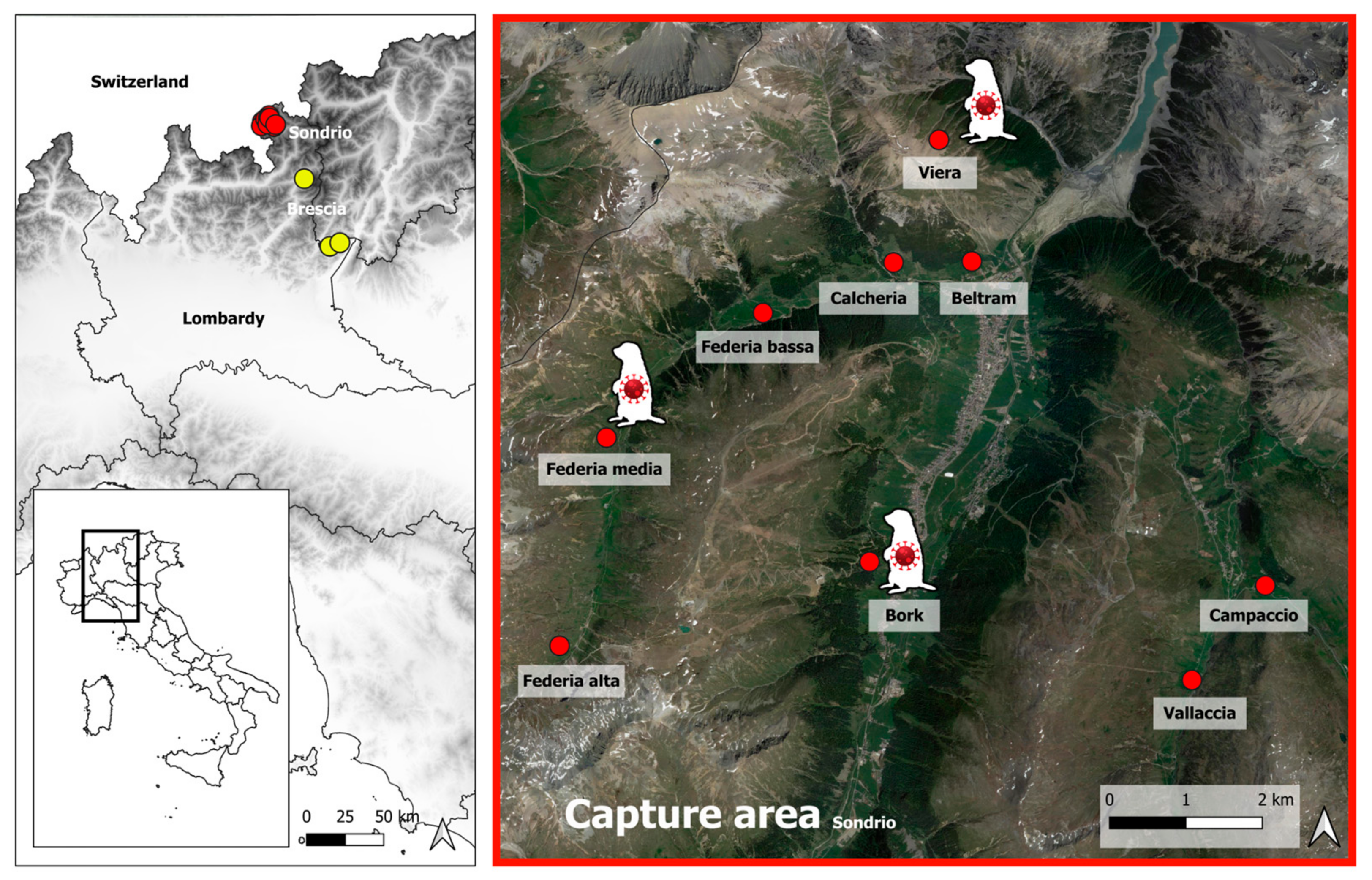
2.1. Sampling
2.2. Diagnostic Examinations
2.3. Virus Isolation
2.4. Metatranscriptome Sequencing and Phylogenetic Analysis
2.5. Antibody Detection against BCoV
3. Results
3.1. Diagnostic Examinations
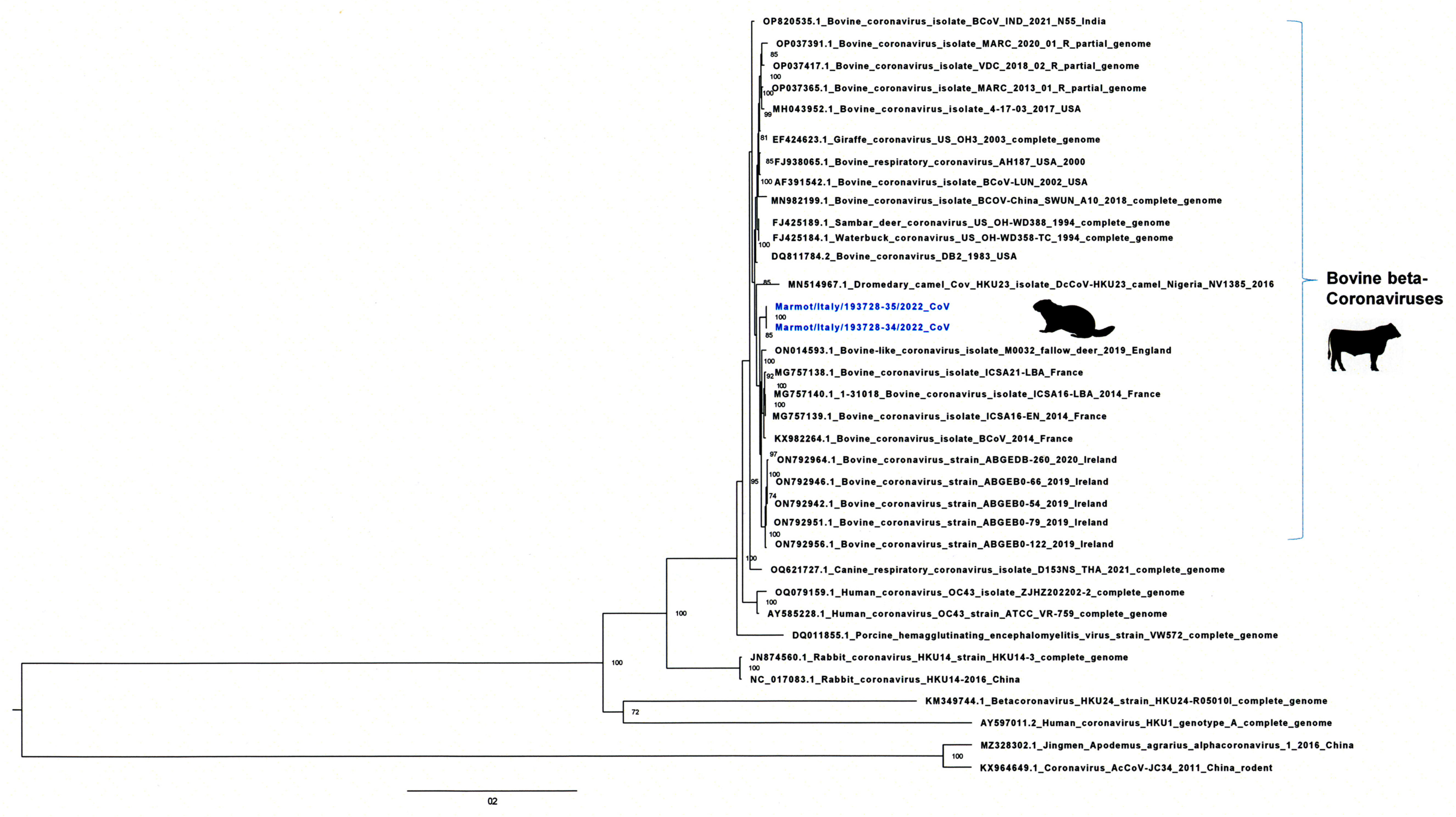
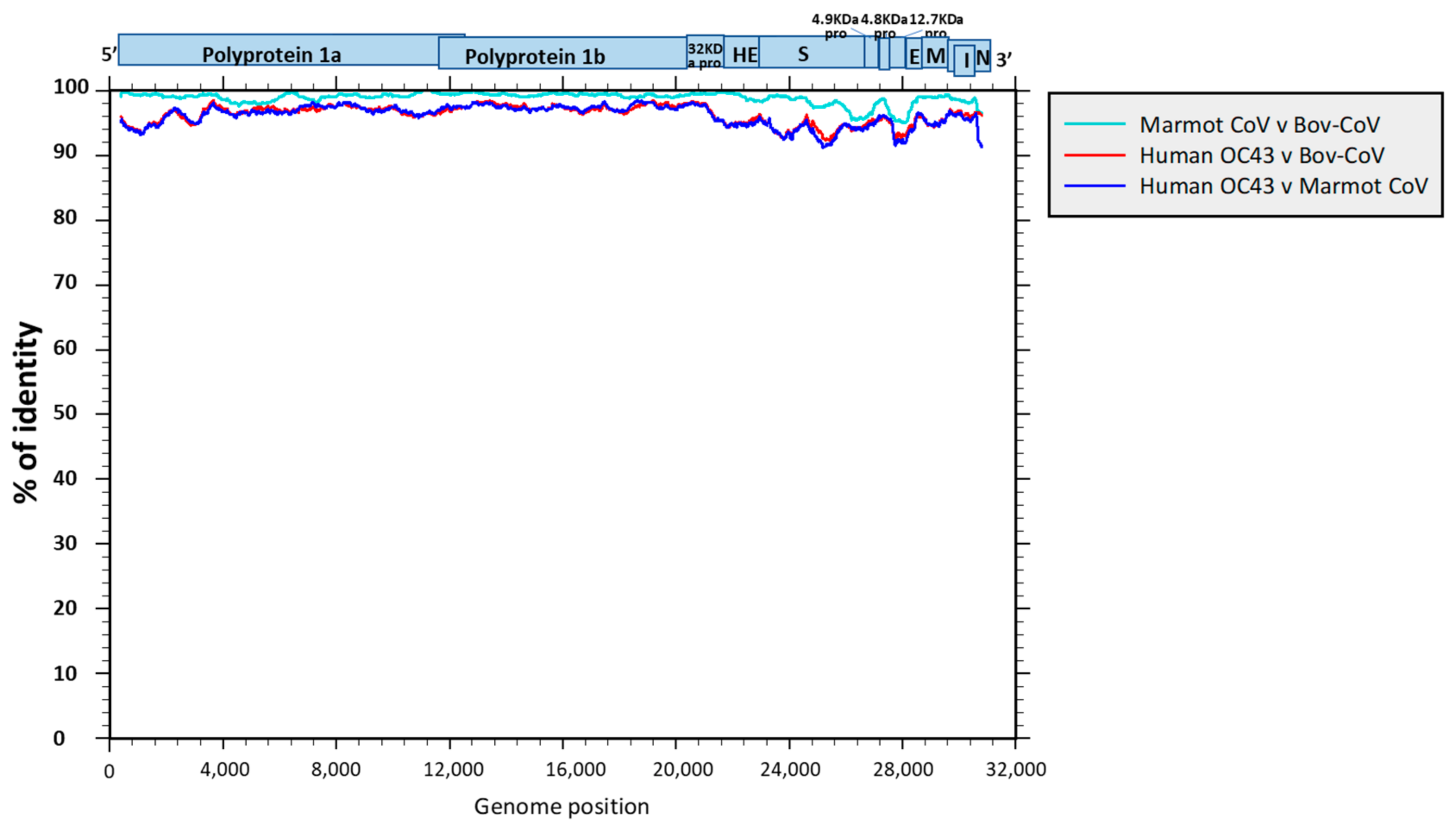
3.2. Genome Sequencing and Phylogenetic and Molecular Analysis
3.3. Serologic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Groot, R.J.; Baker, S.C.; Baric, R.; Enjuanes, L.; Gorbalenya, A.E.; Holmes, K.V.; Perlman, S.; Poon, L.; Rottier, P.J.; Talbot, P.J.; et al. Family Coronaviridae. In Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M., Adams, M.J., Carstens, E.B., Lefkowitz, E., Eds.; Elsevier: Oxford, UK, 2012; pp. 806–828. [Google Scholar] [CrossRef]
- Li, Q.; Shah, T.; Wang, B.; Qu, L.; Wang, R.; Hou, Y.; Baloch, Z.; Xia, X. Cross-species transmission, evolution and zoonotic potential of coronaviruses. Front. Cell. Infect. Microbiol. 2023, 12, 1081370. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lin, X.D.; Guo, W.P.; Zhou, R.H.; Wang, M.R.; Wang, C.Q.; Ge, S.; Mei, S.H.; Li, M.H.; Shi, M.; et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology 2015, 474, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Saif, L.J. Bovine Respiratory Coronavirus. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Desario, C.; Elia, G.; Mari, V.; Lucente, M.S.; Cordioli, P.; Colaianni, M.L.; Martella, V.; Buonavoglia, C. Serological and molecular evidence that canine respiratory coronavirus is circulating in Italy. Vet. Microbiol. 2007, 121, 225–230. [Google Scholar] [CrossRef]
- Zhang, X.M.; Herbst, W.; Kousoulas, K.G.; Storz, J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994, 44, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.R. Intrasexual Territoriality in Woodchucks (Marmota monax). J. Mammal. 2004, 85, 1087–1094. [Google Scholar] [CrossRef]
- Chu, D.K.; Leung, C.Y.; Gilbert, M.; Joyner, P.H.; Ng, E.M.; Tse, T.M.; Guan, Y.; Peiris, J.S.; Poon, L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011, 23, 12815–12820. [Google Scholar] [CrossRef]
- Drzewnioková, P.; Festa, F.; Panzarin, V.; Lelli, D.; Moreno, A.; Zecchin, B.; De Benedictis, P.; Leopardi, S. Best Molecular Tools to Investigate Coronavirus Diversity in Mammals: A Comparison. Viruses 2021, 13, 1975. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Hoffmann, B.; Depner, K.; Schirrmeier, H.; Beer, M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 2006, 136, 200–209. [Google Scholar] [CrossRef]
- Di Francesco, C.E.; Di Francesco, D.; Di Martino, B.; Speranza, R.; Santori, D.; Boari, A.; Marsilio, F. Detection by hemi-nested reverse transcription polymerase chain reaction and genetic characterization of wild type strains of Canine distemper virus in suspected infected dogs. J. Vet. Diagn. Investig. 2012, 24, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Senne, D.A.; Myers, T.J.; Bulaga, L.L.; Garber, L.P.; Perdue, M.L.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002, 40, 3256–3260. [Google Scholar] [CrossRef]
- Faccini, S.; De Mattia, A.; Chiapponi, C.; Barbieri, I.; Boniotti, M.B.; Rosignoli, C.; Franzini, G.; Moreno, A.; Foni, E.; Nigrelli, A.D. Development and evaluation of a new Real-Time RT-PCR assay for detection of proposed influenza D virus. J. Virol. Methods 2017, 243, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P. SSE: A nucleotide and amino acid sequence analysis platform. BMC Res. Notes 2012, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- Fulton, R.W.; Step, D.; Wahrmund, J.; Burge, L.J.; Payton, M.E.; Cook, B.J.; Burken, D.; Richards, C.J.; Confer, A.W. Bovine coronavirus (BCV) infections in transported commingled beef cattle and sole-source ranch calves. Can. J. Vet. Res. 2011, 75, 191–199. [Google Scholar] [PubMed]
- Leopardi, S.; Desiato, R.; Mazzucato, M.; Orusa, R.; Obber, F.; Averaimo, D.; Berjaoui, S.; Canziani, S.; Capucchio, M.T.; Conti, R.; et al. One health surveillance strategy for coronaviruses in Italian wildlife. Epidemiol. Infect. 2023, 151, e96. [Google Scholar] [CrossRef]
- Trogu, T.; Canziani, S.; Tolini, C.; Carrera, M.; Giacomelli, S.; Campagnoli, A.; Cordedda, A.; Pedrotti, R.; Nicoloso, A.; Bianchi, A.; et al. Coronaviruses Detection in Italian Alpine Marmots (Marmota marmota). Int. J. Infect. Dis. 2023, 130, S67–S68. [Google Scholar] [CrossRef]
- Gelinas, A.M.; Sasseville, A.M.; Dea, S. Identification of specific variations within the HE, S1, and ORF4 genes of bovine coronaviruses associated with enteric and respiratory diseases in dairy cattle. Adv. Exp. Med. Biol. 2001, 494, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Maes, P.; Van Reeth, K.; Nauwynck, H.; Pensaert, M.; Van Ranst, M. Evolutionary history of the closely related group 2 coronaviruses: Porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006, 80, 7270–7274. [Google Scholar] [CrossRef] [PubMed]
- Saif, L.J.; Jung, K. Comparative Pathogenesis of Bovine and Porcine Respiratory Coronaviruses in the Animal Host Species and SARS-CoV-2 in Humans. J. Clin. Microbiol. 2020, 58, e01355-20. [Google Scholar] [CrossRef] [PubMed]
- Langereis, M.A.; Zeng, Q.; Heesters, B.A.; Huizinga, E.G.; de Groot, R.J. The murine coronavirus hemagglutinin-esterase receptor-binding site: A major shift in ligand specificity through modest changes in architecture. PLoS Pathog. 2012, 8, e1002492. [Google Scholar] [CrossRef]
- Martínez, N.; Brandão, P.E.; de Souza, S.P.; Barrera, M.; Santana, N.; de Arce, H.D.; Pérez, L.J. Molecular and phylogenetic analysis of bovine coronavirus based on the spike glycoprotein gene. Infect. Genet. Evol. 2012, 12, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Workman, A.M.; McDaneld, T.G.; Harhay, G.P.; Das, S.; Loy, J.D.; Hause, B.M. Recent Emergence of Bovine Coronavirus Variants with Mutations in the Hemagglutinin-Esterase Receptor Binding Domain in U.S. Cattle. Viruses 2022, 14, 2125. [Google Scholar] [CrossRef]

| Protein | % of Identity | Strain Name, Country, Year of Isolation | GenBank Accession Number |
|---|---|---|---|
| 1ab | 99.46 | Bovine/ICSA16-EN, France, 2014 | MG757139 |
| 1a | 99.43 | Bovine/ICSA16-EN, France, 2014 | MG757139 |
| 32KDA | 99.51 | Buffalo, Italy/179/07-11, Italy, 2007 | EU019216 |
| HE | 99.53 | Bovine/S3, Ireland, 2022 | OR271241 |
| S | 98.61 | Bovine/RSVPIV3-221026, Ireland, 2022 | PP156987 |
| 4.9KDA | 96.55 | Bovine, B298, China, 2021 | OP866728 |
| 4.8KDA | 97.62 | Capra hircus, CH-4-22-01, USA, 2022 | OP004056 |
| E | 100 | Bovine-ENT, USA | NC003045 |
| M | 99.13 | Bovine, LSV-94LSS-051, USA | P69599 |
| N | 99.33 | Bovine, EPI/Caen/2008/04, France, 2008 | KT318086 |
| I | 97.75 | Bovine, IND/2021/N77, India, 2021 | OP820539 |
| ID | Protocol Number | BCoV Ab ELISA > 75% | BCoV VNT ≥1/10 |
|---|---|---|---|
| 166 | 179,405/1 | 93 | 1/40 |
| 167 | 179,405/2 | 94 | 1/80 |
| 168 | 179,405/3 | 7 | NEG |
| 169 | 179,405/4 | 94 | 1/80 |
| 170 | 179,405/5 | 92 | 1/20 |
| 171 | 179,405/6 | 93 | 1/40 |
| 172 | 179,405/7 | 94 | 1/80 |
| 174 | 179,405/8 | 13 | NEG |
| 176 | 179,405/9 | 70 | NEG |
| 181 | 179,405/10 | 30 | NEG |
| 182 | 179,405/11 | 30 | NEG |
| 183 | 179,405/12 | 9 | NEG |
| 184 | 179,405/13 | 11 | NEG |
| 185 | 179,405/14 | 75 | NEG |
| 186 | 179,405/15 | 25 | NEG |
| 213 | 179,405/16 | 22 | NEG |
| 214 | 179,405/17 | 94 | 1/10 |
| 215 | 179,405/18 | 51 | NEG |
| 216 | 179,405/19 | 75 | NEG |
| 217 | 179,405/20 | 53 | NEG |
| 218 | 179,405/21 | 23 | NEG |
| 219 | 179,405/22 | 14 | NEG |
| 220 | 179,405/23 | 39 | NEG |
| 221 | 179,405/24 | 18 | NEG |
| 222 | 179,405/25 | 38 | NEG |
| 303 | 179,405/26 | 58 | NEG |
| 306 | 179,405/27 | 81 | NEG |
| 307 | 179,405/28 | 71 | NEG |
| 308 | 179,405/29 | 36 | NEG |
| 309 | 179,405/30 | 33 | NEG |
| 310 | 179,405/31 | 64 | NEG |
| 311 | 179,405/32 | 30 | NEG |
| 312 | 179,405/33 | 45 | NEG |
| 313 | 179,405/34 | 53 | NEG |
| 315 | 179,405/35 | 37 | NEG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno, A.; Canziani, S.; Lelli, D.; Castelli, A.; Bianchi, A.; Bertoletti, I.; Maccarinelli, F.; Carlomagno, M.; Paini, M.; Rossato, M.; et al. Molecular and Serological Detection of Bovine Coronaviruses in Marmots (Marmota marmota) in the Alpine Region. Viruses 2024, 16, 591. https://doi.org/10.3390/v16040591
Moreno A, Canziani S, Lelli D, Castelli A, Bianchi A, Bertoletti I, Maccarinelli F, Carlomagno M, Paini M, Rossato M, et al. Molecular and Serological Detection of Bovine Coronaviruses in Marmots (Marmota marmota) in the Alpine Region. Viruses. 2024; 16(4):591. https://doi.org/10.3390/v16040591
Chicago/Turabian StyleMoreno, Ana, Sabrina Canziani, Davide Lelli, Anna Castelli, Alessandro Bianchi, Irene Bertoletti, Federica Maccarinelli, Marco Carlomagno, Matteo Paini, Marzia Rossato, and et al. 2024. "Molecular and Serological Detection of Bovine Coronaviruses in Marmots (Marmota marmota) in the Alpine Region" Viruses 16, no. 4: 591. https://doi.org/10.3390/v16040591






