Analysis of Replication, Cell Division-Mediated Spread, and HBV Envelope Protein-Dependent Pseudotyping of Three Mammalian Delta-like Agents
Abstract
:1. Introduction
2. Materials and Methods
2.1. HDV and HDV-like Agents Plasmids
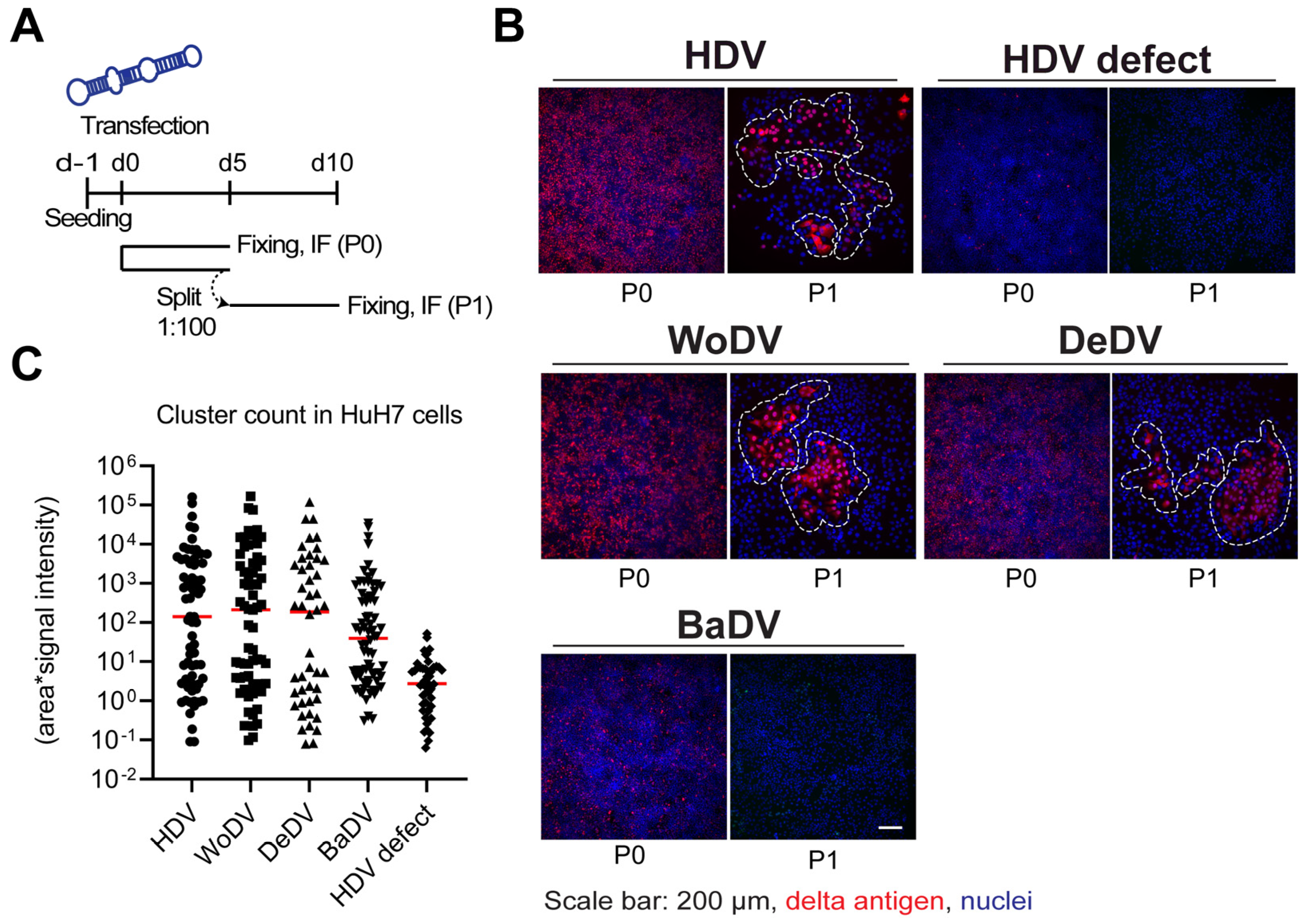
2.2. Cell Division-Mediated Amplification Assay and Cluster Analysis
2.3. HDV, WoDV/HBsAg and DeDV/HBsAg Pseudoparticles Production
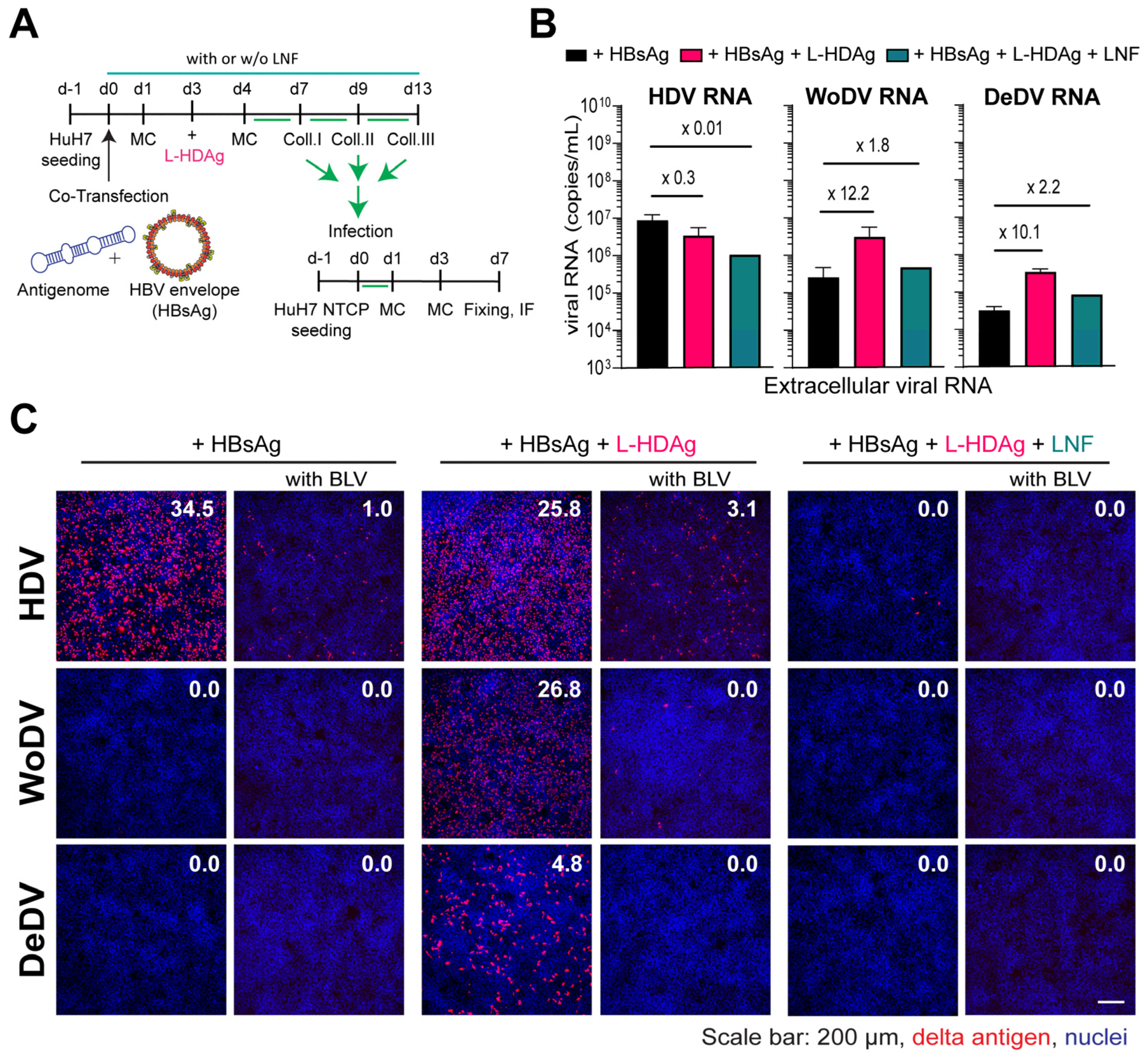
3. Results
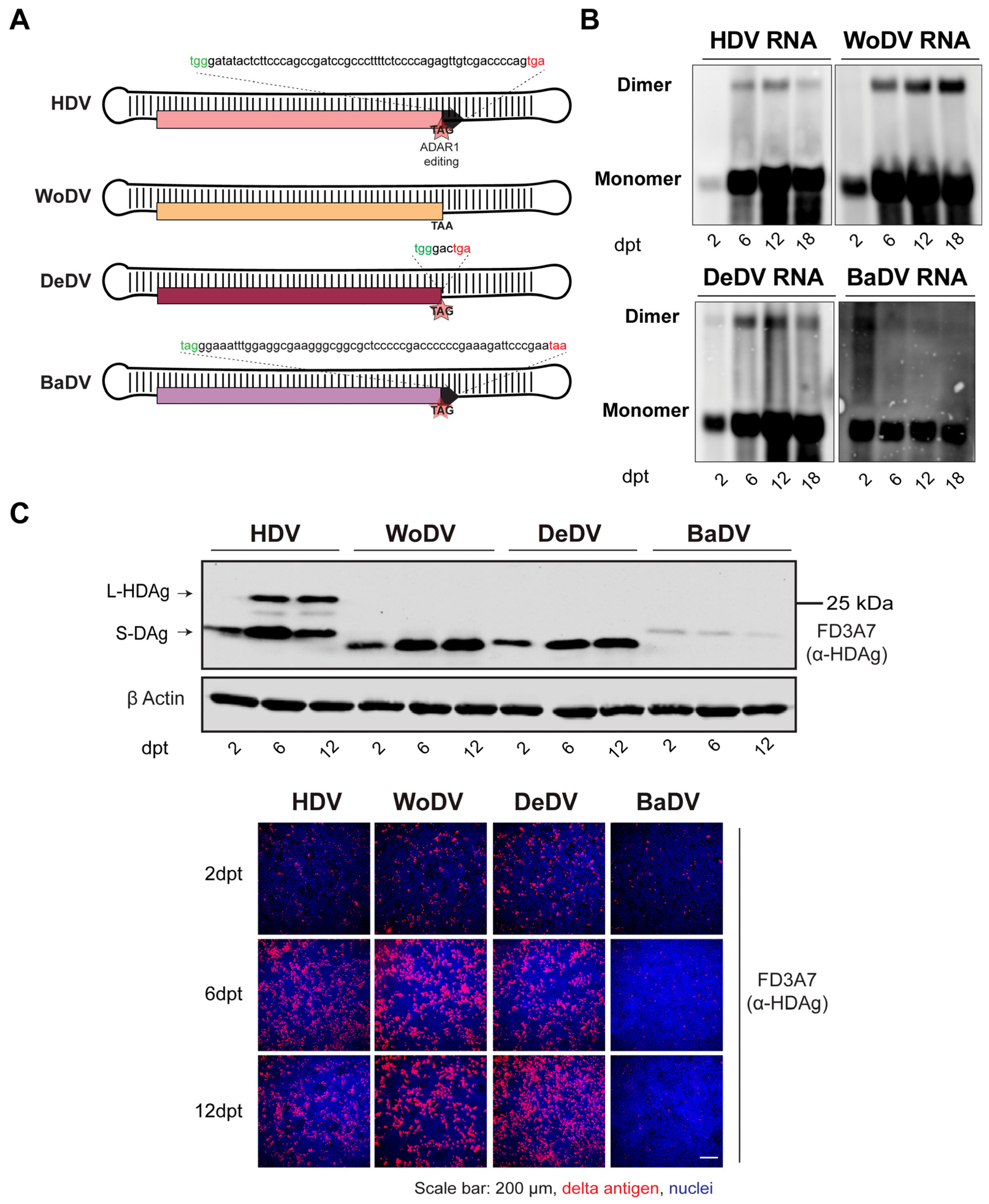
3.1. Establishment and Characterization of HDV-like Agents Infectious Clones
3.2. WoDV and DeDV RNA Can Amplify Efficiently via Cell Division
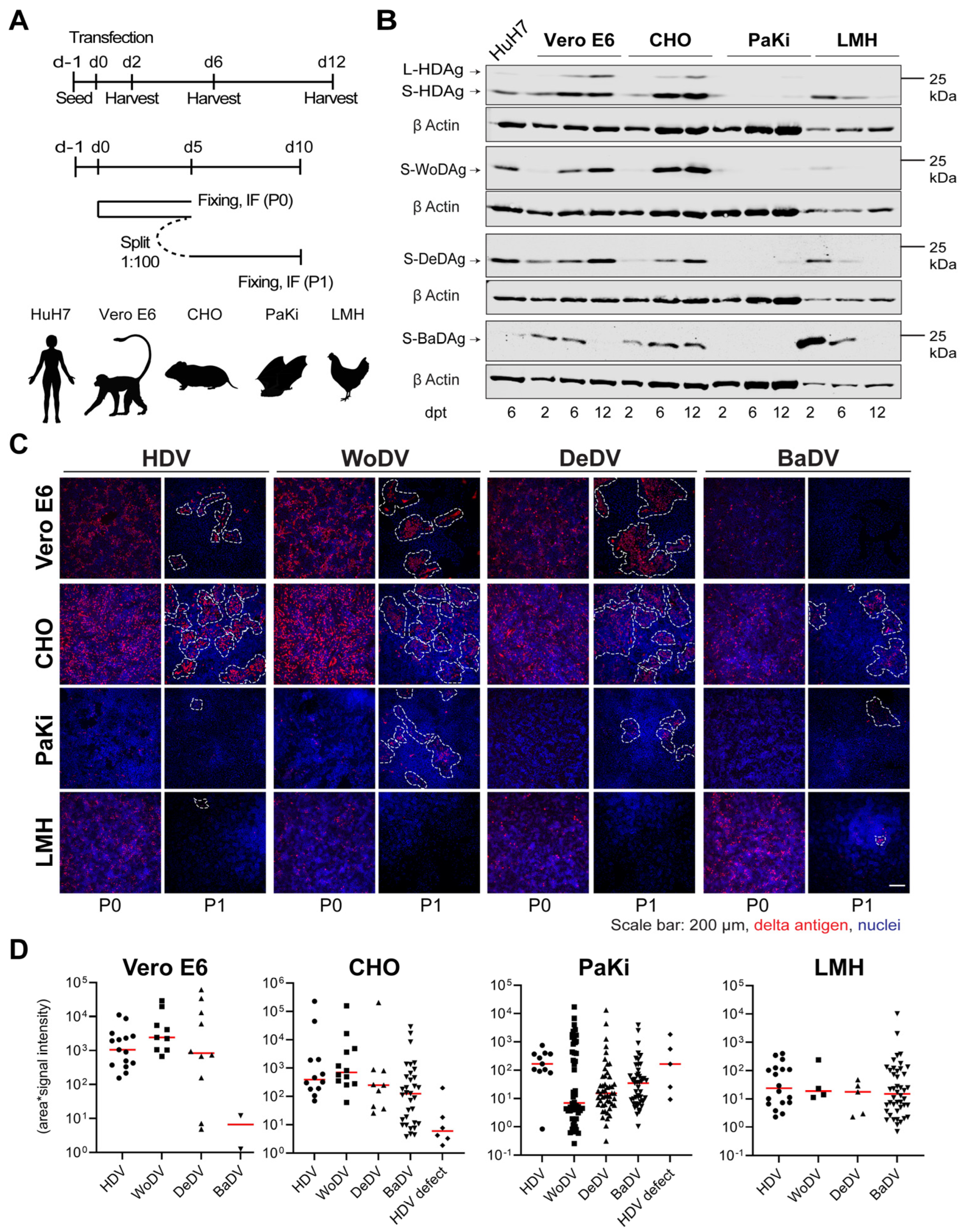
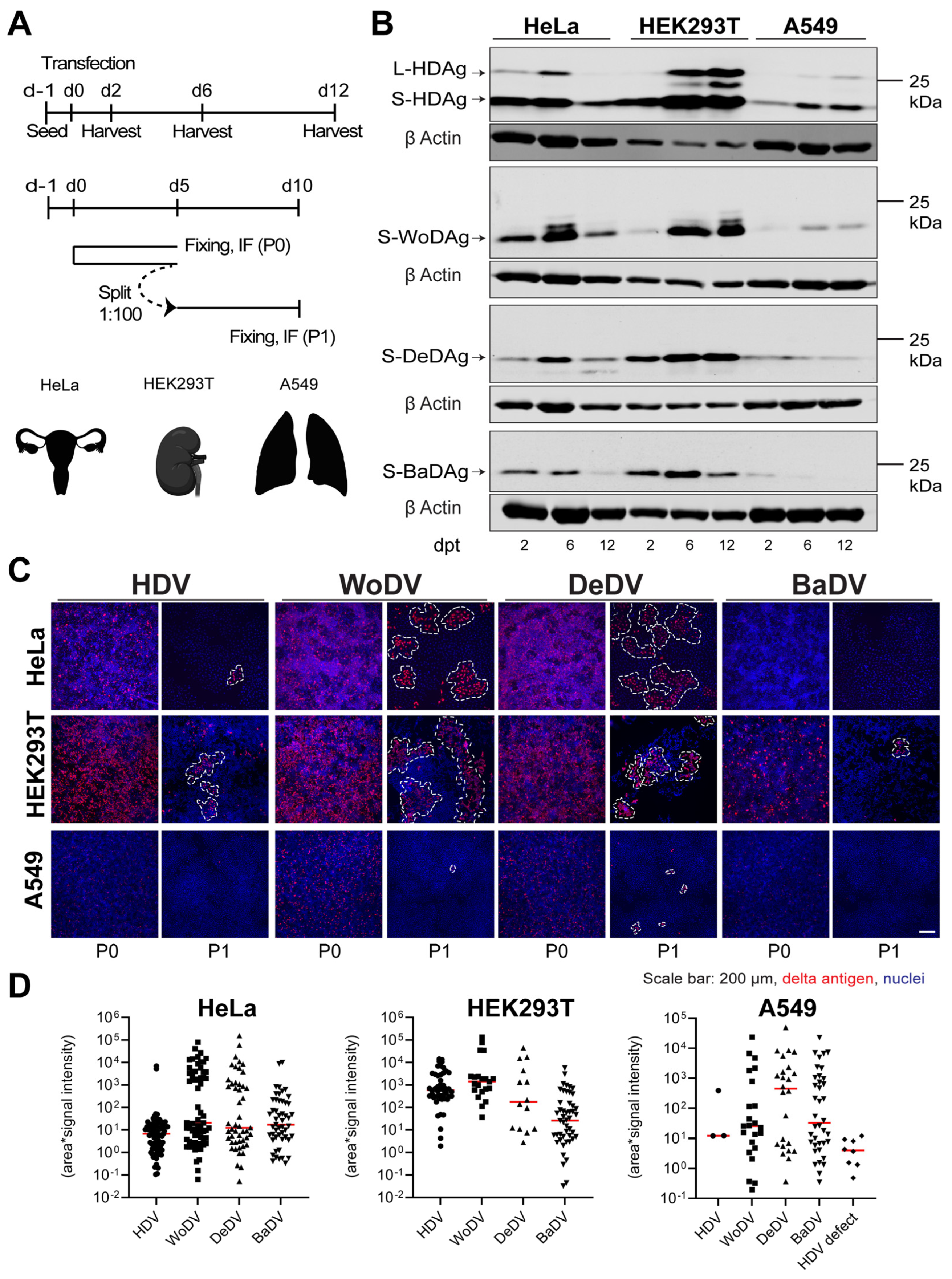
3.3. Delta-like Agents Show a Wide Host Range for Replication
3.4. Cell Division-Mediated Spread of DLA in Non-Hepatic Cell Lines
3.5. WoDV and DeDV Can Exploit HDV L-HDAg for Envelopment and NTCP-Receptor-Mediated Entry into Hepatocytes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rizzetto, M.; Canese, M.G.; Arico, S.; Crivelli, O.; Trepo, C.; Bonino, F.; Verme, G. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut 1977, 18, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Netter, H.J.; Littlejohn, M.; Yuen, L.; Shi, M.; Eden, J.S.; Klaassen, M.; Holmes, E.C.; Hurt, A.C. A Divergent Hepatitis D-Like Agent in Birds. Viruses 2018, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- Hetzel, U.; Szirovicza, L.; Smura, T.; Prahauser, B.; Vapalahti, O.; Kipar, A.; Hepojoki, J. Identification of a Novel Deltavirus in Boa Constrictors. mBio 2019, 10, e00014-19. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.S.; Pettersson, J.H.; Le Lay, C.; Shi, M.; Lo, N.; Wille, M.; Eden, J.S.; Holmes, E.C. Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 2019, 5, vez021. [Google Scholar] [CrossRef] [PubMed]
- Bergner, L.M.; Orton, R.J.; Broos, A.; Tello, C.; Becker, D.J.; Carrera, J.E.; Patel, A.H.; Biek, R.; Streicker, D.G. Diversification of mammalian deltaviruses by host shifting. Proc. Natl. Acad. Sci. USA 2021, 118, e2019907118. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Shibata, Y.; Kawasaki, J.; Kojima, S.; Li, Y.T.; Iwami, S.; Muramatsu, M.; Wu, H.-L.; Wada, K.; Tomonaga, K.; et al. Identification of novel avian and mammalian deltaviruses provides new insights into deltavirus evolution. Virus Evol. 2021, 7, veab003. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, S.; Pirzer, F.; Goldmann, N.; Schmid, J.; Corman, V.M.; Gottula, L.T.; Schroeder, S.; Rasche, A.; Muth, D.; Drexler, J.F.; et al. Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus. Proc. Natl. Acad. Sci. USA 2020, 117, 17977–17983. [Google Scholar] [CrossRef] [PubMed]
- Giersch, K.; Bhadra, O.D.; Volz, T.; Allweiss, L.; Riecken, K.; Fehse, B.; Lohse, A.W.; Petersen, J.; Sureau, C.; Urban, S.; et al. Hepatitis delta virus persists during liver regeneration and is amplified through cell division both in vitro and in vivo. Gut 2018, 68, 150–157. [Google Scholar] [CrossRef]
- Zhang, Z.; Ni, Y.; Lempp, F.A.; Walter, L.; Mutz, P.; Bartenschlager, R.; Urban, S. Hepatitis D virus-induced interferon response and administered interferons control cell division-mediated virus spread. J. Hepatol. 2022, 77, 957–966. [Google Scholar] [CrossRef]
- Jayan, G.C.; Casey, J.L. Increased RNA editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2. J. Virol. 2002, 76, 3819–3827. [Google Scholar] [CrossRef]
- Lee, C.Z.; Chen, P.J.; Lai, M.M.; Chen, D.S. Isoprenylation of large hepatitis delta antigen is necessary but not sufficient for hepatitis delta virus assembly. Virology 1994, 199, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Glenn, J.S.; Watson, J.A.; Havel, C.M.; White, J.M. Identification of a prenylation site in delta virus large antigen. Science 1992, 256, 1331–1333. [Google Scholar] [CrossRef] [PubMed]
- Sureau, C.; Guerra, B.; Lanford, R.E. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J. Virol. 1993, 67, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Szirovicza, L.; Hetzel, U.; Kipar, A.; Hepojoki, J. Short ‘1.2x Genome’ Infectious Clone Initiates Kolmiovirid Replication in Boa constrictor Cells. Viruses 2022, 14, 107. [Google Scholar] [CrossRef]
- Szirovicza, L.; Hetzel, U.; Kipar, A.; Martinez-Sobrido, L.; Vapalahti, O.; Hepojoki, J. Snake Deltavirus Utilizes Envelope Proteins of Different Viruses To Generate Infectious Particles. mBio 2020, 11, e03250-19. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.; Cerikan, B.; Stahl, Y.; Kopp, K.; Magg, V.; Acosta-Rivero, N.; Kim, H.; Klein, K.; Funaya, C.; Haselmann, U.; et al. Enhanced SARS-CoV-2 entry via UPR-dependent AMPK-related kinase NUAK2. Mol. Cell 2023, 83, 2559–2577.e8. [Google Scholar] [CrossRef]
- Wang, W.; Lempp, F.A.; Schlund, F.; Walter, L.; Decker, C.C.; Zhang, Z.; Ni, Y.; Urban, S. Assembly and infection efficacy of hepatitis B virus surface protein exchanges in 8 hepatitis D virus genotype isolates. J. Hepatol. 2021, 75, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, D.; Schutte, C.; Warnecke, J.; Dorn, I.; Hennig, H.; Kirchner, H.; Schlenke, P. The large form of ADAR 1 is responsible for enhanced hepatitis delta virus RNA editing in interferon-alpha-stimulated host cells. J. Viral Hepat. 2006, 13, 150–157. [Google Scholar] [CrossRef]
- Hwang, S.B.; Lai, M.M. Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J. Virol. 1994, 68, 2958–2964. [Google Scholar] [CrossRef]
- Modahl, L.E.; Lai, M.M. The large delta antigen of hepatitis delta virus potently inhibits genomic but not antigenomic RNA synthesis: A mechanism enabling initiation of viral replication. J. Virol. 2000, 74, 7375–7380. [Google Scholar] [CrossRef]
- Flores, R.; Grubb, D.; Elleuch, A.; Nohales, M.A.; Delgado, S.; Gago, S. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: Variations on a theme. RNA Biol. 2011, 8, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Netter, H.J.; Barrios, M.H.; Littlejohn, M.; Yuen, L.K.W. Hepatitis Delta Virus (HDV) and Delta-Like Agents: Insights Into Their Origin. Front. Microbiol. 2021, 12, 652962. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Pelchat, M. Origin of hepatitis delta virus. Future Microbiol. 2010, 5, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Khalfi, P.; Denis, Z.; McKellar, J.; Merolla, G.; Chavey, C.; Ursic-Bedoya, J.; Soppa, L.; Szirovicza, L.; Hetzel, U.; Dufourt, J.; et al. Comparative analysis of human, rodent and snake deltavirus replication. PLoS Pathog. 2024, 20, e1012060. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Fälth, M.; Stindt, J.; Königer, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014, 146, 1070–1083. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.; Morse, S.; Ararat, M.; Graham, F.L. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002, 16, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Ryu, W.S.; Bayer, M.; Taylor, J. Assembly of hepatitis delta virus particles. J. Virol. 1992, 66, 2310–2315. [Google Scholar] [CrossRef]
- Ponzetto, A.; Cote, P.J.; Popper, H.; Hoyer, B.H.; London, W.T.; Ford, E.C.; Bonino, F.; Purcell, R.H.; Gerin, J.L. Transmission of the hepatitis B virus-associated delta agent to the eastern woodchuck. Proc. Natl. Acad. Sci. USA 1984, 81, 2208–2212. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gnouamozi, G.E.; Zhang, Z.; Prasad, V.; Lauber, C.; Seitz, S.; Urban, S. Analysis of Replication, Cell Division-Mediated Spread, and HBV Envelope Protein-Dependent Pseudotyping of Three Mammalian Delta-like Agents. Viruses 2024, 16, 859. https://doi.org/10.3390/v16060859
Gnouamozi GE, Zhang Z, Prasad V, Lauber C, Seitz S, Urban S. Analysis of Replication, Cell Division-Mediated Spread, and HBV Envelope Protein-Dependent Pseudotyping of Three Mammalian Delta-like Agents. Viruses. 2024; 16(6):859. https://doi.org/10.3390/v16060859
Chicago/Turabian StyleGnouamozi, Gnimah Eva, Zhenfeng Zhang, Vibhu Prasad, Chris Lauber, Stefan Seitz, and Stephan Urban. 2024. "Analysis of Replication, Cell Division-Mediated Spread, and HBV Envelope Protein-Dependent Pseudotyping of Three Mammalian Delta-like Agents" Viruses 16, no. 6: 859. https://doi.org/10.3390/v16060859




