A Novel Strain of Fusarium oxysporum Alternavirus 1 Isolated from Fusarium oxysporum f. sp. melonis Strain T-BJ17 Confers Hypovirulence and Increases the Sensitivity of Its Host Fungus to Difenoconazole and Pydiflumetofen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains
2.2. Extraction and Purification of RNA
2.3. Metatranscriptome Sequencing
2.4. Reverse Transcription–Polymerase Chain Reaction (RT-PCR)
2.5. Synthesis and Molecular Cloning of Complementary DNA (cDNA)
2.6. Analysis of Sequences and Phylogenetic Analysis
2.7. Observation of Virions and Confirmation of FoAV1-FOM Assembly into Virus Particles
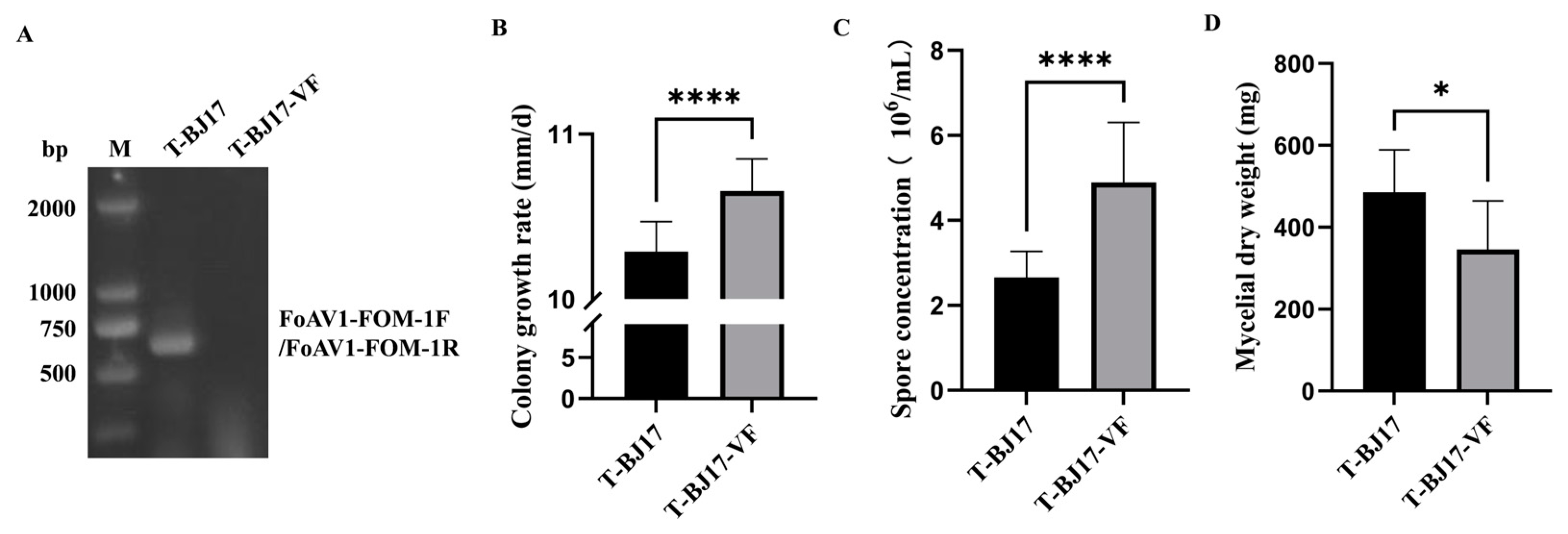
2.8. Elimination of FoAV1-FOM from Strain T-BJ17
2.9. Effect of FoAV1-FOM on the Phenotypes of Its Host Fungus
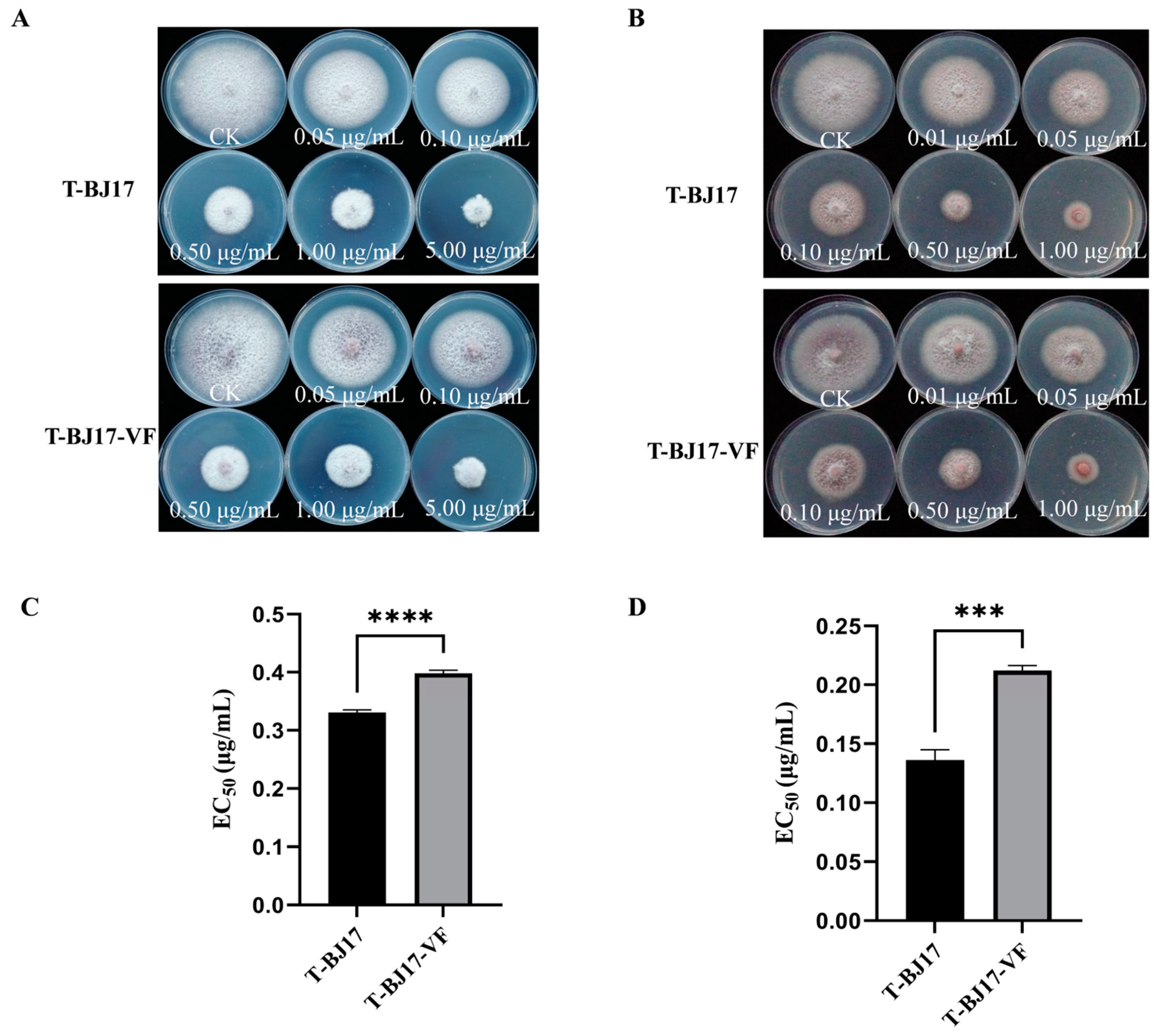
2.10. Sensitivity of T-BJ17 and T-BJ17-VF to Difenoconazole and Pydiflumetofen
2.11. Vertical Transmission Assay
3. Results
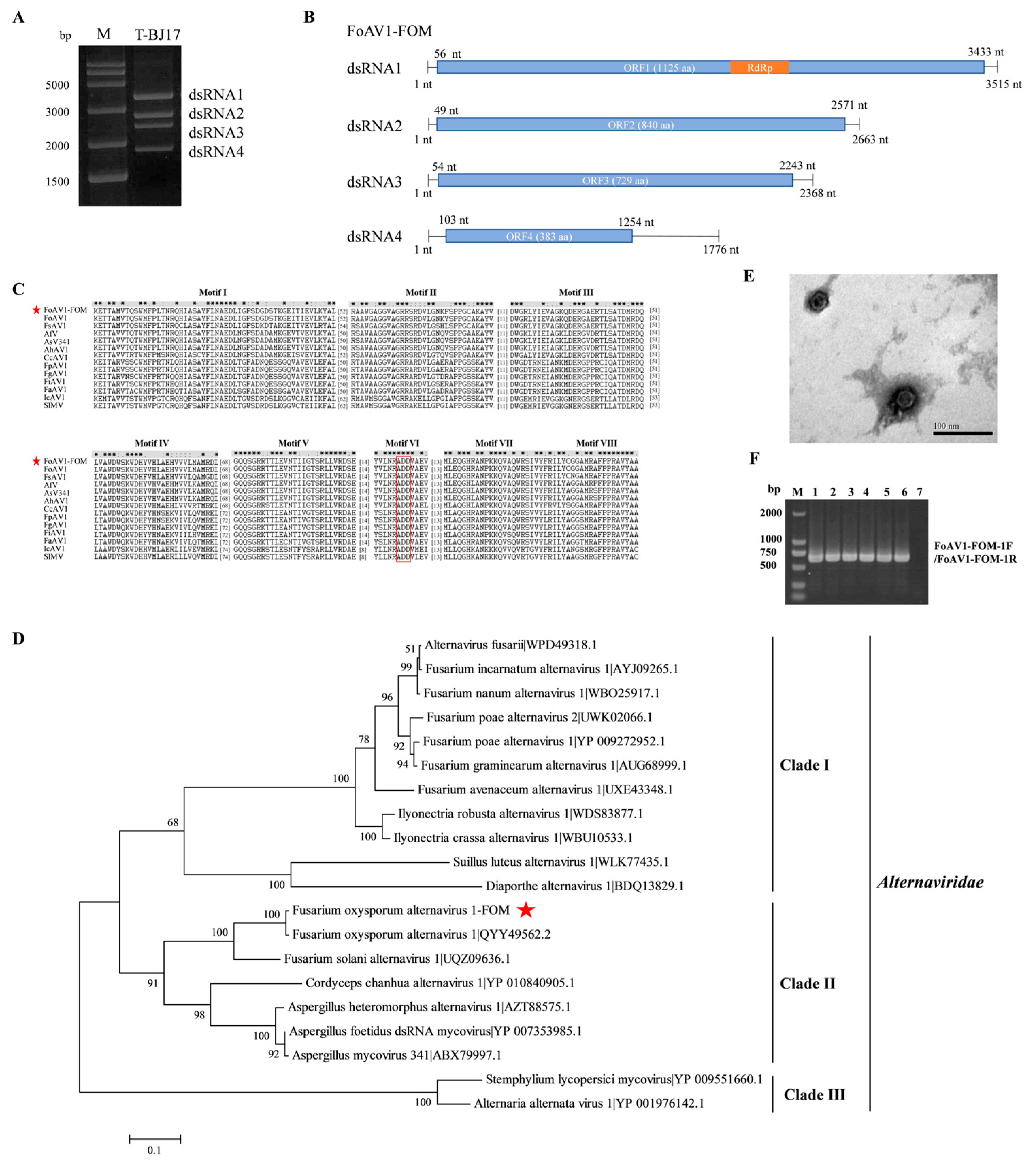
3.1. Metatranscriptomic Identification of Mycoviruses in FOM
3.2. Complete Sequence, Phylogenetic Analysis, and Observation of Virions of FoAV1-FOM
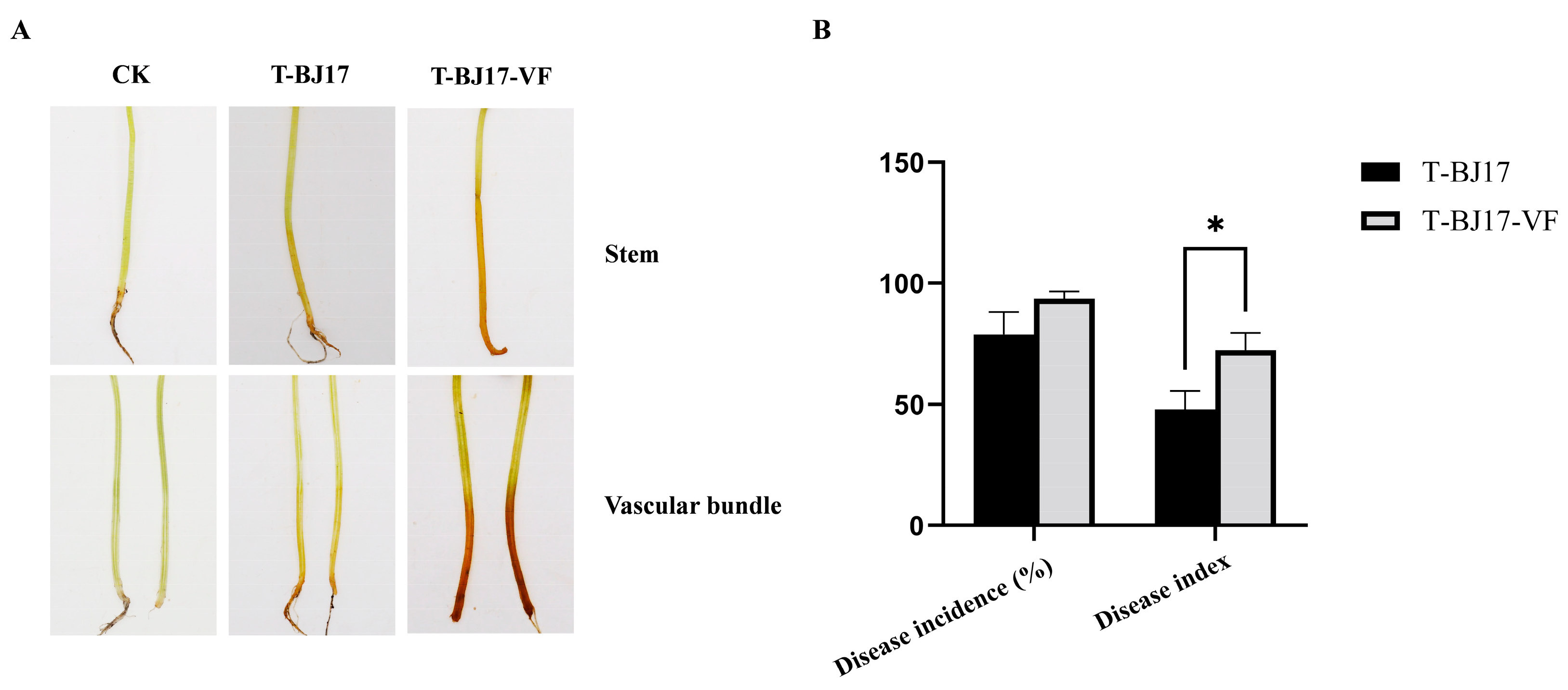
3.3. Effect of FoAV1-FOM on the Phenotypes of Its Host
3.4. Vertical Transmission of FoAV1-FOM
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nag, P.; Paul, S.; Shriti, S.; Das, S. Defence response in plants and animals against a common fungal pathogen, Fusarium oxysporum. Curr. Res. Microb. Sci. 2022, 3, 100135. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.J.; Qiu, R.; Li, X.J.; Zhao, J.; Bai, J.K.; Chen, Y.G.; Li, S.J. Complete genome sequence of a novel mitovirus from the phytopathogenic fungus Fusarium oxysporum. Arch. Virol. 2021, 166, 3211–3216. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.J.; Song, P.Y.; Qiu, R.; Song, R.F.; Li, X.J.; Ni, Y.X.; Zhao, H.; Liu, H.Y.; Li, S.J. Molecular and biological characterization of the first mymonavirus identified in Fusarium oxysporum. Front. Microbiol. 2022, 13, 870204. [Google Scholar] [CrossRef] [PubMed]
- Lemus-Minor, C.G.; Cañizares, M.C.; Garca-Pedrajas, M.D.; Pérez-Artés, E. Fusarium oxysporum f. sp. dianthi virus 1 accumulation is correlated with changes in virulence and other phenotypic traits of its fungal host. Phytopathology 2018, 108, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Lemus-Minor, C.G.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Horizontal and vertical transmission of the hypovirulence-associated mycovirus Fusarium oxysporum f. sp. dianthi virus 1. Eur. J. Plant Pathol. 2019, 153, 645–650. [Google Scholar] [CrossRef]
- Torres-Trenas, A.; Prieto, P.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Mycovirus Fusarium oxysporum f. sp. dianthi virus 1 decreases the colonizing efficiency of its fungal host. Front. Cell. Infect. Microbiol. 2019, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Torres-Trenas, A.; Pérez-Artés, E. Characterization and incidence of the first member of the genus Mitovirus identified in the phytopathogenic species Fusarium oxysporum. Viruses 2020, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- Torres-Trenas, A.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Molecular and biological characterization of the first hypovirus identified in Fusarium oxysporum. Front. Microbiol. 2020, 10, 3131. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.T.; Liu, Y.Y.; Zhang, Y.F.; Wang, X.; Li, H.P.; Li, P.F. Metatranscriptome-based strategy reveals the existence of novel mycoviruses in the plant pathogenic fungus Fusarium oxysporum f. sp. cubense. Front. Microbiol. 2023, 14, 1193714. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.Y.; Wan, X.R.; She, Y.Y.; Li, M.; Xi, H.J.; Xie, J.T.; Wen, C.Y. A novel ourmia-like mycovirus confers hypovirulence-associated traits on Fusarium oxysporum. Front. Microbiol. 2020, 11, 569869. [Google Scholar] [CrossRef]
- Wen, C.Y.; Wan, X.R.; Zhang, Y.Y.; Du, H.Y.; Wei, C.X.; Zhong, R.R.; Zhang, H.; Shi, Y.; Xie, J.T.; Fu, Y.P.; et al. Molecular characterization of the first alternavirus identified in Fusarium oxysporum. Viruses 2021, 13, 2026. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Shamsi, W.; Jamal, A.; Bhatti, M.F.; Kondo, H.; Suzuki, N. Hadaka virus 1: A capsidless eleven-segmented positive-sense single-stranded RNA virus from a phytopathogenic fungus, Fusarium oxysporum. mBio 2020, 11, e00450-20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Wu, C.Y.; Hua, H.H.; Cai, Q.N.; Wu, X.H. Characterization of the first alternavirus identified in Fusarium avenaceum, the causal agent of potato dry rot. Viruses 2023, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Chen, X.G.; Li, P.F.; Qiu, D.W.; Guo, L.H. Complete genome sequence of a Fusarium graminearum double-stranded RNA virus in a newly proposed family, Alternaviridae. Genome Announc. 2018, 6, e00064-18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.T.; Xie, Y.; Zhang, F.; Sun, H.J.; Zhai, Y.Y.; Zhang, S.B.; Yuan, H.X.; Zhou, L.; Gao, F.; Li, H.L. Complete genome sequence of an alternavirus from the phytopathogenic fungus Fusarium incarnatum. Arch. Virol. 2019, 164, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, S.; Ali, A. Identification and complete genome sequence of an alternavirus from a pathogenic fungus, Fusarium nanum, collected by air sampling. PhytoFrontiers 2023, 3, 278–283. [Google Scholar] [CrossRef]
- Osaki, H.; Sasaki, A.; Nomiyama, K.; Tomioka, K. Multiple virus infection in a single strain of Fusarium poae shown by deep sequencing. Virus Genes 2016, 52, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.Y.; Xia, L.L.; Cao, S.L.; Sun, H.Y.; Chen, H.G.; Deng, Q.C.; Li, W. A novel alternavirus with three dsRNA segments from Fusarium pseudograminearum, the pathogen of Fusarium crown rot in wheat. Arch. Virol. 2024, 169, 49. [Google Scholar] [CrossRef] [PubMed]
- Lutz, T.; Japić, E.; Bien, S.; Langer, G.J.; Heinze, C. Characterization of a novel alternavirus infecting the fungal pathogen Fusarium solani. Virus Res. 2022, 317, 198817. [Google Scholar] [CrossRef]
- Li, P.F.; Zhang, H.L.; Chen, X.G.; Qiu, D.W.; Guo, L.H. Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum. Virology 2015, 481, 151–160. [Google Scholar] [CrossRef]
- Darissa, O.; Adam, G.; Schafer, W. A dsRNA mycovirus causes hypovirulence of Fusarium graminearum to wheat and maize. Eur. J. Plant Pathol. 2012, 134, 181–189. [Google Scholar] [CrossRef]
- Chu, Y.M.; Jeon, J.J.; Yea, S.J.; Kim, Y.H.; Yun, S.H.; Lee, Y.W.; Kim, K.H. Double-stranded RNA mycovirus from Fusarium graminearum. Appl. Environ. Microbiol. 2002, 68, 2529–2534. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, Z.F.; Li, K.; Liu, D.W.; Jia, Y.F.; Gao, F.; Dai, J.L.; Zhang, S.B.; Zhang, X.T.; Li, H.L. A megabirnavirus alleviates the pathogenicity of Fusarium pseudograminearum to wheat. Phytopathology 2022, 112, 1175–1184. [Google Scholar] [CrossRef]
- Song, X.S.; Sun, Y.D.; Gao, J.; Gu, K.X.; Hou, Y.P.; Wang, J.X.; Zhou, M.G. Extending the host range of Fusarium poae virus 1 from Fusarium poae to other Fusarium species in the field. Viruses 2022, 14, 2246. [Google Scholar] [CrossRef] [PubMed]
- Fonseka, D.L.; Gudmestad, N.C. Spatial and temporal sensitivity of Alternaria species associated with potato foliar diseases to demethylation inhibiting and anilino-pyrimidine fungicides. Plant Dis. 2016, 100, 1848–1857. [Google Scholar] [CrossRef]
- Sun, C.X.; Li, F.J.; Wei, M.D.; Xiang, Z.X.; Chen, C.J.; Xu, D.L. Detection and biological characteristics of Alternaria alternata resistant to difenoconazole from Paris polyphylla var. chinensis, an indigenous medicinal herb. Plant Dis. 2021, 105, 1546–1554. [Google Scholar] [PubMed]
- Xu, X.M.; Wang, Y.Q.; Lei, T.; Sohail, M.A.; Wang, J.; Wang, H.Y. Synergistic effects of Bacillus amyloliquefaciens SDTB009 and difenoconazole on Fusarium wilt of tomato. Plant Dis. 2022, 106, 2165–2171. [Google Scholar] [CrossRef] [PubMed]
- Acosta-González, U.; Silva-Rojas, H.V.; Fuentes-Aragón, D.; Hernández-Castrejón, J.; Romero-Bautista, A.; Rebollar-Alviter, A. Comparative performance of fungicides and biocontrol products in the management of Fusarium wilt of blackberry. Plant Dis. 2022, 106, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.M.; Zhao, Z.J.; Chen, Q.M.; Shao, H.B. Identification of control agents against melon wilt disease in laboratory and field in NE China. Pak. J. Bot. 2019, 51, 751–754. [Google Scholar] [CrossRef]
- Karki, K.; Grant, J.N.; Biscaia Ribeiro da Silva, A.L.; Petkar, A.; Hajihassani, A.; Coolong, T.W.; Dutta, B. Evaluation of efficacy of Pic-clor 60 [choloropicrin pre-mixed with 1,3 dicholoropropene] and soil-applied fungicides to manage Fusarium wilt in watermelon. Crop Prot. 2021, 154, 105894. [Google Scholar] [CrossRef]
- Wang, H.; Chang, K.F.; Hwang, S.F.; Turnbull, G.D.; Howard, R.J.; Blade, S.F.; Callan, N.W. Fusarium root rot of coneflower seedlings and integrated control using Trichoderma and fungicides. BioControl 2005, 50, 317–329. [Google Scholar] [CrossRef]
- Miller, N.F.; Standish, J.R.; Quesada-Ocampo, L.M. Sensitivity of Fusarium oxysporum f. sp. niveum to prothioconazole and pydiflumetofen in vitro and efficacy for Fusarium wilt management in watermelon. Plant Hlth. Prog. 2020, 21, 13–18. [Google Scholar] [CrossRef]
- Schreuder, W.; Lamprecht, S.C.; Holz, G. Race determination and vegetative compatibility grouping of Fusarium oxysporum f. sp. melonis from south Africa. Plant Dis. 2000, 84, 231–234. [Google Scholar] [CrossRef]
- Namiki, F.; Shiomi, T.; Nishi, K.; Kayamura, T.; Tsuge, T. Pathogenic and genetic variation in the Japanese strains of Fusarium oxysporum f. sp. melonis. Phytopathology 1998, 88, 804–810. [Google Scholar] [CrossRef]
- Perchepied, L.; Pitrat, M. Polygenic inheritance of partial resistance to Fusarium oxysporum f. sp. melonis race 1.2 in melon. Phytopathology 2004, 94, 1331–1336. [Google Scholar] [CrossRef]
- de Cara, M.; Fernández, E.J.; Blanco, R.; Tello Marquina, J.C.; Estrada, F.J.; Montoya, S. Detection of Fusarium oxysporum f. sp. melonis race 1 in soil in Colima State, Mexico. Plant Dis. 2004, 88, 1383. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, S.W.; Ma, Z.H.; Wang, W.J.; Gao, L.H.; Han, C.G.; Yang, A.P.; Wu, X.H. Anastomosis groups and mycovirome of Rhizoctonia isolates causing sugar beet root and crown rot and their sensitivity to flutolanil, thifluzamide, and pencycuron. J. Fungi 2023, 9, 545. [Google Scholar] [CrossRef]
- Froussard, P. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 1992, 20, 2900. [Google Scholar] [CrossRef]
- Li, S.W.; Li, Y.T.; Hu, C.H.; Han, C.G.; Zhou, T.; Zhao, C.; Wu, X.H. Full genome sequence of a new mitovirus from the phytopathogenic fungus Rhizoctonia solani. Arch. Virol. 2020, 165, 1719–1723. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.J.; Wang, J.R.; Cheng, X.H.; Liu, Y.; Zhang, X.Y. Preliminary studies on the effects of oyster mushroom spherical virus China strain on the mycelial growth and fruiting body yield of the edible mushroom Pleurotus ostreatus. Biology 2022, 11, 574. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.J.; Hua, H.H.; Wu, C.Y.; Zhou, T.; Wu, X.H. A botybirnavirus isolated from Alternaria tenuissima confers hypervirulence and decreased sensitivity of its host fungus to difenoconazole. Viruses 2022, 14, 2093. [Google Scholar] [CrossRef] [PubMed]
- Segorbe, D.; Di Pietro, A.; Pérez-Nadales, E.; Turrà, D. Three Fusarium oxysporum mitogen-activated protein kinases (MAPKs) have distinct and complementary roles in stress adaptation and cross-kingdom pathogenicity. Mol. Plant Pathol. 2017, 18, 912–924. [Google Scholar] [CrossRef] [PubMed]
- Aegerter, B.J.; Gordon, T.R.; Davis, R.M. Occurrence and pathogenicity of fungi associated with melon root rot and vine decline in California. Plant Dis. 2000, 84, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Jacquat, A.G.; Theumer, M.G.; Cañizares, M.C.; Debat, H.J.; Iglesias, J.; García Pedrajas, M.D.; Dambolena, J.S. A survey of mycoviral infection in Fusarium spp. Isolated from maize and sorghum in Argentina identifies the first mycovirus from Fusarium verticillioides. Viruses 2020, 12, 1161. [Google Scholar] [CrossRef] [PubMed]
- Aoki, N.; Moriyama, H.; Kodama, M.; Arie, T.; Teraoka, T.; Fukuhara, T. A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata. Virus Res. 2009, 140, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, H.; Liao, X.L.; Gao, B.; Lu, X.; Sun, D.; Gong, W.; Zhong, J.; Zhu, H.; Pan, X.; et al. Mycoviral gene integration converts a plant pathogenic fungus into a biocontrol agent. Proc. Natl. Acad. Sci. USA 2022, 119, e2214096119. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, K.B.; Holcomb, E.E.; Allscheid, R.L.; Carrington, J.C. Hiding in plain sight: New virus genomes discovered via a systematic analysis of fungal public transcriptomes. PLoS ONE 2019, 14, e0219207. [Google Scholar] [CrossRef]
- Hammond, T.M.; Andrewski, M.D.; Roossinck, M.J.; Keller, N.P. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell 2008, 7, 350–357. [Google Scholar] [CrossRef]
- Lutz, T.; Langer, G.; Heinze, C. Complete genome sequence of a novel alternavirus infecting the fungus Ilyonectria crassa. Arch. Virol. 2023, 168, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Shi, N.J.; Wang, P.; Zhu, Q.Y.; Yang, G.G.; Huang, B. Molecular characterization of a novel alternavirus infecting the entomopathogenic fungus Cordyceps chanhua. Arch. Virol. 2022, 167, 1467–1470. [Google Scholar] [CrossRef] [PubMed]
- Pielhop, T.P.; Popp, C.; Fricke, S.; Knierim, D.; Margaria, P.; Maiß, E. Molecular characterization of two new alternaviruses identified in members of the fungal family Nectriaceae. Arch. Microbiol. 2023, 205, 129. [Google Scholar] [CrossRef] [PubMed]
- Kozlakidis, Z.; Herrero, N.; Ozkan, S.; Kanhayuwa, L.; Jamal, A.; Bhatti, M.F.; Coutts, R.H. Sequence determination of a quadripartite dsRNA virus isolated from Aspergillus foetidus. Arch. Virol. 2013, 158, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Z.; Zhang, Y.F.; Liu, Y.Y.; Xiao, J.B.; Huang, Z.J.; Li, Y.F.; Li, H.P.; Li, P.F. Virome analysis of an ectomycorrhizal fungus Suillus luteus revealing potential evolutionary implications. Front. Cell. Infect. Microbiol. 2023, 13, 1229859. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.A.; Baig, D.I.; Bhatti, M.F. An overview of mycoviral curing strategies used in evaluating fungal host fitness. Mol. Biotechnol. 2023, 65, 1547–1564. [Google Scholar] [CrossRef]
- Khan, H.A.; Sato, Y.; Kondo, H.; Jamal, A.; Bhatti, M.F.; Suzuki, N. A second capsidless hadakavirus strain with 10 positive-sense single-stranded RNA genomic segments from Fusarium nygamai. Arch. Virol. 2021, 166, 2711–2722. [Google Scholar] [CrossRef] [PubMed]
- Romeralo, C.; Bezos, D.; Martinez-Alvarez, P.; Diez, J.J. Vertical transmission of Fusarium circinatum mitoviruses FcMV1 and FcMV2-2 via microconidia. Forests 2018, 9, 356. [Google Scholar] [CrossRef]
- Ma, G.P.; Zhang, X.F.; Hua, H.H.; Zhou, T.; Wu, X.H. Molecular and biological characterization of a novel strain of Alternaria alternata chrysovirus 1 identified from the pathogen Alternaria tenuissima causing watermelon leaf blight. Virus Res. 2020, 280, 197904. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, H.; Zhang, X.; Liu, L.; Wu, X. A Novel Strain of Fusarium oxysporum Alternavirus 1 Isolated from Fusarium oxysporum f. sp. melonis Strain T-BJ17 Confers Hypovirulence and Increases the Sensitivity of Its Host Fungus to Difenoconazole and Pydiflumetofen. Viruses 2024, 16, 901. https://doi.org/10.3390/v16060901
Hua H, Zhang X, Liu L, Wu X. A Novel Strain of Fusarium oxysporum Alternavirus 1 Isolated from Fusarium oxysporum f. sp. melonis Strain T-BJ17 Confers Hypovirulence and Increases the Sensitivity of Its Host Fungus to Difenoconazole and Pydiflumetofen. Viruses. 2024; 16(6):901. https://doi.org/10.3390/v16060901
Chicago/Turabian StyleHua, Huihui, Xinyi Zhang, Li Liu, and Xuehong Wu. 2024. "A Novel Strain of Fusarium oxysporum Alternavirus 1 Isolated from Fusarium oxysporum f. sp. melonis Strain T-BJ17 Confers Hypovirulence and Increases the Sensitivity of Its Host Fungus to Difenoconazole and Pydiflumetofen" Viruses 16, no. 6: 901. https://doi.org/10.3390/v16060901





