Mathematical Modeling Suggests That Monocyte Activity May Drive Sex Disparities during Influenza Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Data Collection from Literature
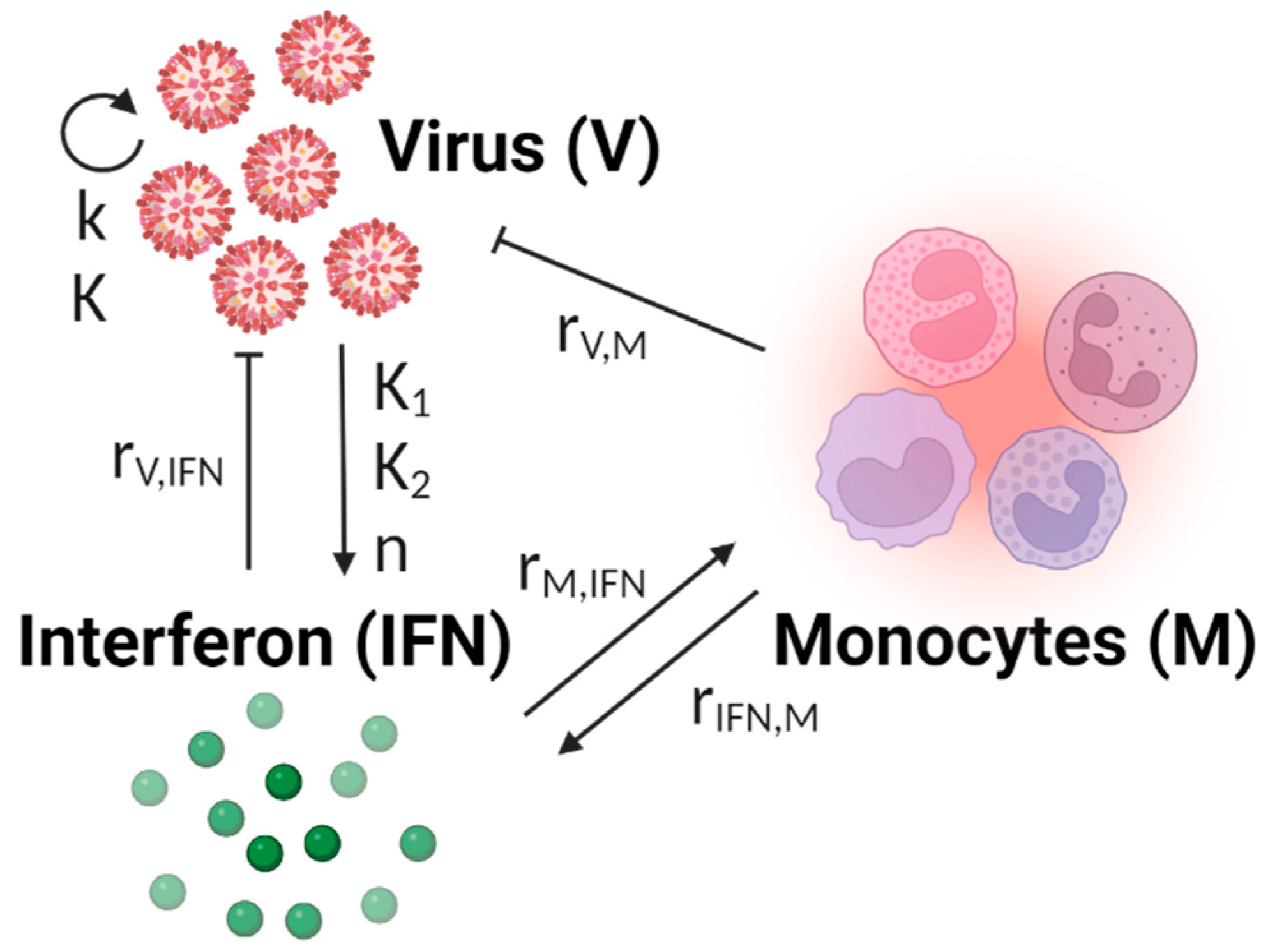
2.2. Model Development
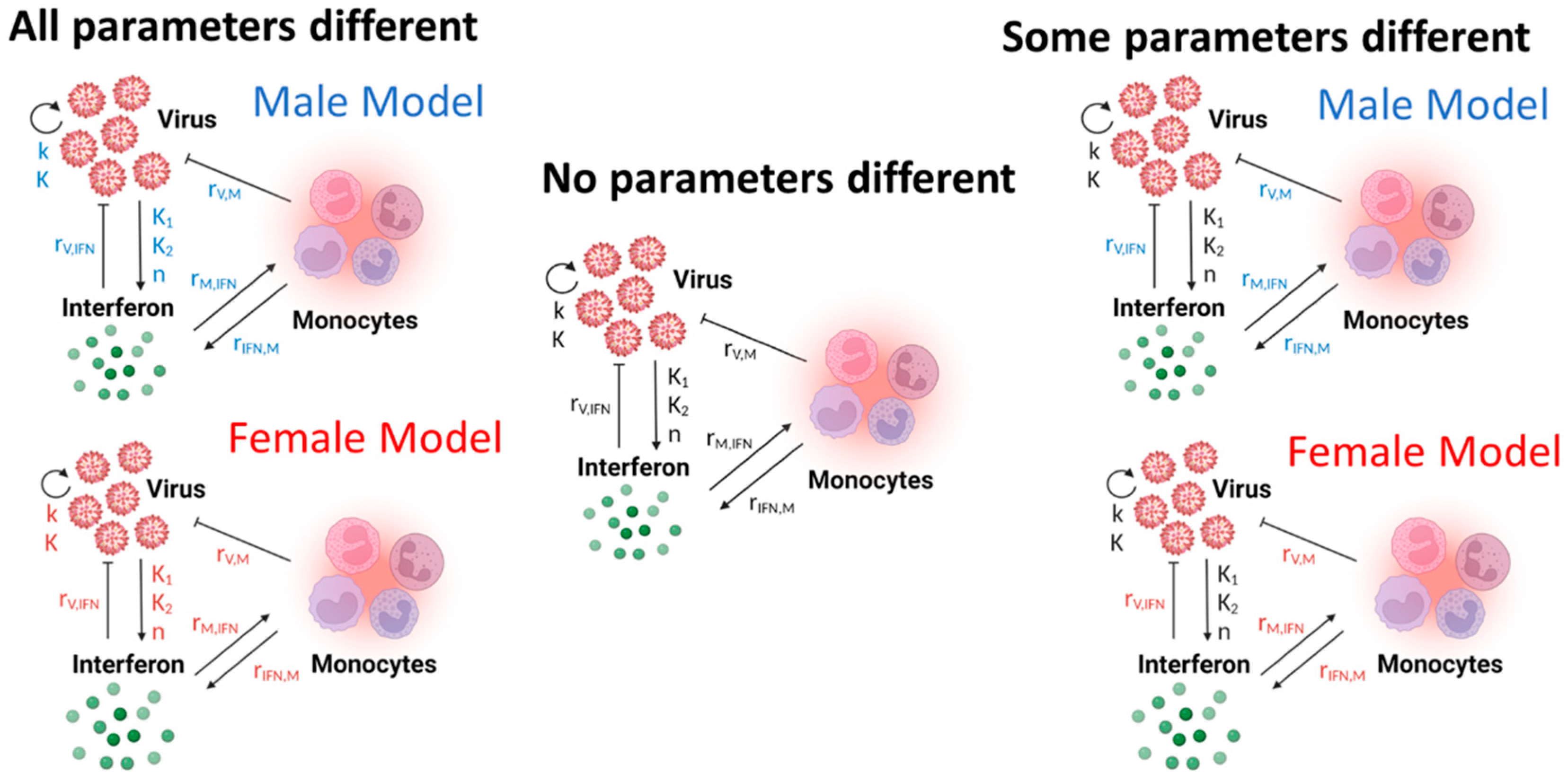
2.3. Parameter Training and Model Selection
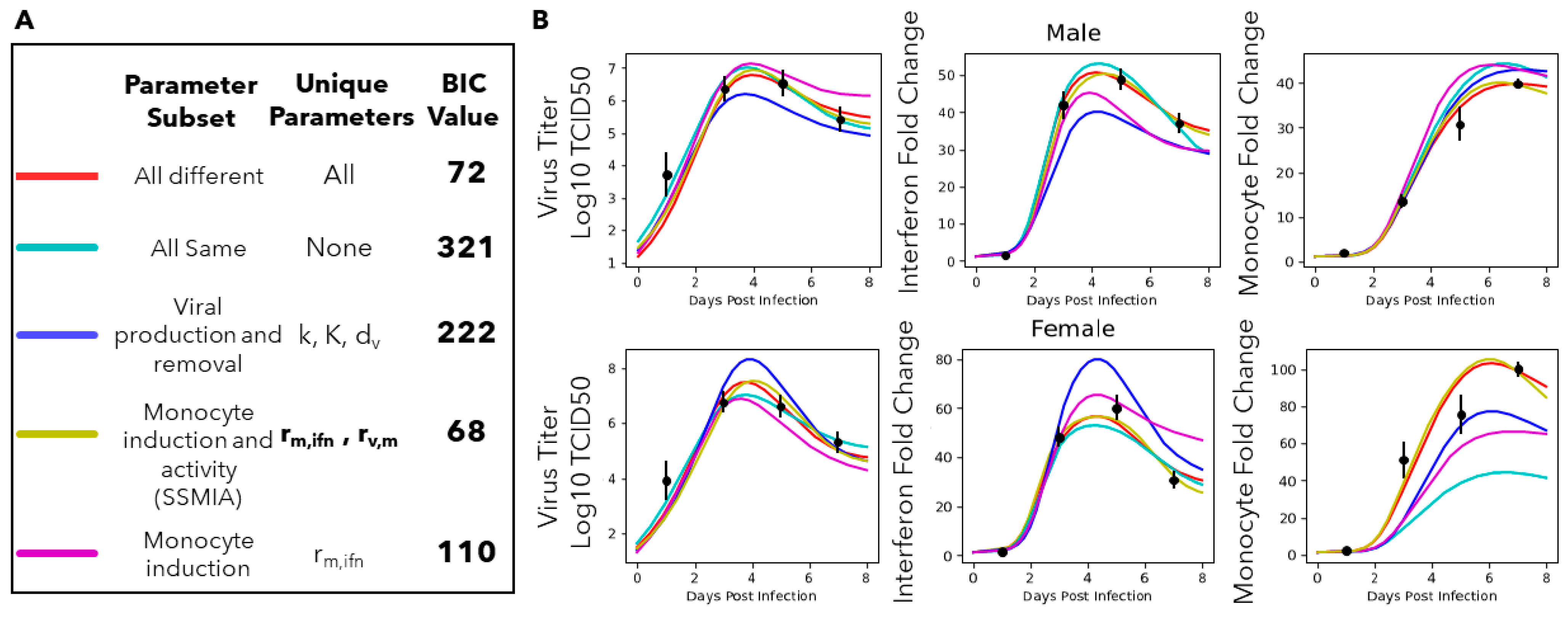
2.4. BIC-Guided Model Selection
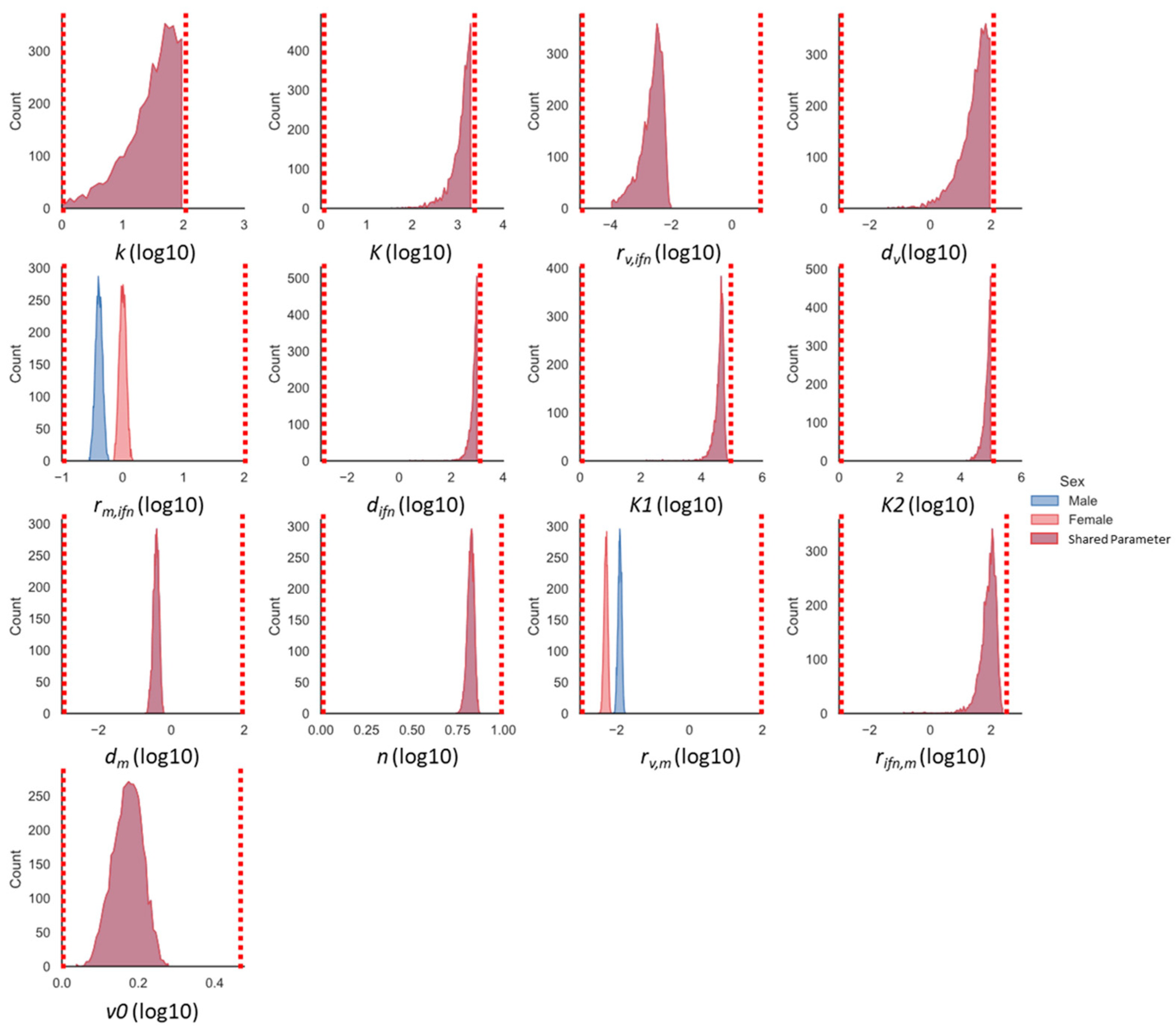
2.5. Markov-Chain Monte Carlo Parameter Exploration
2.6. Sensitivity Analysis
3. Results
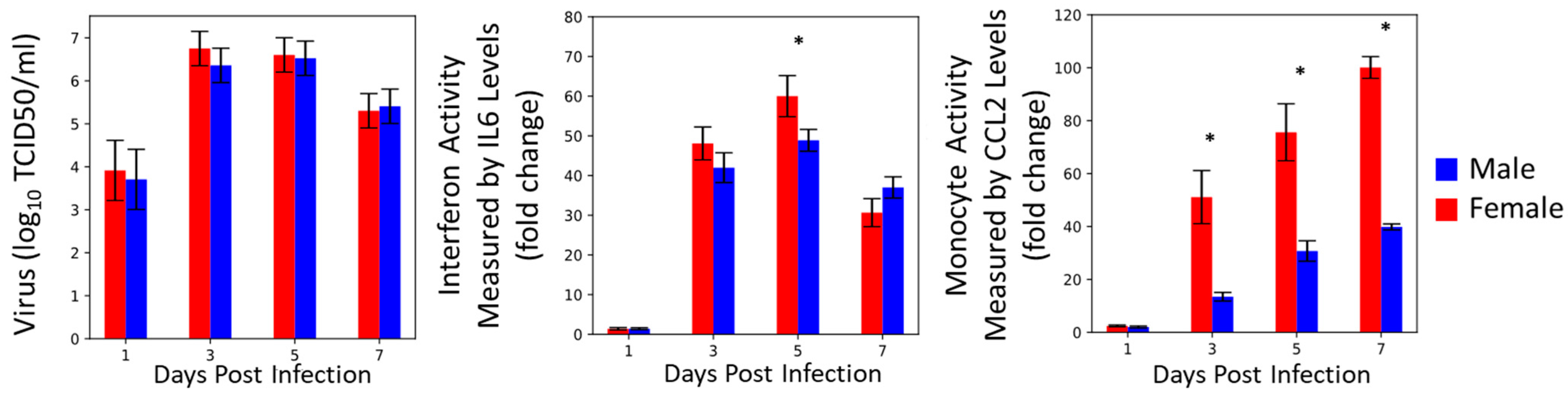
3.1. Reconstructing Prior Data
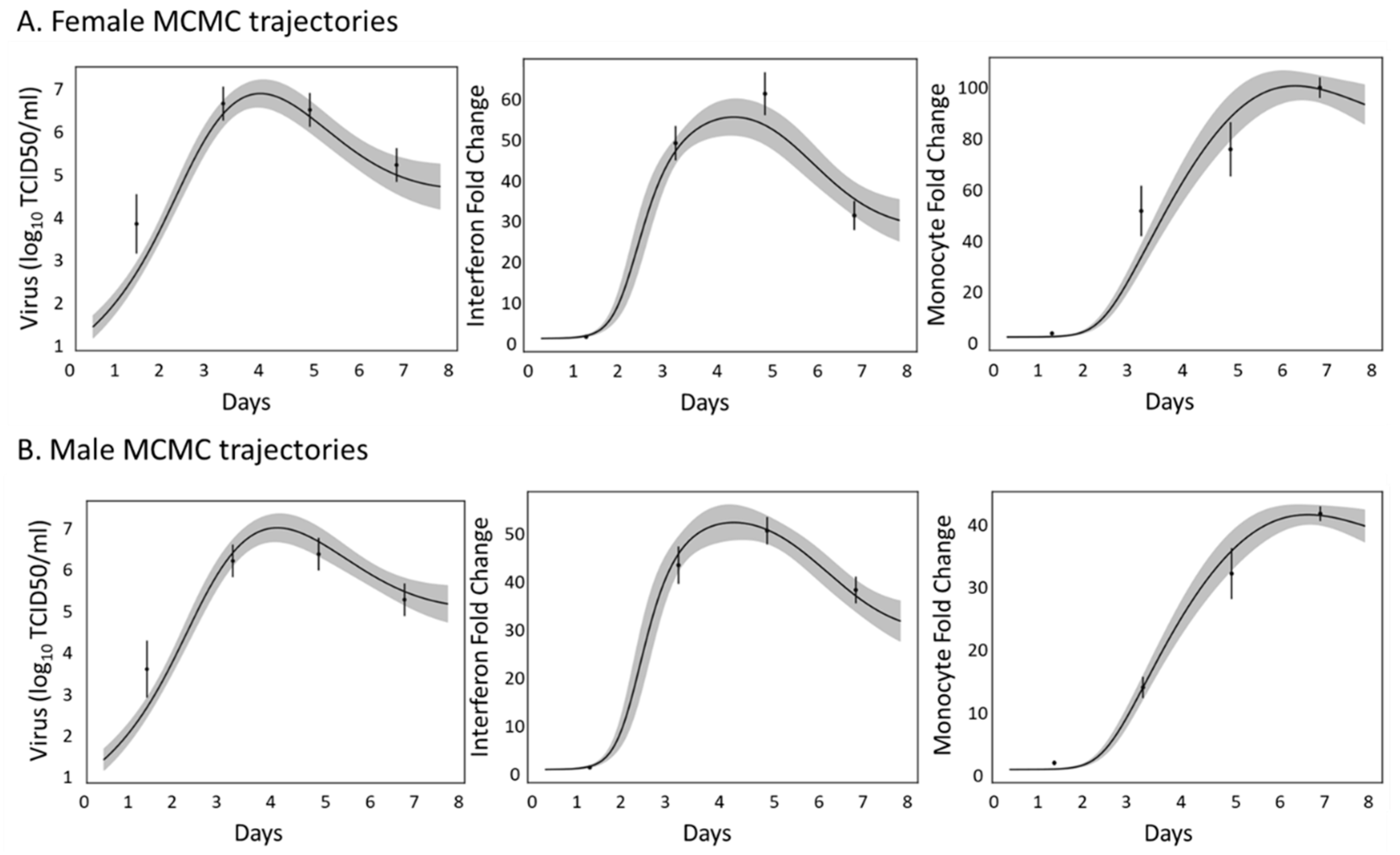
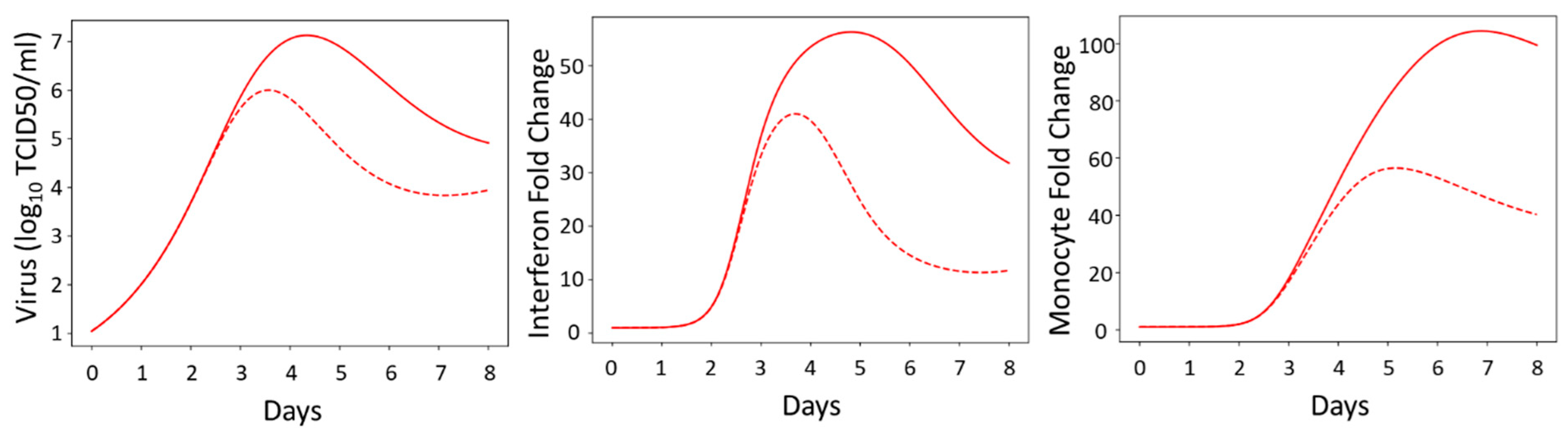
3.2. Innate Immune Mathematical Model Can Fit Immune Response Data from Male and Female Mice
3.3. The Innate Immune Mathematical Model Finds That Male and Female Mice Have Different Rates of Immune Activation to H1N1
3.4. A Computational Screen of Competing Immune Regulation Scenarios Suggests That Monocyte Induction and Activation Are Potential Sex-Specific Parameters
3.5. Monocyte Induction and Activation Are Differentially Regulated in Male and Female Mice
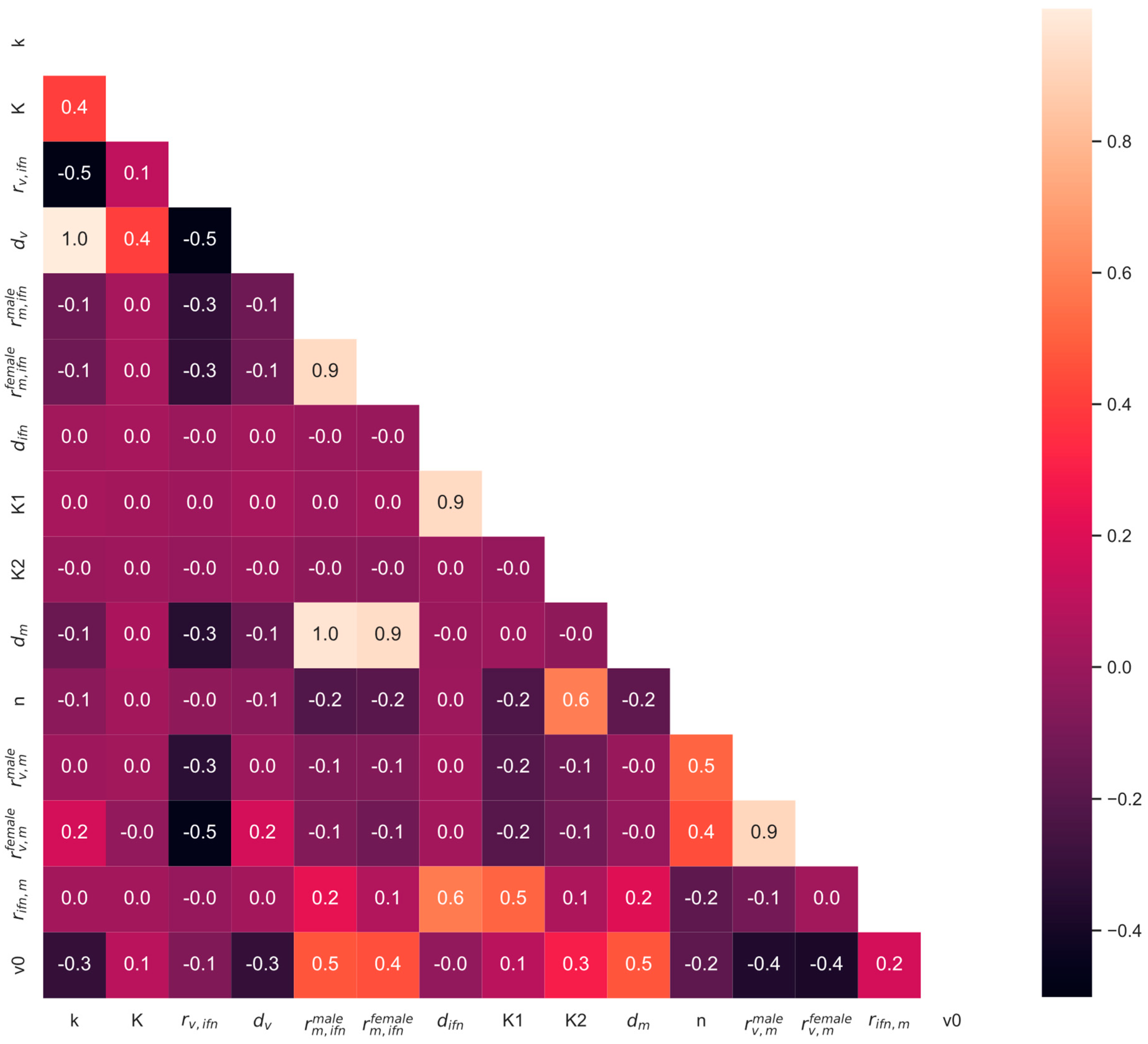
3.6. Global Sensitivity Analysis
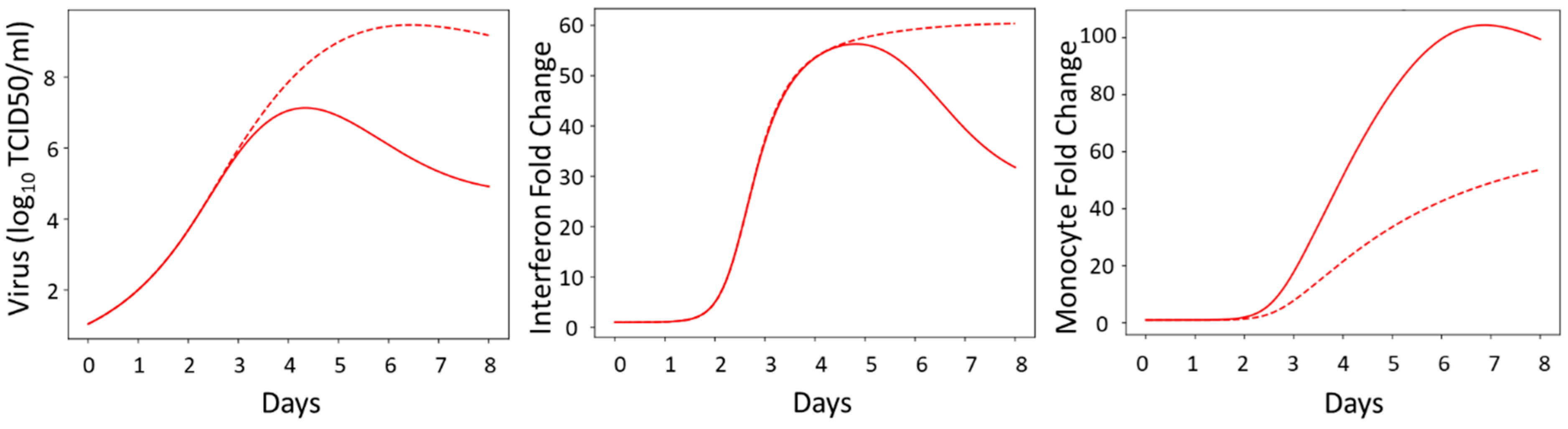
3.7. Model Simulations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Steeg, L.G.V.; Klein, S.L. Sex and sex steroids impact influenza pathogenesis across the life course. Semin. Immunopathol. 2019, 41, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.H.; Archer, B.N.; Baker, M.G.; Cowling, B.J.; Heffernan, R.T.; Mercer, G.; Uez, O.; Hanshaoworakul, W.; Viboud, C.; Schwartz, J.; et al. Searching for sharp drops in the incidence of pandemic A/H1N1 influenza by single year of age. PLoS ONE 2012, 7, e42328. [Google Scholar] [CrossRef] [PubMed]
- WHO. Update on human cases of influenza at the human-animal interface, 2012. Wkly. Epidemiol. Rec. Relev. Épidémiologique Hebd. 2013, 88, 137–144. [Google Scholar]
- Sabra, L.; Klein, A.P.; Catherine, P.; Martha, A.; Peju, O. Sex, Gender and Influenza; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Hoffmann, J.; Otte, A.; Thiele, S.; Lotter, H.; Shu, Y.; Gabriel, G. Sex differences in H7N9 influenza A virus pathogenesis. Vaccine 2015, 33, 6949–6954. [Google Scholar] [CrossRef] [PubMed]
- Dawood, F.S.; Kittikraisak, W.; Patel, A.; Hunt, D.R.; Suntarattiwong, P.; Wesley, M.G.; Thompson, M.G.; Soto, G.; Mundhada, S.; Arriola, C.S.; et al. Incidence of influenza during pregnancy and association with pregnancy and perinatal outcomes in three middle-income countries: A multisite prospective longitudinal cohort study. Lancet Infect. Dis. 2021, 21, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, P.; Deane, J.A.; Difilippantonio, M.J.; Tarasenko, T.; Satterthwaite, A.B.; Bolland, S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science 2006, 312, 1669–1672. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Sarda, C.; Palma, P.; Rello, J. Severe influenza: Overview in critically ill patients. Curr. Opin. Crit. Care 2019, 25, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Romagnani, S. Th1/th2 cells. Inflamm. Bowel Dis. 1999, 5, 285–294. [Google Scholar] [CrossRef]
- Koyama, S.; Ishii, K.J.; Coban, C.; Akira, S. Innate immune response to viral infection. Cytokine 2008, 43, 336–341. [Google Scholar] [CrossRef]
- Berghöfer, B.; Frommer, T.; Haley, G.; Fink, L.; Bein, G.; Hackstein, H. TLR7 ligands induce higher IFN-α production in females. J. Immunol. 2006, 177, 2088–2096. [Google Scholar] [CrossRef] [PubMed]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (MCP-1): An overview. J. Interferon. Cytokine Res. 2009, 29, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, E.E.; Weaver, J.J.A.; Shoemaker, J.E. Mathematical Modeling Finds Disparate Interferon Production Rates Drive Strain-Specific Immunodynamics during Deadly Influenza Infection. Viruses 2022, 14, 906. [Google Scholar] [CrossRef] [PubMed]
- Saenz, R.A.; Quinlivan, M.; Elton, D.; MacRae, S.; Blunden, A.S.; Mumford, J.A.; Daly, J.M.; Digard, P.; Cullinane, A.; Grenfell, B.T.; et al. Dynamics of Influenza Virus Infection and Pathology. J. Virol. 2010, 84, 3974–3983. [Google Scholar] [CrossRef]
- Baccam, P.; Beauchemin, C.; Macken, C.A.; Hayden, F.G.; Perelson, A.S. Kinetics of Influenza A Virus Infection in Humans. J. Virol. 2006, 80, 7590–7599. [Google Scholar] [CrossRef] [PubMed]
- Hancioglu, B.; Swigon, D.; Clermont, G. A dynamical model of human immune response to influenza A virus infection. J. Theor. Biol. 2007, 246, 70–86. [Google Scholar] [CrossRef]
- Pawelek, K.A.; Huynh, G.T.; Quinlivan, M.; Cullinane, A.; Rong, L.; Perelson, A.S. Modeling within-host dynamics of influenza virus infection including immune responses. PLoS Comput. Biol. 2012, 8, e1002588. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Adler, F.R.; Ribeiro, R.M.; Gutenkunst, R.N.; McAuley, J.L.; McCullers, J.A.; Perelson, A.S. Kinetics of coinfection with influenza A virus and Streptococcus pneumoniae. PLoS Pathog. 2013, 9, e1003238. [Google Scholar] [CrossRef]
- Pawelek, K.A.; Dor, D.; Salmeron, C.; Handel, A. Within-host models of high and low pathogenic influenza virus infections: The role of macrophages. PLoS ONE 2016, 11, e0150568. [Google Scholar] [CrossRef]
- Price, I.; Mochan-Keef, E.D.; Swigon, D.; Ermentrout, G.B.; Lukens, S.; Toapanta, F.R.; Ross, T.M.; Clermont, G. The inflammatory response to influenza A virus (H1N1): An experimental and mathematical study. J. Theor. Biol. 2015, 374, 83–93. [Google Scholar] [CrossRef]
- Gregg, R.W.; Sarkar, S.N.; Shoemaker, J.E. Mathematical modeling of the cGAS pathway reveals robustness of DNA sensing to TREX1 feedback. J. Theor. Biol. 2019, 462, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Gregg, R.W.; Shabnam, F.; Shoemaker, J.E. Agent-based modeling reveals benefits of heterogeneous and stochastic cell populations during cGAS-mediated IFNβ production. Bioinformatics 2021, 37, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Aponte-Serrano, J.O.; Weaver, J.J.A.; Sego, T.J.; Glazier, J.A.; Shoemaker, J.E. Multicellular spatial model of RNA virus replication and interferon responses reveals factors controlling plaque growth dynamics. PLoS Comput. Biol. 2021, 17, e1008874. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Vargas, E.A.; Wilk, E.; Canini, L.; Toapanta, F.R.; Binder, S.C.; Uvarovskii, A.; Ross, T.M.; Guzmán, C.A.; Perelson, A.S.; Meyer-Hermann, M. Effects of aging on influenza virus infection dynamics. J. Virol. 2014, 88, 4123–4131. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.P.; Lorenzo, M.E.; Jian, W.; Klein, S.L. Elevated 17β-estradiol protects females from influenza A virus pathogenesis by suppressing inflammatory responses. PLoS Pathog. 2011, 7, e1002149. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Salinas, L.; Verdugo-Rodriguez, A.; Rodriguez, L.L.; Borca, M.V. The Role of Interleukin 6 During Viral Infections. Front. Microbiol. 2019, 10, 1057. [Google Scholar] [CrossRef] [PubMed]
- Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N.; Teller, A.H.; Teller, E. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953, 21, 1087–1092. [Google Scholar] [CrossRef]
- Hastings, W.K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 1970, 57, 97–109. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Dawson, T.C.; Beck, M.A.; Kuziel, W.A.; Henderson, F.; Maeda, N. Contrasting Effects of CCR5 and CCR2 Deficiency in the Pulmonary Inflammatory Response to Influenza A Virus. Am. J. Pathol. 2000, 156, 1951–1959. [Google Scholar] [CrossRef]
- Kash, J.C.; Taubenberger, J.K. The role of viral, host, and secondary bacterial factors in influenza pathogenesis. Am. J. Pathol. 2015, 185, 1528–1536. [Google Scholar] [CrossRef] [PubMed]
- Uiprasertkul, M.; Puthavathana, P.; Sangsiriwut, K.; Pooruk, P.; Srisook, K.; Peiris, M.; Nicholls, J.M.; Chokephaibulkit, K.; Vanprapar, N.; Auewarakul, P. Influenza A H5N1 replication sites in humans. Emerg. Infect. Dis. 2005, 11, 1036. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.P.; Hall, O.J.; Nilles, T.L.; Bream, J.H.; Klein, S.L. 17β-estradiol protects females against influenza by recruiting neutrophils and increasing virus-specific CD8 T cell responses in the lungs. J. Virol. 2014, 88, 4711–4720. [Google Scholar] [CrossRef] [PubMed]
- Sophia, D.; Maini, M.K.; Wack, A. Pathogenic potential of interferon αβ in acute influenza infection. Nat. Commun. 2014, 5, 3864. [Google Scholar]

| Parameter | Lower Bound | Upper Bound | Units | Parameter Name |
|---|---|---|---|---|
| k | 1 | 100 | days−1 | Maximum viral growth rate |
| K | 1 | 2000 | log10(TCID50/mL) | Viral carrying capacity in lungs |
| rv,ifn | 1 × 10−5 | 10 | days−1 | Interferon-regulated inhibition of virus replication |
| dv | 1 × 10−3 | 500 | days−1 | Nonspecific viral decay rate |
| rm,ifn | 1 × 10−1 | 100 | days−1 | Monocyte induction via interferon |
| difn | 1 × 10−3 | 1000 | days−1 | Nonspecific interferon decay rate |
| K1 | 1 | 100,000 | days−1 | Interferon production from virus |
| K2 | 1 | 100,000 | unitless | Apparent dissociation constant |
| dm | 1 × 10−3 | 100 | days−1 | Nonspecific monocyte decay rate |
| N | 1 | 10 | unitless | Hill coefficient, activation threshold of virus needed for interferon production |
| rv,m | 1 × 10−3 | 100 | days−1 | Monocyte activity on viral removal |
| rifn,m | 1 × 10−3 | 250 | days−1 | Monocyte-regulated interferon production |
| v0 | 1 | 3 | log10(TCID50/mL) | Initial Virus Titer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liparulo, T.S.; Shoemaker, J.E. Mathematical Modeling Suggests That Monocyte Activity May Drive Sex Disparities during Influenza Infection. Viruses 2024, 16, 837. https://doi.org/10.3390/v16060837
Liparulo TS, Shoemaker JE. Mathematical Modeling Suggests That Monocyte Activity May Drive Sex Disparities during Influenza Infection. Viruses. 2024; 16(6):837. https://doi.org/10.3390/v16060837
Chicago/Turabian StyleLiparulo, Tatum S., and Jason E. Shoemaker. 2024. "Mathematical Modeling Suggests That Monocyte Activity May Drive Sex Disparities during Influenza Infection" Viruses 16, no. 6: 837. https://doi.org/10.3390/v16060837






