Host Immune Response Modulation in Avian Coronavirus Infection: Tracheal Transcriptome Profiling In Vitro and In Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Propagation and Titration
2.2. cTEC Preparation
2.3. Infection of cTECs with IBV
2.4. Chickens
2.5. Infections of Chickens with IBV
2.6. Quantification of IBV Viral Genome Load and Host mRNA Gene Expression
2.7. RNA Isolation, cDNA Library Preparations and High-Throughput Sequencing
2.8. RNA-Seq Differential Expression, Gene Ontology (GO), and Pathway Analysis
2.9. Statistical Analysis
3. Results
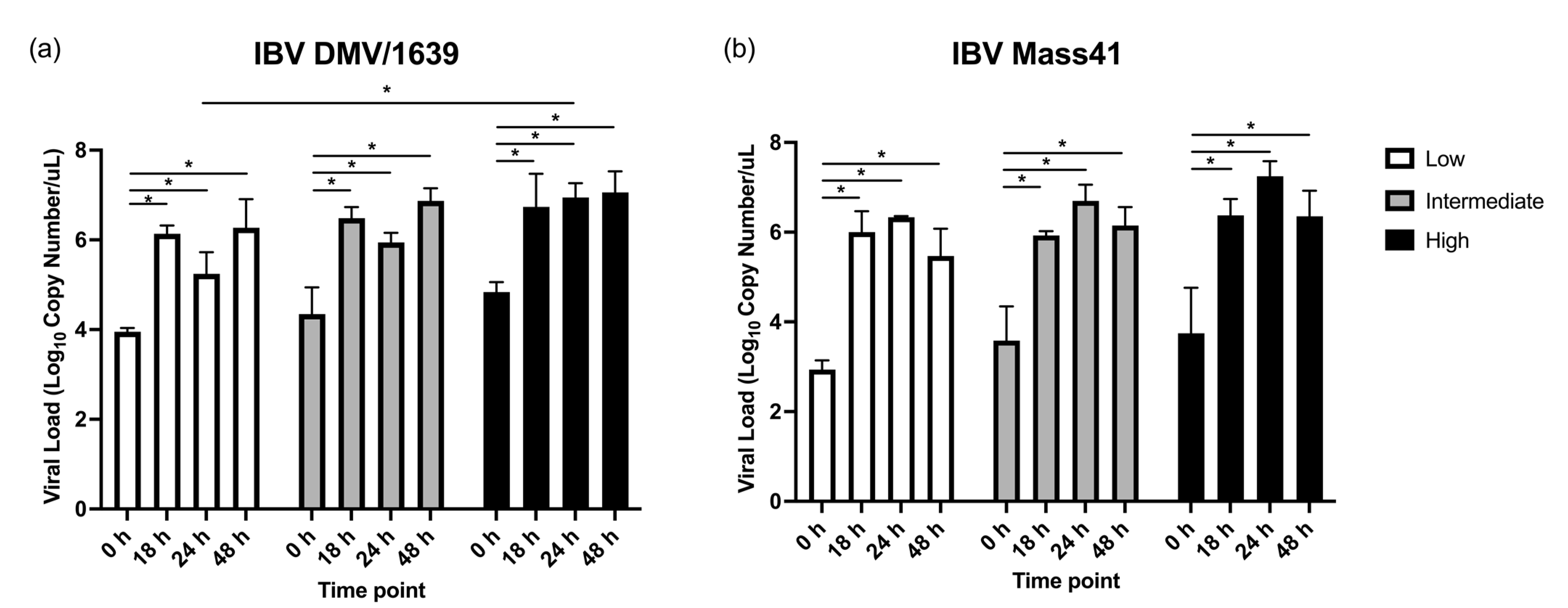
3.1. IBV Genome Load in cTEC Supernatants and the Trachea
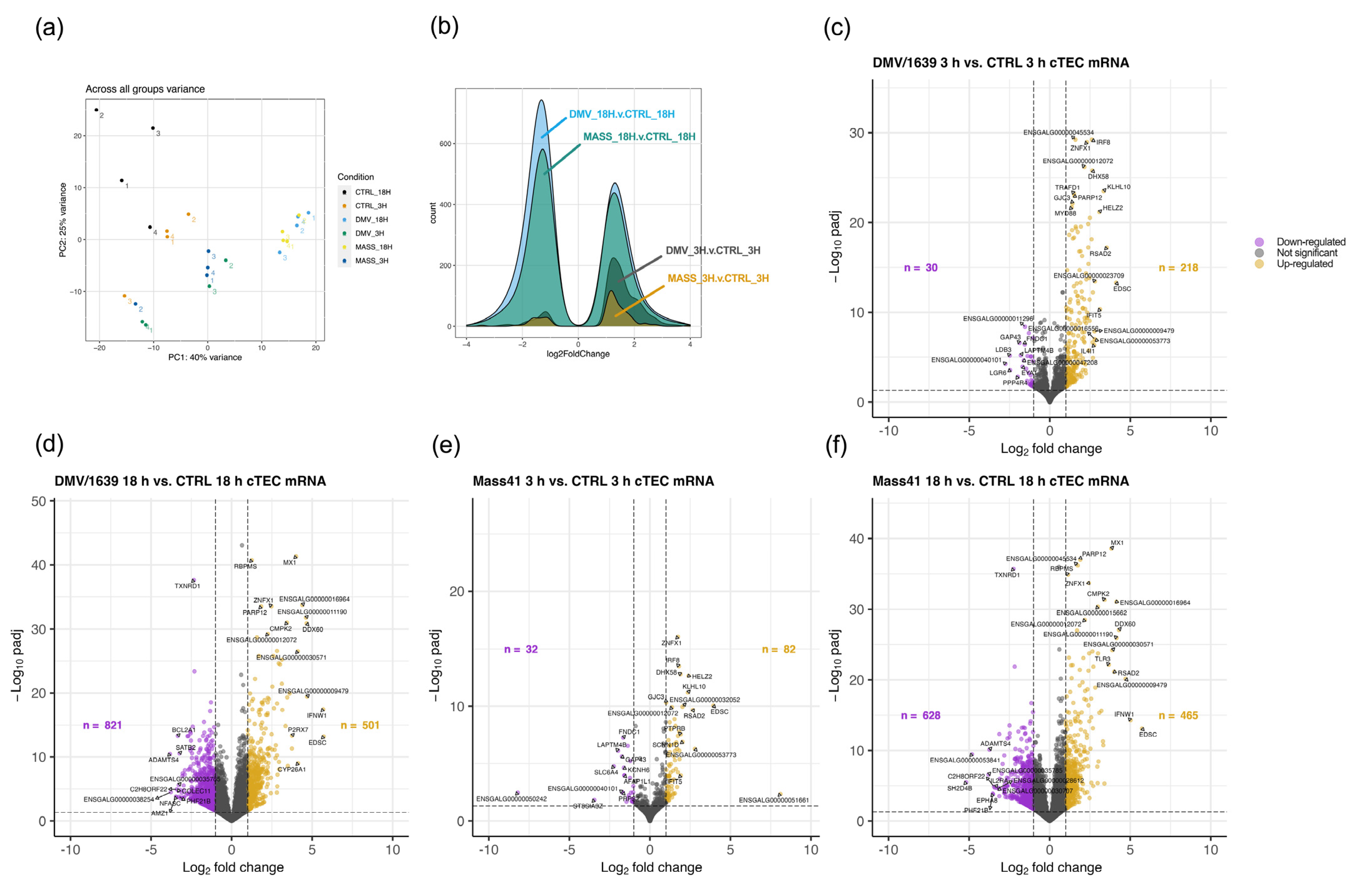
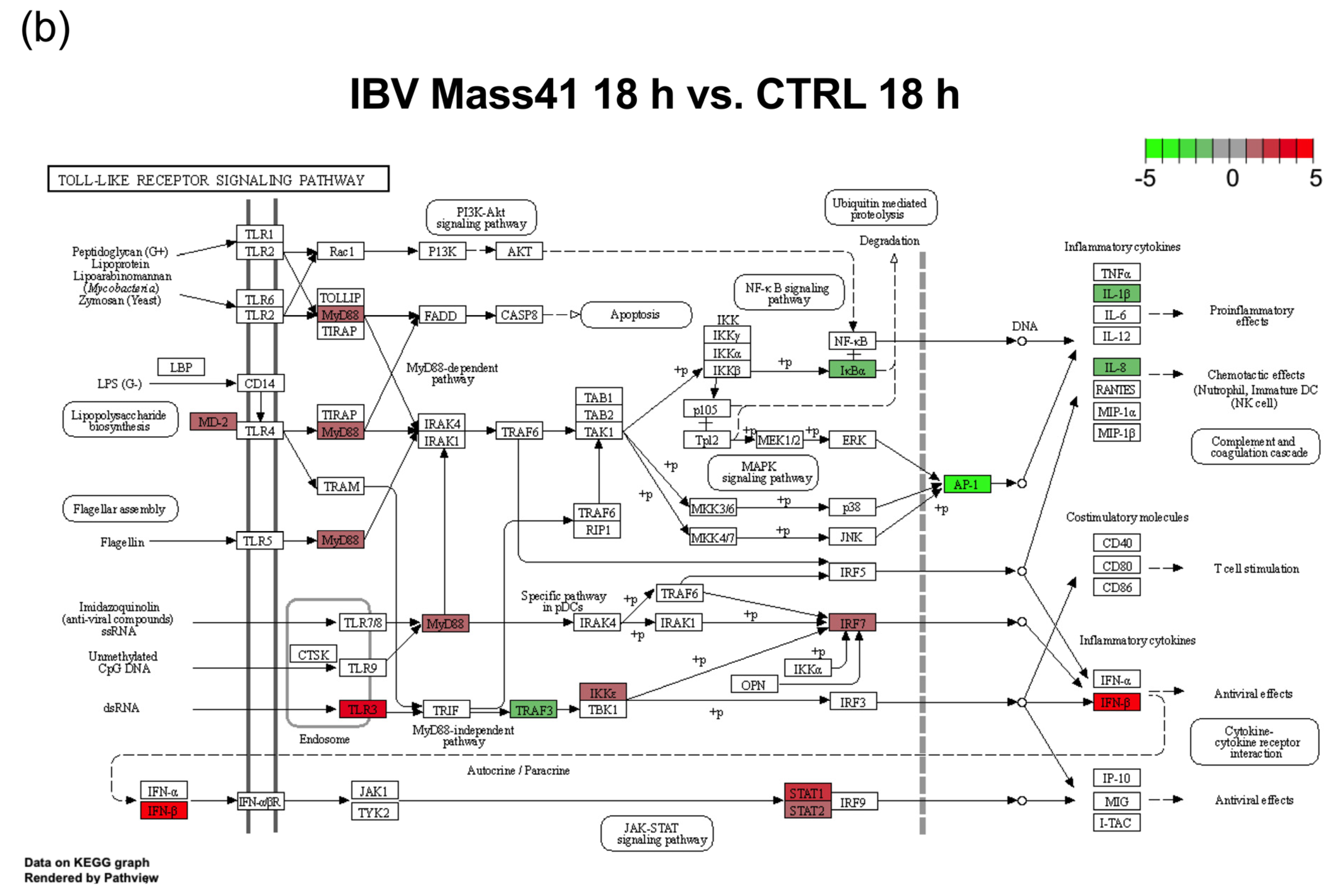
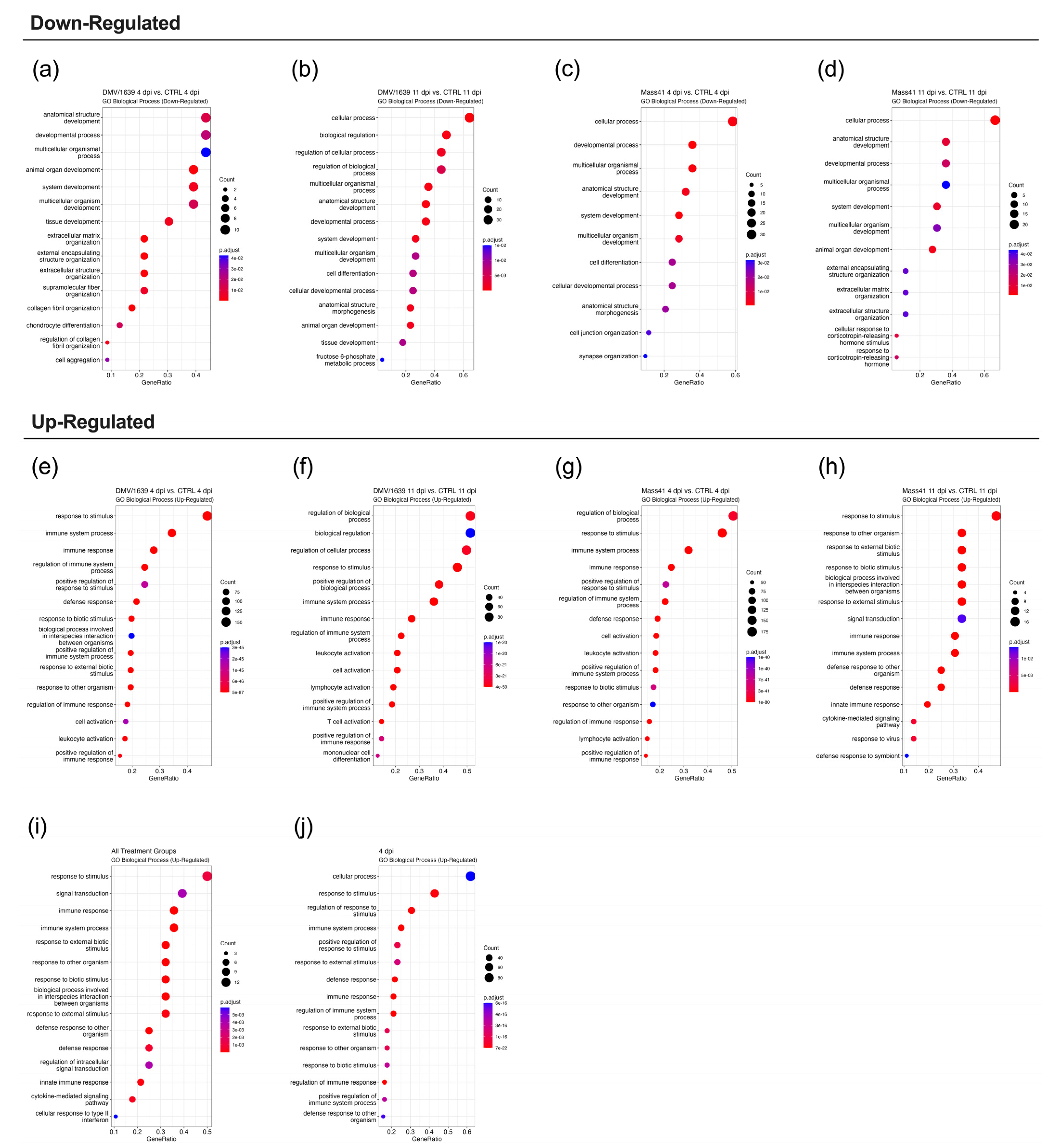
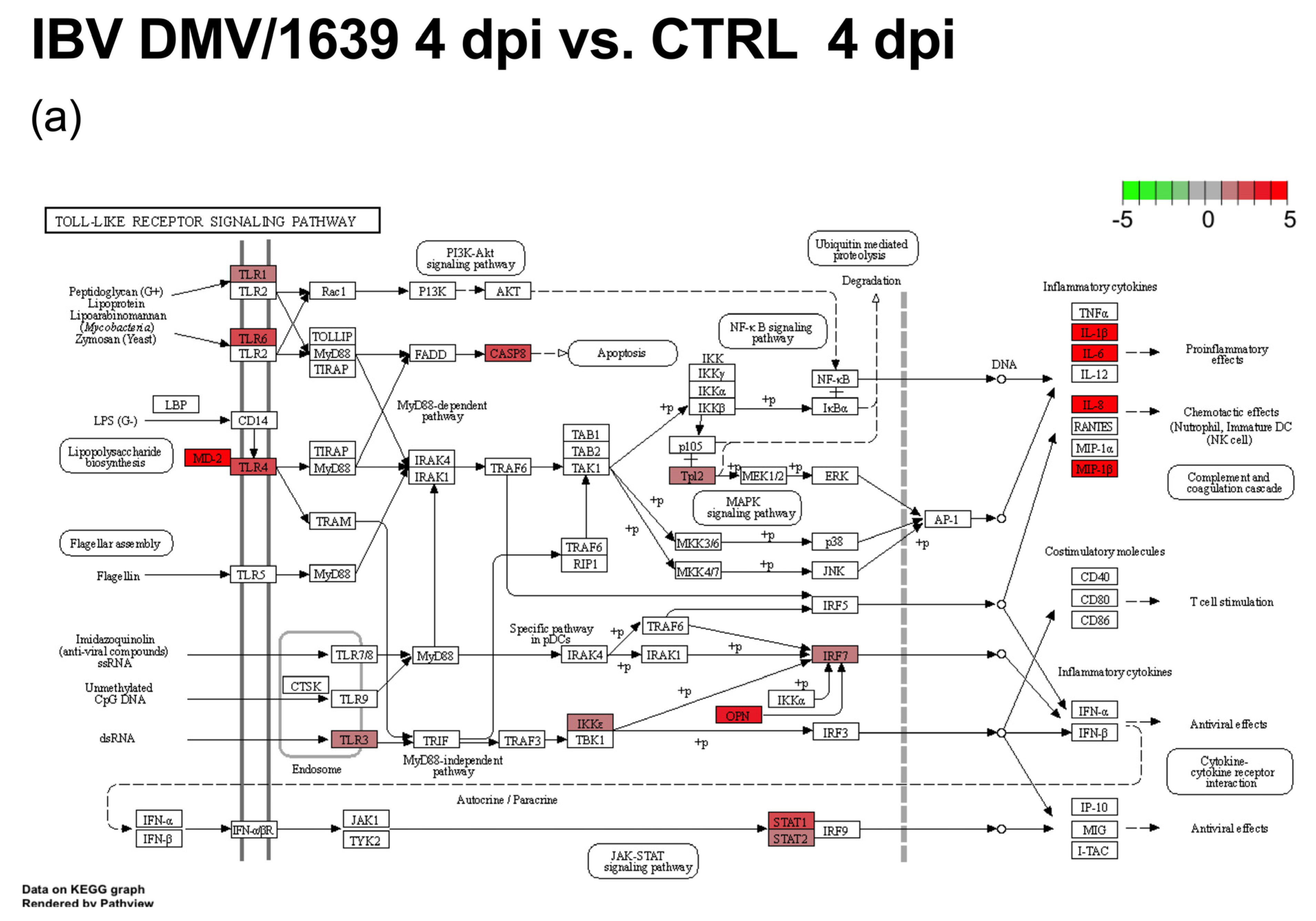
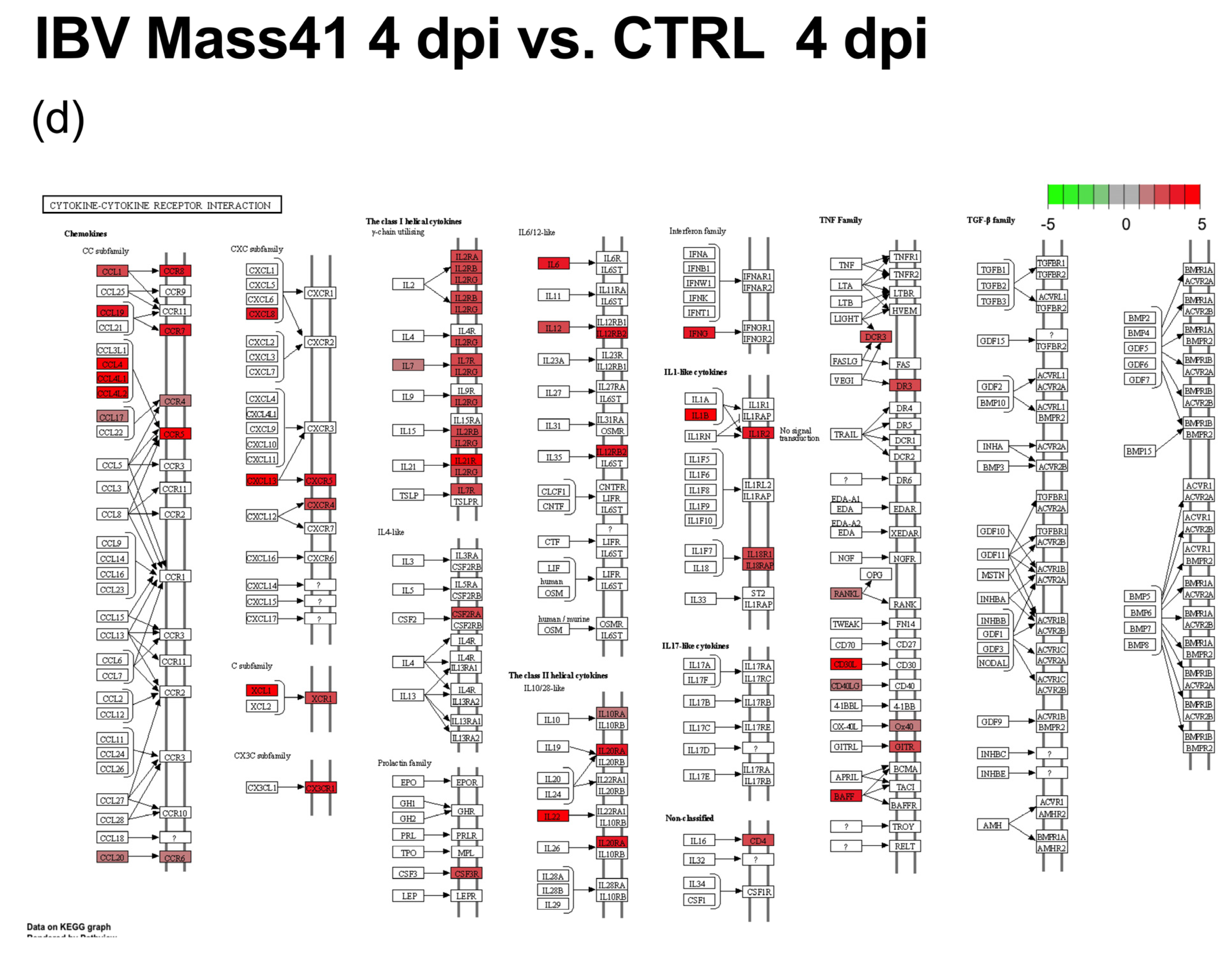
3.2. mRNA Expression and Functional Profiles from cTECs Infected with Different IBV Strains
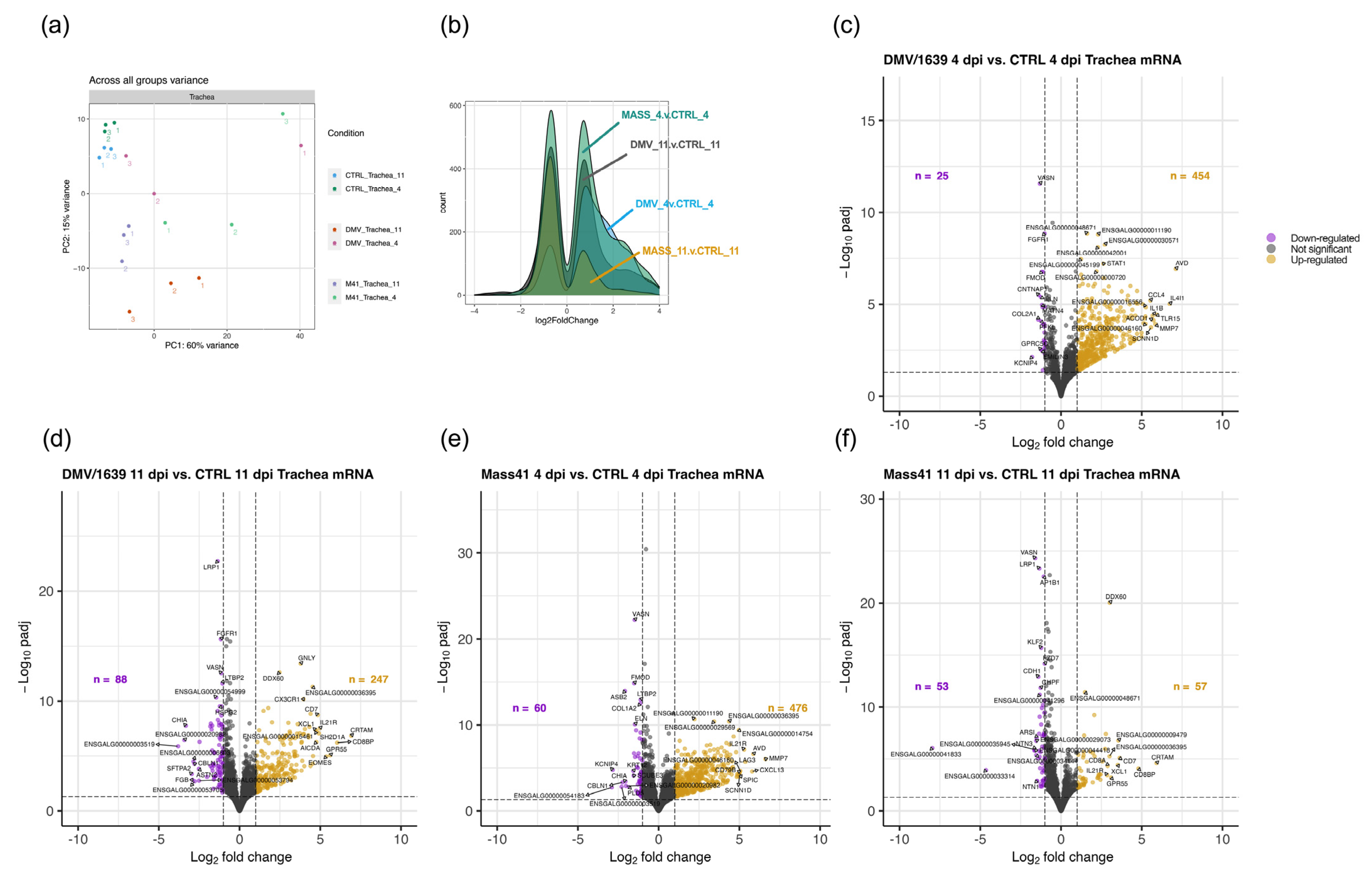
3.3. mRNA Expression and Functional Profiles in the Tracheal Tissues of IBV DMV/1639- and IBV Mass41-Infected Chickens
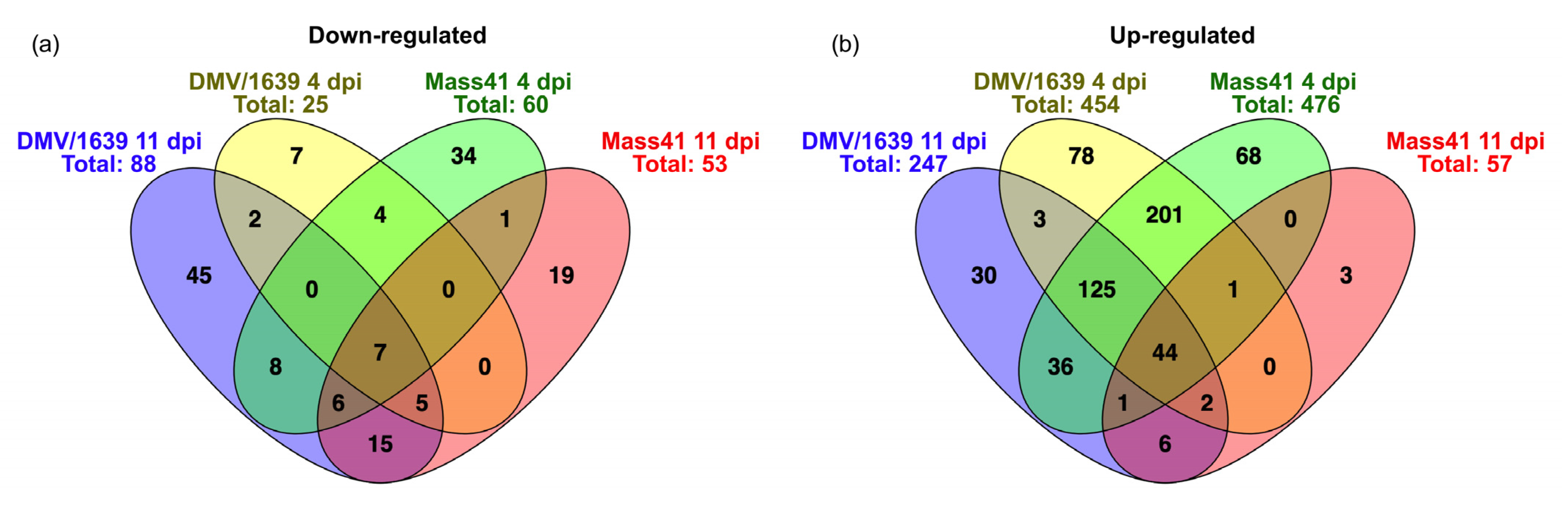
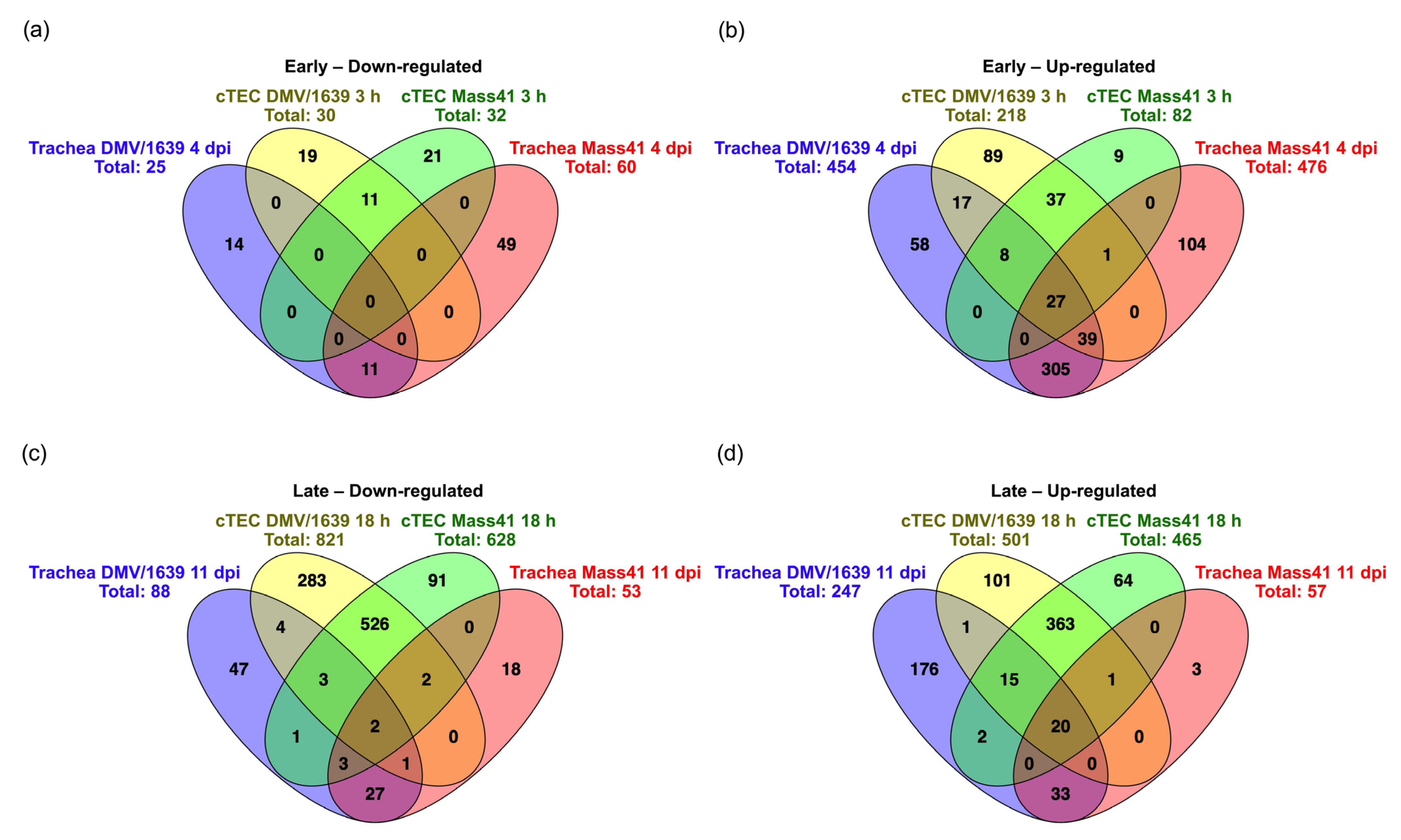
3.4. Comparisons of DE mRNAs in In Vitro and In Vivo Infection Models
3.5. Host mRNA Gene Expression Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jackwood, M.W.; de Wit, S. Infectious Bronchitis. In Diseases of Poultry; Wiley-Blackwell: Hoboken, NJ, USA, 2020; pp. 167–188. [Google Scholar]
- Ignjatovic, J.; Sapats, S. Avian infectious bronchitis virus. Rev. Sci. Tech. 2000, 19, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Moneim, A.S. Coronaviridae: Infectious Bronchitis Virus. In Emerging and Re-Emerging Infectious Diseases of Livestock; Springer International Publishing: Cham, Switzerland, 2017; pp. 133–166. [Google Scholar]
- Hofstad, M.S.; Yoder, H.W., Jr. Avian infectious bronchitis: Virus distribution in tissues of chicks. Avian Dis. 1966, 10, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007, 38, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Najimudeen, S.M.; Hassan, M.S.H.; Cork, S.C.; Abdul-Careem, M.F. Infectious Bronchitis Coronavirus Infection in Chickens: Multiple System Disease with Immune Suppression. Pathogens 2020, 9, 779. [Google Scholar] [CrossRef] [PubMed]
- Najimudeen, S.M.; Abd-Elsalam, R.M.; Ranaweera, H.A.; Isham, I.M.; Hassan, M.S.H.; Farooq, M.; Abdul-Careem, M.F. Replication of infectious bronchitis virus (IBV) Delmarva (DMV)/1639 variant in primary and secondary lymphoid organs leads to immunosuppression in chickens. Virology 2023, 587, 109852. [Google Scholar] [CrossRef] [PubMed]
- Ambali, A.G.; Jones, R.C. Early Pathogenesis in Chicks of Infection with an Enterotropic Strain of Infectious Bronchitis Virus. Avian Dis. 1990, 34, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.Q.; Wang, H.N.; Zhang, Y.; Guan, Z.B.; Wang, T.; Xu, C.W.; Zhang, A.Y.; Yang, X. Comparative dynamic distribution of avian infectious bronchitis virus M41, H120, and SAIBK strains by quantitative real-time RT-PCR in SPF chickens. Biosci. Biotechnol. Biochem. 2012, 76, 2255–2260. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.K.A. Duration of Experimental Infectious Bronchitis in Chickens. Res. Vet. Sci. 1968, 9, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, A.; Abdul-Cader, M.S.; Nazir, S.; De Silva Senapathi, U.; van der Meer, F.; Cork, S.C.; Gomis, S.; Abdul-Careem, M.F. Infectious bronchitis corona virus establishes productive infection in avian macrophages interfering with selected antimicrobial functions. PLoS ONE 2017, 12, e0181801. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.R.; Trus, I.; Desmarets, L.M.; Li, Y.; Theuns, S.; Nauwynck, H.J. Productive replication of nephropathogenic infectious bronchitis virus in peripheral blood monocytic cells, a strategy for viral dissemination and kidney infection in chickens. Vet. Res. 2016, 47, 70. [Google Scholar] [CrossRef] [PubMed]
- Chong, K.T.; Apostolov, K. The pathogenesis of nephritis in chickens induced by infectious bronchitis virus. J. Comp. Pathol. 1982, 92, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Sevoian, M.; Levine, P.P. Effects of Infectious Bronchitis on the Reproductive Tracts, Egg Production, and Egg Quality of Laying Chickens. Avian Dis. 1957, 1, 136–164. [Google Scholar] [CrossRef]
- McMartin, D.A. The pathogenicity of an infectious bronchitis virus for laying hens, with observations on pathogenesis. Br. Vet. J. 1968, 124, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.K.A.; Huggins, M.B. Newly isolated Serotypes of infectious bronchitis virus: Their role in disease. Avian Pathol. 1986, 15, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Sjaak de Wit, J.J.; Cook, J.K.; van der Heijden, H.M. Infectious bronchitis virus variants: A review of the history, current situation and control measures. Avian Pathol. 2011, 40, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Jackwood, M.W.; Hall, D.; Handel, A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012, 12, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Jordan, B. Vaccination against infectious bronchitis virus: A continuous challenge. Vet. Microbiol. 2017, 206, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Gelb, J., Jr.; Ladman, B.S.; Pope, C.R.; Ruano, J.M.; Brannick, E.M.; Bautista, D.A.; Coughlin, C.M.; Preskenis, L.A. Characterization of nephropathogenic infectious bronchitis virus DMV/1639/11 recovered from Delmarva broiler chickens in 2011. Avian Dis. 2013, 57, 65–70. [Google Scholar] [CrossRef]
- Hassan, M.S.H.; Ojkic, D.; Coffin, C.S.; Cork, S.C.; van der Meer, F.; Abdul-Careem, M.F. Delmarva (DMV/1639) Infectious Bronchitis Virus (IBV) Variants Isolated in Eastern Canada Show Evidence of Recombination. Viruses 2019, 11, 1054. [Google Scholar] [CrossRef]
- Martin, E.; Brash, M.; Stalker, M.; Ojkić, D. Using phylogenetic analysis to examine the changing strains of infectious bronchitis virus infections in Ontario over time. In Proceedings of the 16th Annual Meeting of the Canadian Animal Health Laboratorians Network, Guelph, ON, Canada, 4–7 June 2017. [Google Scholar]
- Brochu Morin, M.-E.; Chénier, S.; Bergeron, N. La Bronchite Infectieuse Aviaire au Québec (Avian Infectious Bronchitis in Quebec). Bulletin Zoosanitaire: Réseau d’alerte et D’information Zoosanitaire (RAIZO); Ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec: Québec, QC, Canada, 2018. [Google Scholar]
- Hassan, M.S.H.; Ali, A.; Buharideen, S.M.; Goldsmith, D.; Coffin, C.S.; Cork, S.C.; van der Meer, F.; Boulianne, M.; Abdul-Careem, M.F. Pathogenicity of the Canadian Delmarva (DMV/1639) Infectious Bronchitis Virus (IBV) on Female Reproductive Tract of Chickens. Viruses 2021, 13, 2488. [Google Scholar] [CrossRef]
- Hassan, M.S.H.; Najimudeen, S.M.; Ali, A.; Altakrouni, D.; Goldsmith, D.; Coffin, C.S.; Cork, S.C.; van der Meer, F.; Abdul-Careem, M.F. Immunopathogenesis of the Canadian Delmarva (DMV/1639) infectious bronchitis virus (IBV): Impact on the reproductive tract in layers. Microb. Pathog. 2022, 166, 105513. [Google Scholar] [CrossRef] [PubMed]
- Mcmartin, D.A.; Macleod, H. Abnormal Oviducts in Laying Hens Following Natural Infection with Infectious Bronchitis at an Early Age. Brit Vet. J. 1972, 128, xix–xxi. [Google Scholar]
- Jones, R.C.; Jordan, F.T. The exposure of day-old chicks to infectious bronchitis and the subsequent development of the oviduct. Vet. Rec. 1970, 87, 504–505. [Google Scholar] [CrossRef] [PubMed]
- Crinion, R.A.P.; Hofstad, M.S. Pathogenicity of Two Embryo-Passage Levels of Avian Infectious Bronchitis Virus for the Oviduct of Young Chickens of Various Ages. Avian Dis. 1972, 16, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.A. Respiratory epithelial cells as master communicators during viral infections. Curr. Clin. Microbiol. Rep. 2019, 6, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, R.J.; Lloyd, C.M. Regulation of immune responses by the airway epithelial cell landscape. Nat. Rev. Immunol. 2021, 21, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.E.; Ward, A.C.; Lowenthal, J.W.; Bean, A.G. Chicken interferons, their receptors and interferon-stimulated genes. Dev. Comp. Immunol. 2013, 41, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Santhakumar, D.; Rubbenstroth, D.; Martinez-Sobrido, L.; Munir, M. Avian Interferons and Their Antiviral Effectors. Front. Immunol. 2017, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Li, M.M.H. All About the RNA: Interferon-Stimulated Genes That Interfere With Viral RNA Processes. Front. Immunol. 2020, 11, 605024. [Google Scholar] [CrossRef] [PubMed]
- Kint, J.; Fernandez-Gutierrez, M.; Maier, H.J.; Britton, P.; Langereis, M.A.; Koumans, J.; Wiegertjes, G.F.; Forlenza, M. Activation of the chicken type I interferon response by infectious bronchitis coronavirus. J. Virol. 2015, 89, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.; Munir, S.; Vishwanathan, S.; Manuja, A.; Griebel, P.; Tikoo, S.; Townsend, H.; Potter, A.; Kapur, V.; Babiuk, L.A. Transcriptional analysis of avian embryonic tissues following infection with avian infectious bronchitis virus. Virus Res. 2005, 110, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Rosa, A.J.M.; Oliverira, H.N.; Rosa, G.J.M.; Guo, X.; Travnicek, M.; Girshick, T. Transcriptome of Local Innate and Adaptive Immunity during Early Phase of Infectious Bronchitis Viral Infection. Viral Immunol. 2006, 19, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Rosa, A.J.; Chen, D.G.; Wang, X. Molecular mechanisms of primary and secondary mucosal immunity using avian infectious bronchitis virus as a model system. Vet. Immunol. Immunopathol. 2008, 121, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Hamzić, E.; Kjaerup, R.B.; Mach, N.; Minozzi, G.; Strozzi, F.; Gualdi, V.; Williams, J.L.; Chen, J.; Wattrang, E.; Buitenhuis, B.; et al. RNA sequencing-based analysis of the spleen transcriptome following infectious bronchitis virus infection of chickens selected for different mannose-binding lectin serum concentrations. BMC Genom. 2016, 17, 82. [Google Scholar] [CrossRef] [PubMed]
- Kemp, V.; Laconi, A.; Cocciolo, G.; Berends, A.J.; Breit, T.M.; Verheije, M.H. miRNA repertoire and host immune factor regulation upon avian coronavirus infection in eggs. Arch. Virol. 2020, 165, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Chuwatthanakhajorn, S.; Chang, C.S.; Ganapathy, K.; Tang, P.C.; Chen, C.F. Comparison of Immune-Related Gene Expression in Two Chicken Breeds Following Infectious Bronchitis Virus Vaccination. Animals 2023, 13, 1642. [Google Scholar] [CrossRef]
- Smith, J.; Sadeyen, J.R.; Cavanagh, D.; Kaiser, P.; Burt, D.W. The early immune response to infection of chickens with Infectious Bronchitis Virus (IBV) in susceptible and resistant birds. BMC Vet. Res. 2015, 11, 256. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.P.D.; Hauck, R.; Kern, C.; Wang, Y.; Zhou, H.; Gallardo, R.A. Effects of Chicken MHC Haplotype on Resistance to Distantly Related Infectious Bronchitis Viruses. Avian Dis. 2019, 63, 310–317. [Google Scholar] [CrossRef]
- Ghobadian Diali, H.; Hosseini, H.; Fallah Mehrabadi, M.H.; Yahyaraeyat, R.; Ghalyanchilangeroudi, A. Evaluation of viral load and transcriptome changes in tracheal tissue of two hybrids of commercial broiler chickens infected with avian infectious bronchitis virus: A comparative study. Arch. Virol. 2022, 167, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.X.; Yang, B.; Fung, T.S.; Chen, R.A.; Liu, D.X. Transcriptomic analysis reveals crucial regulatory roles of immediate-early response genes and related signaling pathways in coronavirus infectious bronchitis virus infection. Virology 2022, 575, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cong, F.; Liu, X.; Han, Z.; Shao, Y.; Kong, X.; Liu, S. Transcriptome analysis of chicken kidney tissues following coronavirus avian infectious bronchitis virus infection. BMC Genom. 2013, 14, 743. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Liu, P.; Zhou, C.; Shi, Y.; Wu, Q.; Yang, Y.; Li, G.; Hu, G.; Guo, X. A Multi-Omics Study of Chicken Infected by Nephropathogenic Infectious Bronchitis Virus. Viruses 2019, 11, 1070. [Google Scholar] [CrossRef] [PubMed]
- Dinan, A.M.; Keep, S.; Bickerton, E.; Britton, P.; Firth, A.E.; Brierley, I. Comparative Analysis of Gene Expression in Virulent and Attenuated Strains of Infectious Bronchitis Virus at Subcodon Resolution. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, X.; Zhang, Z.; Li, J.; Zou, W.; Zeng, F.; Wang, H. Comparative transcriptome analysis reveals induction of apoptosis in chicken kidney cells associated with the virulence of nephropathogenic infectious bronchitis virus. Microb. Pathog. 2017, 113, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Jung, J.S.; Yeo, J.I.; Kwon, H.M.; Park, J. Transcriptome analysis of primary chicken cells infected with infectious bronchitis virus strain K047-12 isolated in Korea. Arch. Virol. 2021, 166, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, Z.; Wang, J.; Yang, Q. Microarray analysis of infectious bronchitis virus infection of chicken primary dendritic cells. BMC Genom. 2019, 20, 557. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Cao, Y.; Wang, Z.; Shah, A.U.; Wang, W.; Dai, C.; Chen, M.; Lin, J.; Yang, Q. The mechanism of antigen-presentation of avian bone marrowed dendritic cells suppressed by infectious bronchitis virus. Genomics 2021, 113, 1719–1732. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cui, P.; Fu, X.; Zhang, L.; Yan, W.; Zhai, Y.; Lei, C.; Wang, H.; Yang, X. Identification and analysis of long non-coding RNAs and mRNAs in chicken macrophages infected with avian infectious bronchitis coronavirus. BMC Genom. 2021, 22, 67. [Google Scholar] [CrossRef]
- Yang, Q.; Gong, H.; Liu, S.; Huang, S.; Yan, W.; Wang, K.; Li, H.; Lei, C.-W.; Wang, H.-N.; Yang, X. Differential analysis of IBV-infected primary chicken embryonic fibroblasts and immortalized DF-1. Microbiol. Spectr. 2024, 12, e02402–e02423. [Google Scholar] [CrossRef] [PubMed]
- Jackwood, M.W.; Jordan, B.J. Molecular Evolution of Infectious Bronchitis Virus and the Emergence of Variant Viruses Circulating in the United States. Avian Dis. 2021, 65, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Moneim, A.S.; Zlotowski, P.; Veits, J.; Keil, G.M.; Teifke, J.P. Immunohistochemistry for detection of avian infectious bronchitis virus strain M41 in the proventriculus and nervous system of experimentally infected chicken embryos. Virol. J. 2009, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, A.; Abdul-Cader, M.S.; Almatrouk, Z.; van der Meer, F.; Cork, S.C.; Gomis, S.; Abdul-Careem, M.F. Induction of innate host responses characterized by production of interleukin (IL)-1beta and recruitment of macrophages to the respiratory tract of chickens following infection with infectious bronchitis virus (IBV). Vet. Microbiol. 2018, 215, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Abd-Elsalam, R.M.; Ratcliff, N.; Hassan, M.S.H.; Najimudeen, S.M.; Cork, S.C.; Checkley, S.; Niu, Y.D.; Abdul-Careem, M.F. Comparative pathogenicity of infectious bronchitis virus Massachusetts and Delmarva (DMV/1639) genotypes in laying hens. Front. Vet. Sci. 2023, 10, 1329430. [Google Scholar] [CrossRef] [PubMed]
- Mueller Slay, A.; Franca, M.; Jackwood, M.; Jordan, B. Infection with IBV DMV/1639 at a Young Age Leads to Increased Incidence of Cystic Oviduct Formation Associated with False Layer Syndrome. Viruses 2022, 14, 852. [Google Scholar] [CrossRef] [PubMed]
- Kint, J.; Maier, H.J.; Jagt, E. Quantification of infectious bronchitis coronavirus by titration in vitro and in ovo. Methods Mol. Biol. 2015, 1282, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Payment, P.; Trudel, M. Isolement et identification des virus. In Manuel de Techniques Virologiques, 1st ed.; Presses de l’Université du Québec: Québec, QC, Canada, 1989; pp. 21–44. [Google Scholar]
- Reed, L.J.; Muench, H. A Simple Method for Estimating Fifty Per Cent Endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Barjesteh, N.; Taha-Abdelaziz, K.; Kulkarni, R.R.; Sharif, S. Innate antiviral responses are induced by TLR3 and TLR4 ligands in chicken tracheal epithelial cells: Communication between epithelial cells and macrophages. Virology 2019, 534, 132–142. [Google Scholar] [CrossRef]
- Shen, C.I.; Wang, C.H.; Liao, J.W.; Hsu, T.W.; Kuo, S.M.; Su, H.L. The infection of primary avian tracheal epithelial cells with infectious bronchitis virus. Vet. Res. 2010, 41, 6. [Google Scholar] [CrossRef] [PubMed]
- Zaffuto, K.M.; Estevez, C.N.; Afonso, C.L. Primary chicken tracheal cell culture system for the study of infection with avian respiratory viruses. Avian Pathol. 2008, 37, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Pajtler, K.; Bohrer, A.; Maurer, J.; Schorle, H.; Schramm, A.; Eggert, A.; Schulte, J.H. Production of chick embryo extract for the cultivation of murine neural crest stem cells. J. Vis. Exp. 2010, 45, e2380. [Google Scholar] [CrossRef]
- Kameka, A.M.; Haddadi, S.; Kim, D.S.; Cork, S.C.; Abdul-Careem, M.F. Induction of innate immune response following infectious bronchitis corona virus infection in the respiratory tract of chickens. Virology 2014, 450–451, 114–121. [Google Scholar] [CrossRef] [PubMed]
- St Paul, M.; Mallick, A.I.; Haq, K.; Orouji, S.; Abdul-Careem, M.F.; Sharif, S. In vivo administration of ligands for chicken toll-like receptors 4 and 21 induces the expression of immune system genes in the spleen. Vet. Immunol. Immunopathol. 2011, 144, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yin, Y.; Lin, Z.; Wen, H.; Chen, J.; Luo, W. Transcriptome profile analysis reveals KLHL30 as an essential regulator for myoblast differentiation. Biochem. Biophys. Res. Commun. 2021, 559, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Cui, C.; Han, S.; Chen, Y.; Zhao, J.; He, H.; Li, D.; Zhu, Q. Fibromodulin Modulates Chicken Skeletal Muscle Development via the Transforming Growth Factor-beta Signaling Pathway. Animals 2020, 10, 1477. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.I.; Kulkarni, R.R.; Sharif, S. Synthetic double-stranded RNA oligonucleotides are immunostimulatory for chicken spleen cells. Dev. Comp. Immunol. 2011, 35, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Huang, M.; Qiu, X.; Zhi, X.; Liang, L.; Sun, Y.; Liao, Y.; Song, C.; Ren, T.; Ding, C. Chicken-derived MERTK protein inhibits Newcastle disease virus replication by increasing STAT1 phosphorylation in DF-1 cells. Virus Res. 2023, 326, 199065. [Google Scholar] [CrossRef] [PubMed]
- Sarson, A.J.; Parvizi, P.; Lepp, D.; Quinton, M.; Sharif, S. Transcriptional analysis of host responses to Marek’s disease virus infection in genetically resistant and susceptible chickens. Anim. Genet. 2008, 39, 232–240. [Google Scholar] [CrossRef]
- Giotis, E.S.; Robey, R.C.; Skinner, N.G.; Tomlinson, C.D.; Goodbourn, S.; Skinner, M.A. Chicken interferome: Avian interferon-stimulated genes identified by microarray and RNA-seq of primary chick embryo fibroblasts treated with a chicken type I interferon (IFN-alpha). Vet. Res. 2016, 47, 75. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bourgey, M.; Dali, R.; Eveleigh, R.; Chen, K.C.; Letourneau, L.; Fillon, J.; Michaud, M.; Caron, M.; Sandoval, J.; Lefebvre, F.; et al. GenPipes: An open-source framework for distributed and scalable genomic analyses. Gigascience 2019, 8, giz037. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing. 2021. Available online: https://www.R-project.org/ (accessed on 4 January 2022).
- Team, R. RStudio: Integrated Development for R. 2020. Available online: http://www.rstudio.com/ (accessed on 4 January 2022).
- Xie, Y. knitr: A General-Purpose Package for Dynamic Report Generation in R. 2021. Available online: https://yihui.org/knitr/ (accessed on 4 January 2022).
- Slowikowski, K. ggrepel: Automatically Position Non-Overlapping Text Labels with ‘ggplot2’. 2023. Available online: https://github.com/slowkow/ggrepel (accessed on 26 August 2023).
- Müller, K.; Wickham, H. tibble: Simple Data Frames. 2023. Available online: https://github.com/tidyverse/tibble (accessed on 26 August 2023).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Bache, S.; Wickham, H.; Henry, L.; Henry, M.L. magrittr: A Forward-Pipe Operator for R. 2022. Available online: https://magrittr.tidyverse.org (accessed on 26 August 2023).
- Sjoberg, D. Hablar: Non-Astonishing Results in R. 2023. Available online: https://davidsjoberg.github.io/ (accessed on 26 August 2023).
- Zhu, H. kableExtra: Construct Complex Table with ‘kable’ and Pipe Syntax. 2019. Available online: http://haozhu233.github.io/kableExtra/ (accessed on 26 August 2023).
- DeLuca, D.S.; Levin, J.Z.; Sivachenko, A.; Fennell, T.; Nazaire, M.D.; Williams, C.; Reich, M.; Winckler, W.; Getz, G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012, 28, 1530–1532. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M. False discovery rates: A new deal. Biostatistics 2016, 18, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Mangiola, S.; Papenfuss, A. tidyHeatmap: An R package for modular heatmap production based on tidy principles. J. Open Source Softw. 2020, 5, 2472. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. R Package Version 1.18.0. 2023. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 26 August 2023).
- Venny, J.C.O. An interactive tool for comparing lists with Venn’s diagrams. 2007–2015. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 9 January 2024).
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. gprofiler2—An R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res 2020, 9, ELIXIR-709. [Google Scholar] [CrossRef] [PubMed]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Yu, G. Enrichplot: Visualization of Functional Enrichment Result. R Package Version 1.21.3. 2023. Available online: https://bioconductor.org/packages/enrichplot (accessed on 26 August 2023).
- Yu, G.; Wang, L.G.; Yan, G.R.; He, Q.Y. DOSE: An R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015, 31, 608–609. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Grote, S. GOfuncR: Gene ontology enrichment using FUNC. R package version 1.20.0. 2023. Available online: https://bioconductor.org/packages/release/bioc/html/GOfuncR.html (accessed on 26 August 2023).
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M. org.Gg.eg.db: Genome wide annotation for Chicken. R package version 3.8.2. 2019. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Gg.eg.db.html (accessed on 26 August 2023).
- Cook, J.K.; Darbyshire, J.H.; Peters, R.W. The use of chicken tracheal organ cultures for the isolation and assay of avian infectious bronchitis virus. Arch. Virol. 1976, 50, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.K.; Darbyshire, J.H.; Peters, R.W. Growth kinetic studies of avian infectious bronchitis virus in tracheal organ cultures. Res. Vet. Sci. 1976, 20, 348–349. [Google Scholar] [CrossRef] [PubMed]
- Holmes, H.C. The resistance to reinfection of tracheal organ cultures from chickens previously infected with avian infections bronchitis virus. Res. Vet. Sci. 1978, 25, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Colwell, W.M.; Lukert, P.D. Effects of Avian Infectious Bronchitis Virus (IBV) on Tracheal Organ Cultures. Avian Dis. 1969, 13, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Weerts, E.; Bouwman, K.M.; Paerels, L.; Grone, A.; Jan Boelm, G.; Verheije, M.H. Interference between avian corona and influenza viruses: The role of the epithelial architecture of the chicken trachea. Vet. Microbiol. 2022, 272, 109499. [Google Scholar] [CrossRef] [PubMed]
- Steyn, A.; Keep, S.; Bickerton, E.; Fife, M. The Characterization of chIFITMs in Avian Coronavirus Infection In Vivo, Ex Vivo and In Vitro. Genes 2020, 11, 918. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cai, H.; Li, Q.; Fang, C.; Peng, L.; Lan, J.; Zhou, J.; Liao, M. Identification of Host Proteins Interacting with IBV S1 Based on Tracheal Organ Culture. Viruses 2023, 15, 1216. [Google Scholar] [CrossRef] [PubMed]
- Chandra, M. Comparative nephropathogenicity of different strains of infectious bronchitis virus in chickens. Poult. Sci. 1987, 66, 954–959. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Huo, C.; Zhao, J.; Liu, T.; Li, X.; Yan, S.; Wang, Z.; Hu, Y.; Zhang, G. Pathogenicity differences between QX-like and Mass-type infectious bronchitis viruses. Vet. Microbiol. 2018, 213, 129–135. [Google Scholar] [CrossRef] [PubMed]
- De Wit, J.J. Detection of infectious bronchitis virus. Avian Pathol. 2000, 29, 71–93. [Google Scholar] [CrossRef] [PubMed]
- Barjesteh, N.; Alkie, T.N.; Hodgins, D.C.; Nagy, E.; Sharif, S. Local Innate Responses to TLR Ligands in the Chicken Trachea. Viruses 2016, 8, 207. [Google Scholar] [CrossRef]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef] [PubMed]
- Barjesteh, N.; O’Dowd, K.; Vahedi, S.M. Antiviral responses against chicken respiratory infections: Focus on avian influenza virus and infectious bronchitis virus. Cytokine 2020, 127, 154961. [Google Scholar] [CrossRef] [PubMed]
- Keestra, A.M.; de Zoete, M.R.; Bouwman, L.I.; Vaezirad, M.M.; van Putten, J.P. Unique features of chicken Toll-like receptors. Dev. Comp. Immunol. 2013, 41, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Karpala, A.J.; Lowenthal, J.W.; Bean, A.G. Activation of the TLR3 pathway regulates IFNbeta production in chickens. Dev. Comp. Immunol. 2008, 32, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Park, Y.H.; Chungu, K.; Woo, S.J.; Han, S.T.; Choi, H.J.; Rengaraj, D.; Han, J.Y. Targeted Knockout of MDA5 and TLR3 in the DF-1 Chicken Fibroblast Cell Line Impairs Innate Immune Response Against RNA Ligands. Front. Immunol. 2020, 11, 678. [Google Scholar] [CrossRef]
- Keestra, A.M.; de Zoete, M.R.; Bouwman, L.I.; van Putten, J.P. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J. Immunol. 2010, 185, 460–467. [Google Scholar] [CrossRef]
- Brownlie, R.; Zhu, J.; Allan, B.; Mutwiri, G.K.; Babiuk, L.A.; Potter, A.; Griebel, P. Chicken TLR21 acts as a functional homologue to mammalian TLR9 in the recognition of CpG oligodeoxynucleotides. Mol. Immunol. 2009, 46, 3163–3170. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.; Potter, A.; Tikoo, S.; Gerdts, V.; Lai, K.; Babiuk, L.A.; Mutwiri, G. CpG oligodeoxynucleotides activate innate immune response that suppresses infectious bronchitis virus replication in chicken embryos. Avian Dis. 2009, 53, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zheng, X.; Zhang, Y.; Wang, R.; Wei, X. Comparative proteomics analysis of kidney in chicken infected by infectious bronchitis virus. Poult. Sci. 2023, 103, 103259. [Google Scholar] [CrossRef] [PubMed]
- Santhakumar, D.; Rohaim, M.; Hussein, H.A.; Hawes, P.; Ferreira, H.L.; Behboudi, S.; Iqbal, M.; Nair, V.; Arns, C.W.; Munir, M. Chicken Interferon-induced Protein with Tetratricopeptide Repeats 5 Antagonizes Replication of RNA Viruses. Sci. Rep. 2018, 8, 6794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, X.; Chen, W.; Chen, L. IFIT5 potentiates anti-viral response through enhancing innate immune signaling pathways. Acta Biochim. Biophys. Sin. 2013, 45, 867–874. [Google Scholar] [CrossRef]
- Jang, H.; Koo, B.S.; Jeon, E.O.; Lee, H.R.; Lee, S.M.; Mo, I.P. Altered pro-inflammatory cytokine mRNA levels in chickens infected with infectious bronchitis virus. Poult. Sci. 2013, 92, 2290–2298. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Qi, J.; Sun, L.; Zhao, J.; Zhang, G.; Zhao, Y. Pathological effect of different avian infectious bronchitis virus strains on the bursa of Fabricius of chickens. Avian Pathol. 2022, 51, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Najimudeen, S.M.; Barboza-Solis, C.; Ali, A.; Buharideen, S.M.; Isham, I.M.; Hassan, M.S.; Ojkic, D.; Van Marle, G.; Cork, S.C.; van der Meer, F.; et al. Pathogenesis and host responses in lungs and kidneys following Canadian 4/91 infectious bronchitis virus (IBV) infection in chickens. Virology 2022, 566, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, D.; Jia, Z.; Chang, J.; Hou, X. Cellular immune response in chickens infected with avian infectious bronchitis virus (IBV). Eur. J. Inflamm. 2017, 15, 35–41. [Google Scholar] [CrossRef]
- Ramirez, H.; Patel, S.B.; Pastar, I. The Role of TGFbeta Signaling in Wound Epithelialization. Adv. Wound Care 2014, 3, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Kitsis, R.N.; Leinwand, L.A. Discordance between gene regulation in vitro and in vivo. Gene Expr. 1992, 2, 313–318. [Google Scholar] [PubMed]
- McMahon, M.; Ye, S.; Pedrina, J.; Dlugolenski, D.; Stambas, J. Extracellular Matrix Enzymes and Immune Cell Biology. Front. Mol. Biosci. 2021, 8, 703868. [Google Scholar] [CrossRef] [PubMed]
- Kozyrina, A.N.; Piskova, T.; Di Russo, J. Mechanobiology of Epithelia From the Perspective of Extracellular Matrix Heterogeneity. Front. Bioeng. Biotechnol. 2020, 8, 596599. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.Y.; Solanki, H.S.; Advani, J.; Khan, A.A.; Keshava Prasad, T.S.; Gowda, H.; Thiyagarajan, S.; Chatterjee, A. Comprehensive network map of interferon gamma signaling. J. Cell Commun. Signal 2018, 12, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Gu, K.; Li, H.; Zhao, Y.; Li, C.; Wen, R.; Zhou, C.; Lei, C.; Yang, X.; Wang, H. Infectious Bronchitis Virus Nsp14 Degrades JAK1 to Inhibit the JAK-STAT Signaling Pathway in HD11 Cells. Viruses 2022, 14, 1045. [Google Scholar] [CrossRef] [PubMed]
- Hanukoglu, I.; Hanukoglu, A. Epithelial sodium channel (ENaC) family: Phylogeny, structure-function, tissue distribution, and associated inherited diseases. Gene 2016, 579, 95–132. [Google Scholar] [CrossRef]
- Hoerr, F.J. The Pathology of Infectious Bronchitis. Avian Dis. 2021, 65, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Stannard, W.; O’Callaghan, C. Ciliary Function and the Role of Cilia in Clearance. J. Aerosol Med. 2006, 19, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Di Matteo, A.M.; Soñez, M.C.; Plano, C.M.; von Lawzewitsch, I. Morphologic Observations on Respiratory Tracts of Chickens after after Hatchery Infectious Bronchitis Vaccination and Formaldehyde Fumigation. Avian Dis. 2000, 44, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Verhelst, J.; Parthoens, E.; Schepens, B.; Fiers, W.; Saelens, X. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 2012, 86, 13445–13455. [Google Scholar] [CrossRef] [PubMed]
- Haller, O.; Stertz, S.; Kochs, G. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 2007, 9, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Feng, Y.; Liu, W.; Tan, L.; Sun, Y.; Song, C.; Liao, Y.; Xu, C.; Ren, T.; Ding, C.; et al. A Role for the Chicken Interferon-Stimulated Gene CMPK2 in the Host Response Against Virus Infection. Front. Microbiol. 2022, 13, 874331. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Lv, J.; Wang, W.; Guo, R.; Zhong, C.; Antia, A.; Zeng, Q.; Li, J.; Liu, Q.; Zhou, J.; et al. CMPK2 is a host restriction factor that inhibits infection of multiple coronaviruses in a cell-intrinsic manner. PLoS Biol. 2023, 21, e3002039. [Google Scholar] [CrossRef] [PubMed]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, A.; Popowich, S.; De Silva Senapathi, U.; Abdul-Cader, M.S.; Marshall, F.; van der Meer, F.; Cork, S.C.; Gomis, S.; Abdul-Careem, M.F. Shell-Less Egg Syndrome (SES) Widespread in Western Canadian Layer Operations Is Linked to a Massachusetts (Mass) Type Infectious Bronchitis Virus (IBV) Isolate. Viruses 2018, 10, 437. [Google Scholar] [CrossRef] [PubMed]





| IBV DMV/1639 4 dpi | IBV DMV/1639 11 dpi | ||||
|---|---|---|---|---|---|
| Gene | RNA-Seq FC | qPCR FC | RNA-Seq FC | qPCR FC | |
| Down-regulated | KLHL30 | −1.012 | −3.230 | −1.147 | −1.826 |
| FMOD | −2.283 | −2.987 | −2.181 | −1.401 | |
| NR4A1 | −1.005 | −3.131 | −2.516 | −2.829 | |
| Up-regulated | SOCS1 | 6.538 | 3.030 | 1.057 | 1.633 |
| TLR3 | 2.192 | 1.697 | 1.704 | 1.061 | |
| STAT1 | 6.167 | 1.636 | 2.635 | 1.168 | |
| STAT2 | 2.749 | 4.224 | 1.141 | 2.269 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Dowd, K.; Isham, I.M.; Vatandour, S.; Boulianne, M.; Dozois, C.M.; Gagnon, C.A.; Barjesteh, N.; Abdul-Careem, M.F. Host Immune Response Modulation in Avian Coronavirus Infection: Tracheal Transcriptome Profiling In Vitro and In Vivo. Viruses 2024, 16, 605. https://doi.org/10.3390/v16040605
O’Dowd K, Isham IM, Vatandour S, Boulianne M, Dozois CM, Gagnon CA, Barjesteh N, Abdul-Careem MF. Host Immune Response Modulation in Avian Coronavirus Infection: Tracheal Transcriptome Profiling In Vitro and In Vivo. Viruses. 2024; 16(4):605. https://doi.org/10.3390/v16040605
Chicago/Turabian StyleO’Dowd, Kelsey, Ishara M. Isham, Safieh Vatandour, Martine Boulianne, Charles M. Dozois, Carl A. Gagnon, Neda Barjesteh, and Mohamed Faizal Abdul-Careem. 2024. "Host Immune Response Modulation in Avian Coronavirus Infection: Tracheal Transcriptome Profiling In Vitro and In Vivo" Viruses 16, no. 4: 605. https://doi.org/10.3390/v16040605
APA StyleO’Dowd, K., Isham, I. M., Vatandour, S., Boulianne, M., Dozois, C. M., Gagnon, C. A., Barjesteh, N., & Abdul-Careem, M. F. (2024). Host Immune Response Modulation in Avian Coronavirus Infection: Tracheal Transcriptome Profiling In Vitro and In Vivo. Viruses, 16(4), 605. https://doi.org/10.3390/v16040605







