Abstract
HPV16 is responsible for approximately 60% and 90% of global HPV–induced cervical and oropharyngeal cancers, respectively. HPV16 intratype variants have been identified by HPV genome sequencing and classified into four phylogenetic lineages (A–D). Our understanding of HPV16 variants mostly derives from epidemiological studies on cervical cancer (CC) in which HPV16 B, C, and D lineages (previously named “non-European” variants) were mainly associated with high-grade cervical lesions and cancer. Although a predominance of HPV16 lineage A (previously named “European variants”) has been observed in head and neck squamous cell carcinoma (HNSCC), epidemiological and in vitro biological studies are still limited for this tumor site. Next Generation Sequencing (NGS) of the entire HPV genome has deepened our knowledge of the prevalence and distribution of HPV variants in CC and HNSCC. Research on cervical cancer has shown that certain HPV16 sublineages, such as D2, D3, A3, and A4, are associated with an increased risk of cervical cancer, and sublineages A4, D2, and D3 are linked to a higher risk of developing adenocarcinomas. Additionally, lineage C and sublineages D2 or D3 of HPV16 show an elevated risk of developing premalignant cervical lesions. However, it is still crucial to conduct large-scale studies on HPV16 variants in different HPV–related tumor sites to deeply evaluate their association with disease development and outcomes. This review discusses the current knowledge and updates on HPV16 phylogenetic variants distribution in HPV–driven anogenital and head and neck cancers.
1. Introduction
Human papillomaviruses (HPVs) are non-enveloped DNA viruses that infect both cutaneous and mucosal epithelia. To date, over 400 papillomaviruses (PVs) have been identified and over 200 are HPV genotypes classified into alpha, beta, gamma, mu, and nu genera [1] (www.hpvcenter.se) (accessed on 13 March 2024). A clear association with human cancers has been established for the following mucosal alphapapillomavirus HPV types, namely HPV51, grouped in the alpha-5 species; HPV 56, in the alpha-6; HPV18, 39, 45, and 59, in the alpha-7; and HPV16, 31, 35, 33, 52, and 58, grouped as alpha-9 species. Therefore, they have been classified by the International Agency for Research on Cancer (IARC) monograph as carcinogenic (Group 1) or high risk (HR) HPV genotypes [2]. Among them, approximately 60% of cervical cancer and 90% of oropharyngeal HPV–driven cancers are attributable to HPV16.
In particular, the HPV16 genotype belongs to the Alphapapillomavirus genus, and it is included in species 9. Its circular double-stranded DNA genome comprises the following regions: (i) the long control region (LCR), containing genetic elements involved in viral replication and transcription; (ii) the early (E) region encoding for the non-structural proteins E1, E2, E4, E5, E6, and E7 that are involved in fundamental viral processes, with E6 and E7 responsible for HPV oncogenicity; and (iii) the late (L) region, encoding the structural proteins L1 and L2, which are, respectively, the major and minor viral capsid proteins (Figure 1A–C).
The classification of HPV into genera, species, and genotypes is primarily based on the nucleotide sequence of the L1 ORF, the most conserved gene, which encodes the major viral capsid protein [3,4]. A minimum of 60% identity in the L1 nucleotide (nt) sequence defines HPV types belonging to the same genus. Viruses with L1 nt sequence identity between 71% and 89% belong to the same species (e.g., Alphapapillomavirus species 9), while viruses showing more than 90% L1 nt identity are defined as distinct genotypes (e.g., HPV16 or HPV18).
Additionally, HPV intra-genotype variants have been identified by sequencing and phylogenetic analysis of the entire viral genome and classified into lineages when the genome variability ranges from 1% to 10% (HPV16 lineages A–D), and into sublineages when the variability ranges from 0.5% to 1% (HPV16 sublineages A1–4, B1–4, C1–4, and D1–4) [3,5] (pave.niaid.nih.gov accessed on 13 March 2024). HPV16 variant classification and their phylogenetic stratification into lineages and sublineages are reported in Table 1 and Figure 2.

Table 1.
Alphanumeric and geographical classification systems of the HPV16 variants. The known HPV16 variants, stratified into lineages and sublineages, and the respective GenBank numbers are listed in the table and indicated in the text. The HPV16 variants named B3 (KU053915), B4 (KU053914), C2 (HQ644244), C3 (KU053921), C4 (KU053922), and D4 (KU053933) were only classified with the alphanumeric system [5] (https://pave.niaid.nih.gov/explore/variants/variant_genomes) (accessed on 13 March 2024).
Studies on the role of HPV16 variants were mainly focused on cervical cancer, highlighting a link between specific phylogenetic HPV variants and a higher cancer risk. Some HPV16 lineages/sublineages were reported to be preferentially associated with an increased risk of cancer, such as HPV16 sublineages D2, D3, or A4. They are also found to be associated with an increased risk of adenocarcinoma. Moreover, the role of single nucleotide polymorphisms (SNPs) in the HPV16 genome, such as the E6 T350G that leads to the amino acid substitution L83V, has been investigated to understand their role in cervical disease progression and viral persistence. Thus, investigating HPV genome variability could be beneficial to further exploring the possible association with HPV–related tumors and/or with the disease outcome.
HPV does not encode its own DNA polymerase. Instead, it recruits high-fidelity host enzymes for viral genome synthesis, resulting in a low mutation rate across the HPV genome. DNA mutations occur differently in coding and non-coding viral genomic regions. In coding regions, the estimated mutation rate ranges from 2 × 10−8 to 5 × 10−9 substitutions per site/year, whereas in non-coding regions, the rate is twice as fast, as reviewed in [6].
Moreover, during viral infection, as a part of the innate immune response of the host, the HPV genome can be targeted by cytosine deaminases of the apolipoprotein B mRNA editing catalytic polypeptide-like 3 (APOBEC3 or A3) family, which includes A3A, A3C, A3H, A3B, A3D, A3F, and A3G [7,8]. The APOBEC A3 enzymes catalyze the cytosine-to-uracil conversion, leading to a thymidine substitution during the viral replication. They act as viral restriction factors to clear the infection. Human APOBEC3 enzymes can inhibit a broad spectrum of viruses, such as HIV, HBV, HHV-1, and HHV-4 [7,9,10,11]. Some studies have reported that a group of viruses, including HR HPV types, have evolved mechanisms to induce the upregulation of some APOBEC3 family members [11,12,13]. Thus, HPV variants may arise due to mutations driven by the APOBEC activity, which targets the viral genome during the infection and may accidentally contribute to viral evolution. Alternatively, HPV variants may be selected and evolve to evade APOBEC activity by reducing APOBEC3 target sequences in their genome [14].
Here, the role of HPV16 phylogenetic variants in HPV–related cancers is reviewed and the implications of variants in anogenital and head and neck cancers are also discussed based on recent NGS findings.
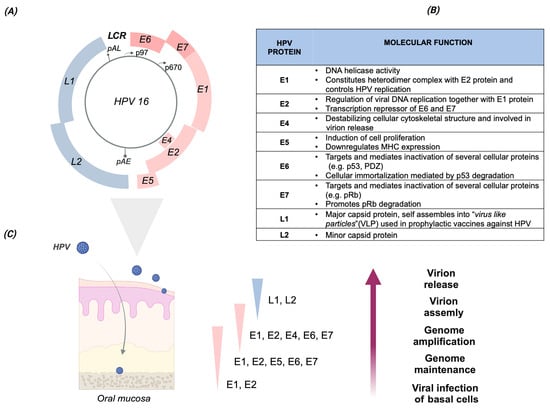
Figure 1.
HPV16 genome organization, protein functions, and viral life cycle during productive infection. (A) Genome organization of the HPV16 genotype. The HPV genome is a double-stranded DNA (indicated by the gray circle), with a size of about 7900 bp. The major six early (E) open reading frames (ORFs), namely E6, E7, E1, E2, E4, and E5, and the two late (L) ORFs, namely L1 and L2, are indicated by different colors, with E6 and E7 shown in dark pink. Furthermore, the HR HPVs express an additional early protein, E8ˆE2C, by spliced mRNA. The major early p97 and late p670 promoters are indicated by arrows. The early and late polyadenylation sites, pAE and pAL, respectively, are also indicated by grey bar lines. The long control region, LCR (alternatively named the upstream regulatory region, or URR), comprises the replication origin and sequences involved in transcription. (B) HPV16 proteins. List of the HPV16 proteins and their principal functions. (C) HPV life cycle during productive infection. The viral life cycle during productive HPV infection in the host epithelial tissue (schematically represented on the left) is characterized by a specific pattern of viral gene expression across the epithelial layers. The viral life cycle is strictly regulated and linked to the host cell epithelial differentiation process. As reported for the cervical epithelium, HPV gains access to the basal layer through the epithelial transition zones (TZ) of the uterine cervix, in the presence of microlesions, wounds, or cuts. After the infection of basal cells, the HPV genome is maintained in the nucleus in an episomal state at a relatively low copy number. The expression of E6 and E7, through the p97 promoter, is necessary to start the viral life cycle. As the infected cells migrate to the upper epithelial layers, the viral proteins E1, E2, E4, and E5 are upregulated via the p670 promoter to facilitate viral genome amplification. In the upper epithelial layers, the expression of the late viral capsid proteins, L1 and L2, promotes capsid assembly and subsequent release of the new virion from the epithelial surface. Text and figures are based on the following manuscript: [15,16,17,18,19]. Created with BioRender.com.
Figure 1.
HPV16 genome organization, protein functions, and viral life cycle during productive infection. (A) Genome organization of the HPV16 genotype. The HPV genome is a double-stranded DNA (indicated by the gray circle), with a size of about 7900 bp. The major six early (E) open reading frames (ORFs), namely E6, E7, E1, E2, E4, and E5, and the two late (L) ORFs, namely L1 and L2, are indicated by different colors, with E6 and E7 shown in dark pink. Furthermore, the HR HPVs express an additional early protein, E8ˆE2C, by spliced mRNA. The major early p97 and late p670 promoters are indicated by arrows. The early and late polyadenylation sites, pAE and pAL, respectively, are also indicated by grey bar lines. The long control region, LCR (alternatively named the upstream regulatory region, or URR), comprises the replication origin and sequences involved in transcription. (B) HPV16 proteins. List of the HPV16 proteins and their principal functions. (C) HPV life cycle during productive infection. The viral life cycle during productive HPV infection in the host epithelial tissue (schematically represented on the left) is characterized by a specific pattern of viral gene expression across the epithelial layers. The viral life cycle is strictly regulated and linked to the host cell epithelial differentiation process. As reported for the cervical epithelium, HPV gains access to the basal layer through the epithelial transition zones (TZ) of the uterine cervix, in the presence of microlesions, wounds, or cuts. After the infection of basal cells, the HPV genome is maintained in the nucleus in an episomal state at a relatively low copy number. The expression of E6 and E7, through the p97 promoter, is necessary to start the viral life cycle. As the infected cells migrate to the upper epithelial layers, the viral proteins E1, E2, E4, and E5 are upregulated via the p670 promoter to facilitate viral genome amplification. In the upper epithelial layers, the expression of the late viral capsid proteins, L1 and L2, promotes capsid assembly and subsequent release of the new virion from the epithelial surface. Text and figures are based on the following manuscript: [15,16,17,18,19]. Created with BioRender.com.
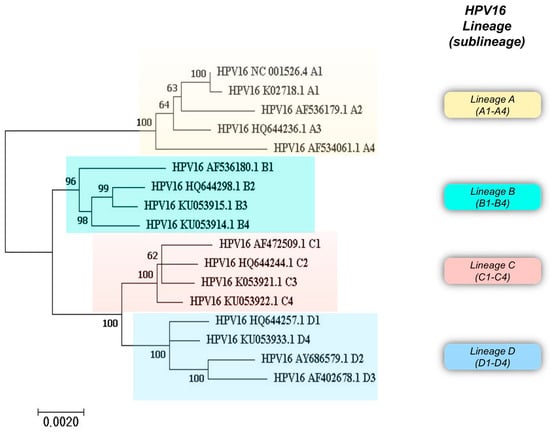
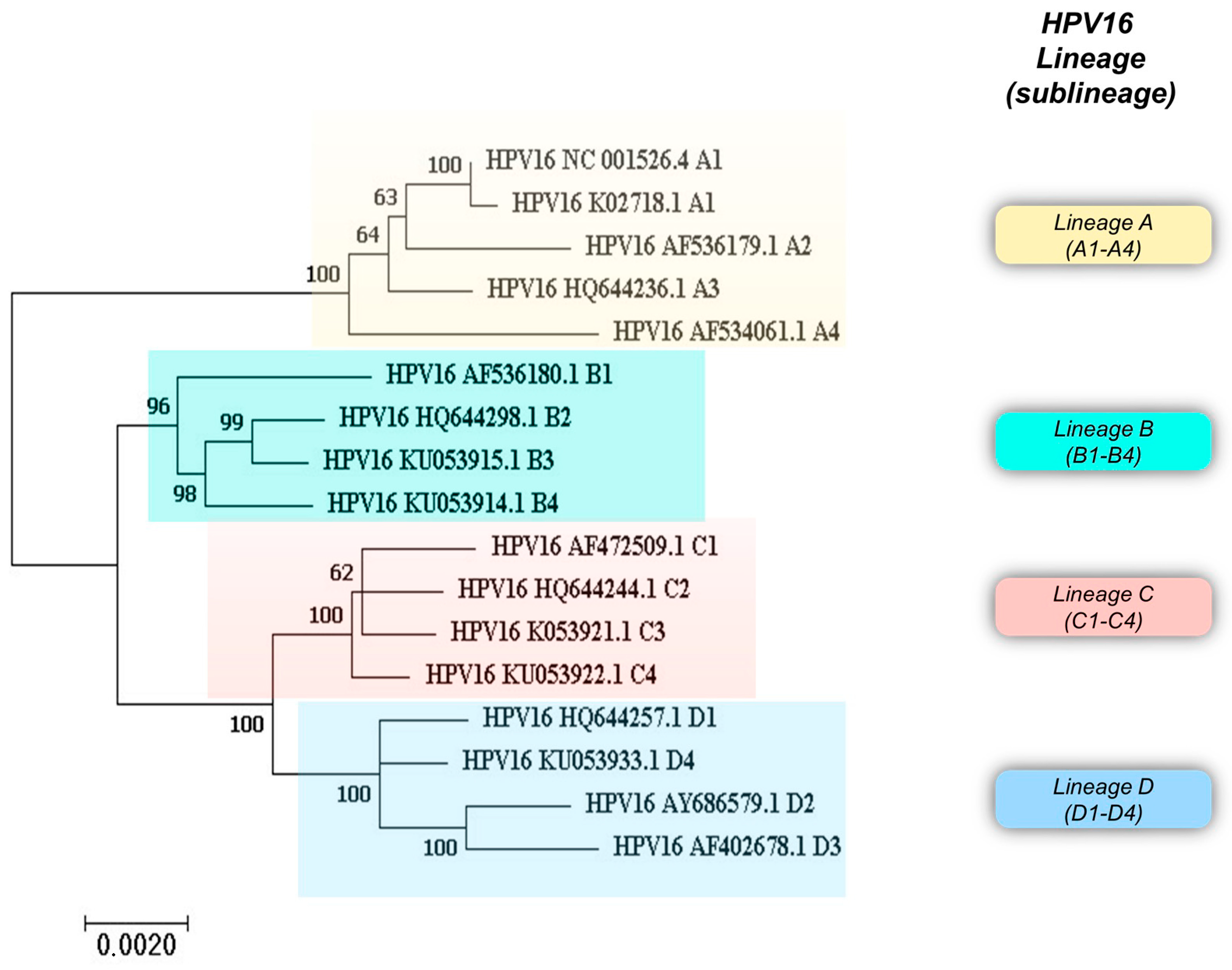
Figure 2.
Phylogenetic analysis of HPV16 variant reference sequences with the corresponding GenBank accession number. Phylogenetic analysis of HPV16 variant sequences was performed with MEGA 7.0.26 software. The tree was built using the Maximum Likelihood method.
Figure 2.
Phylogenetic analysis of HPV16 variant reference sequences with the corresponding GenBank accession number. Phylogenetic analysis of HPV16 variant sequences was performed with MEGA 7.0.26 software. The tree was built using the Maximum Likelihood method.
2. Methodology
The aim of the present review was to give an overview of the most investigated HPV16 SNPs, variant lineages, and sublineages and their geographic distribution, and to describe their possible link with HPV–related cancers. Moreover, the comprehensive review presented here will be helpful in exploring the role of HPV16 phylogenetic variants and their contribution to cancer progression. The literature search strategy relied only on the use of PubMed with the following keywords: “HPV16 variants”, “HPV16 lineages”, “HPV16 sublineages”, “HPV16 SNPs”, “HPV16 variants and cervical cancer, “HPV16 variants and head and neck cancer”, “HPV16 variants and oropharyngeal cancer”, “HPV16 variants and HPV-associated cancers”, and “HPV16 variants and next generation sequencing”. In the present manuscript, peer-reviewed reviews, meta-analyses, and original research articles were included. Non-peer-reviewed sources, articles not available in English, and studies not including the HPV16 genotype were not included in this review.
3. HPV16 Variants, Lineages, and Their Classification
Overall, analysis of the whole genome sequence of HPV16 has revealed characteristic mutations that categorize HPV16 isolates into four phylogenetic lineages, namely A, B, C, and D, subdivided into 16 sublineages: A1 to A4, B1 to B4, C1 to C4, and D1 to D4. Figure 2 shows a phylogenetic tree built using HPV16 variant reference sequences retrieved from the Pave database (https://pave.niaid.nih.gov) (accessed on 13 March 2024) [5].
The HPV16 variants have been geographically classified depending on the place of their initial identification; in fact, they were named European (E), Asian American (AA), African-1, and African-2 (Af-1 and Af-2) variants [20]. According to this first classification, the European lineages appear to be the most prevalent worldwide [21], showing a large diffusion in Europe, North and South America (sublineages A1–3), and Asia (sublineage A4), and with a minor representation in Africa, where B and C variants predominate [22,23]. Recently, the novel A5 subvariant was identified in cervical samples from Japanese women [24], although its corresponding sequence has yet to be reported in the Pave database (https://pave.niaid.nih.gov) (accessed on 13 March 2024).
An alphanumeric system is now used to classify HPV16 variants [22], while geographic classification is no longer recommended, as reported by Burk et al. and Mirabello et al. [5,25] (Table 1). Indeed, to avoid misleading geographic nomenclatures of HPV variants and facilitate cross-study comparisons, adherence to the new classification based on lineages (A up to D) and sublineages (A1–up to D–4) is recommended [25].
Additionally, Cornet et al. (2012) [23] have reported a combination of SNPs located in the E6 and LCR regions specific to each of the HPV16 sublineages, which they proposed as “diagnostic” SNPs. Yet, some non-lineage-specific SNPs were also found in the HPV16 sublineages, such as the most studied T350G nt variation [23]. Moreover, a larger spectrum of SNPs occurring in the viral early genes, namely E1, E2, E4, E5, E6, and E7, and in the LCR was reviewed by Bletsa et al. (2021) [26]. Focusing on E6, a pattern of SNPs in this viral ORF was found in each of the HPV16 sublineages, such as the A1 and A2 sublineages (T350G), the A4 sublineage (T178G), the B lineage (G132C, C143G, G145T, T286A, A289G, and C335T), the C lineage (T109C, G132T, C143G, G145T, T286A, A289G, C335T, and G403G), and the D lineage (G145T, T286A, A289G, C335T, T350G, and A532G) reviewed in [26]. Also, the nt sequence analysis of E7 revealed specific nucleotide variations related to the following lineages: A4 sublineage (A647G), B lineage (T789C and T795G), C lineage (A647G, T789C, and T795G), and D lineage (T732C, T789C, and T795G), reviewed in [26]. Most of the SNPs located in the E6 and E7 genes are described in the following paragraphs and in Table 2. Mainly SNPs in HPV E6 and E7 ORFs have been investigated, as these genes encode for oncoproteins that facilitate the degradation of essential tumor suppressor cellular proteins, such as p53 and pRb, involved in the cell cycle, apoptosis, and cellular proliferation pathways [15] (Figure 1B). Therefore, E6 and E7 are recognized as the major viral oncoproteins, since they are the main drivers of HPV–mediated carcinogenicity.

Table 2.
Main findings from studies reporting (A) HPV phylogenetic variants distribution in head and neck tumors from 2000 to 2023, including those exploring also (B) specific SNPs in E6 and E7 viral oncogenes.
4. HPV16 Variants and Cervical Cancer
HR HPV genotypes are associated with various anogenital cancers, i.e., cervical, vaginal, vulvar, penile, and anal cancers, as well as oropharyngeal squamous cell carcinoma (OPSCC). HR HPV genotypes display different oncogenicity, with HPV16 being the most prevalent type detected in premalignant and malignant lesions [2,41] and responsible for the majority of cervical cancers worldwide [42,43]. Epidemiological studies conducted in patients with cervical cancer have provided evidence that HPV16 variants differ in (i) geographical distribution, (ii) persistence in the infected host, and (iii) ability to favor progression to cancer [44,45,46].
In the next subsections, data from studies exploring HPV variant distribution and persistence in the host are discussed; the sturdiness and advancement of research studies are highlighted together with the connection between viral variants and disease outcome.
4.1. HPV Variant Distribution and Persistence in the Host
Focusing on geographical variant distribution, the HPV16 sublineages A1, A2, and A3 are the most prevalent worldwide and are responsible for the majority of HPV16 infections as observed by analyzing cancer samples and healthy controls [23,47,48]. The persistence of HPV16 infection in the host is a key step for viral-mediated carcinogenesis. Previous studies have often compared “European” to “non-European” HPV16 variants and found that non-European variants exhibit an increased risk of viral persistence [46,49,50,51,52]. As reported in a 3-year longitudinal follow-up study based on HPV16 whole-genome Sanger sequencing, infections with variants other than A1–A2 (the most common “European” sublineages) are preferentially cleared by the host. Moreover, single nucleotide polymorphisms (SNPs) across the HPV16 genome did not affect clearance or viral persistence, suggesting that the progression to tumor could be host-related [53]. The risk of HPV16 persistence in the infected host has been extensively studied, focusing on specific viral regions; e.g., E6 nucleotide polymorphism 350T versus 350G. As described above, this nucleotide mutation results in AA change from valine to a leucine (L83V) at position 83 in the E6 viral protein. Recently, the presence of the T350G variant has been associated with progression to high-grade lesions and with an elevated risk of developing CC in a study conducted in Argentina [54].
The prevalence of the E6 variation in HPV16 sublineages A1–A3 has shown a higher mutation rate of T350G in Central/South America compared to a European cohort, particularly in cervical and penile cancers [47]. In vitro biological studies supported the epidemiological findings [55,56,57,58,59]. However, it has become evident that the oncogenicity of HPV16 E6 variants could be population-dependent [60,61], underlining a role played by the host’s genetic background [60,62,63,64]. In addition, a two-fold increase in the risk of HPV persistence has been observed for the European E6 350T prototype in a European cohort [64,65].
Recently, E6 AA changes have been studied by using several machine-learning approaches to predict the development of high-grade cervical lesions (H-SIL) [66]. These in silico findings indicate that D32E and H85Y AA mutations in the E6 protein result in an increased ability to degrade p53 when compared to the E6 prototype [66].
4.2. Sturdiness and Advancement
To better discriminate the variability within the HPV genome sequence, large-scale studies based on NGS techniques have been designed [52,63,67,68]. This sequencing methodology allows for high throughput testing and facilitates full analysis of the entire HPV genome to unravel mutations.
Mirabello and collaborators assessed HPV16 variant lineages and their association with the risk of developing cervical precancer and cancer in 3200 enrolled women [63]. They showed that sublineage A4 was associated with an increased risk of developing adenocarcinoma, whereas lineage C showed an elevated risk of developing cervical premalignant lesion CIN3, as did the D2–D3 sublineages, which are also associated with an increased cancer risk compared to the A1–A2 sublineages [63]. Furthermore, the study showed an increased risk of developing precancerous or cancerous lesions when the ethnicity of the patients matched the geographical origin of the infecting HPV16 variants [63]. In another study conducted by Clifford et al. (2019), the HPV16 D2 and D3 sublineages, along with A3 and A4, showed an increased risk of cervical cancer compared to the A1 sublineage [48]. In a study conducted in Guatemala by Lou et al. (2020), the HPV16 D2 and D3 sublineages were frequently observed in cervical tumors and in adenocarcinoma histological type [69]. Moreover, in cancers harboring the HPV16 D2 sublineages, the authors reported a higher rate of viral DNA integration into the host genome [69].
In a study conducted in Japan by whole genome sequencing (WGS) on HPV16 isolates from cervical samples collected from women with and without cervical malignancies and invasive cervical cancer, Hirose et al. (2019) reported a prevalent clustering of HPV16 isolates mainly in sublineages A4 (52%) and A1 (21%) [24]. Sublineage A4 showed a significantly higher risk for cervical cancer development compared to other A sublineages [24]. In addition, the A4 sublineage was frequently detected in invasive CC (73.2% of cases), and a higher risk of progression from premalignant lesions (CIN2-3) to cervical squamous cell carcinoma was found for this sublineage compared to HPV16 clades A1, A2, and A3 [24].
4.3. Disease Outcome
Exploring the role of HPV variants in premalignant and malignant cervical lesions, HPV16 B, C, and D lineages (known as the “non-European” variants) have been shown to be prevalent in high-grade lesions and cervical squamous cell carcinoma [44,50,51,70,71]; however, some studies have reported a lack of association with cervical disease [72,73,74]. In a large study across Europe, Asia, and Central/South America, HPV16 sublineages A1–A3 were shown to be the most prevalent in cervical, vaginal, and penile cancers, regardless of the geographical origin of patients, while sublineage A4 was mainly associated with anal cancer in Asian cohorts [47]. Recently, in the context of the HPV Infection in Men (HIM) studies, a high prevalence of the HPV16 A1 sublineage was found in the anal swabs from men with anal cancer from Brazil, Mexico, and the United States [75], with no significant differences observed between variant lineages and HPV16 persistence [75].
The association between HPV16 sublineages and cancer histology showed an increased risk of developing an adenocarcinoma for the A4 sublineage, while the D2–D3 HPV16 sublineages were strongly associated with an increased risk of developing premalignant CIN3 lesions and cervical cancer, with the strongest risk of adenocarcinomas linked to the D2 sublineage [25,63]. The HPV16 D clade was also prevalent in adenocarcinomas [76,77] from South/Central and North American patients [48], although these findings were not confirmed by De Boer et al. when E6 and L1 were sequenced in a relatively small group of HPV16–positive adenocarcinoma samples [78].
Genomic characterization of 228 primary cervical cancers within “The Cancer Genome Atlas (TCGA)” program showed a predominance of European HPV16 variants (primarily the A1 variant), whereas non-European variants (sublineages A4, B1, C1, D2, and D3) were significantly associated with cervical adenocarcinomas [79].
5. HPV16 Variants and Head and Neck Tumor
Head and neck cancers rank as the sixth most common cancer type globally [80], with an increasing incidence of HPV–driven OPSCC in developed countries, particularly among men [81]. Alcohol, smoking, and persistent HR HPV infections are the major risk factors for HNSCC. A recent systematic review and meta-analysis has provided evidence that artificial intelligence (AI) using image-based analysis can be a promising tool for predicting HPV status in HNSCC [82]. However, its accuracy is still lower compared to the p16INK4a immunohistochemistry, a reference diagnostic method in HPV-related OPC [82]. Among HR HPVs, the majority (up to 90%) of HPV–driven OPSCCs are related to HPV16 infection. The prevalence of HPV–induced OPSCCs varies geographically, ranging from 22 to more than 74% [83], whereas only 2.0–3.9% of oral and 2.0–3.1% of laryngeal cancers are attributed to HR HPV infections [83,84,85]. Although HPV16 variants have been extensively studied in cervical cancer, less is known about their significance in head and neck cancers [86]. A list of studies conducted on HNSCC and published in the last 23 years is reported in Table 2.
A study conducted in Greece showed that 85% of HPV16 sequences detected in 40 specimens from subjects with HNSCC were clustered into the European sublineage A3, while the remaining 15% of HPV16 variants were related to sublineage D1 [35]. In another study on HPV16–positive OPSCCs performed in the USA, the A1 sublineage was associated with poor recurrence-free survival (RFS) [87]. Conversely, variants other than the HPV16 A1 sublineage are correlated with improved RFS, particularly in moderate or low tobacco smokers [87].
A recent systematic review of studies on head and neck cancers and HPV16 variants using geographical nomenclature and including studies from the USA (n = 3), Germany (n = 1), Italy, (n = 2) Brazil (n = 1), Japan (n = 1), and Iran (n = 1) revealed a predominance of European variants in HNSCCs, followed by Asian American and African lineages [86]. Therefore, even though a predominance of European strains has been reported, no correlation with patient prognosis has been made [86].
In a comparative study conducted in the USA on HPV16 variant distribution in CC and OPSCC by grouping both European and Asian (E plus A) variants, the authors showed that this combined variant group was more prevalent in OPSCC compared to CC [32]. Conversely, the group of Asian American (AA1 plus AA2) HPV16 variants prevailed in cervical samples. In addition, non-synonymous mutations in the E6 protein showed significantly higher prevalence rates in OPSCCs, while E7 nucleotide sequences showed fewer mutations in both cancer types [32].
Conversely, an Italian study reported a prevalence of 19.6% for African HPV16 variants in HNSCC samples, highlighting their relevance in the head and neck anatomical tumor site [30]. As seen before in CC studies, when focusing on specific viral genome regions, such as the E6 gene, the most frequent polymorphism reported in a small cohort of HNSCCs was T350G [35,37]. However, this finding needs further investigation to determine its impact on clinical outcomes. Others have compared the frequency of the E6 polymorphisms in tonsillar squamous cell carcinoma (TSCC), CC, and cervical samples from Swedish patients [39], and reported that the R10G amino acid change (nt A131G) was frequent in TSCC, rare in cervical samples, and absent in CC, with no significant differences found in 3-year disease-free survivors among patients. In addition, European E6 variants carrying the L83V (nt change T350G) mutation were detected across all cancer types, with no significant correlation found with disease-free survivors among the patients [39].
Finally, a comparative study performed in the USA based on E6 sequencing data from oral rinse samples and matched tumor tissues showed that the most frequent variants were European [29], with the E6 E-350T prototype (n = 6) being the most prevalent in oral rinse samples, followed by the E6 variant E-350G (n = 4).
To date, very few NGS–based studies on HPV variants in HNSCC specimens have been conducted [33,34]. Thus, further studies are needed to elucidate the possible impact of HPV16 phylogenetic variants on HNSCC and particularly in OPC. In a large USA–based study focused on HPV16–positive OPSCC, whole-genome NGS sequencing identified A1 as the most prevalent sublineage, although no correlation between HPV variant lineages and histological subtypes was reported [33]. In a recent study performed in the USA on a cohort of OPC patients, the majority of the HPV16 variants belonged to the A lineage (90.3% of cases) [34]. Among them, A1 was the most common sublineage, being detected in more than a half of the cases, followed by A2 (27.8%), D3 (6.5%), and A4 (5.7%) [34]. The most important findings in this study include eight SNPs, observed in some HPV genes, significantly associated with reduced patient survival. These polymorphisms were found in the viral E1 gene (nt position 1053), with four in the L2 gene (nt positions 4410, 4539, 5050, and 5254), two in the L1 gene (nt position 5962 and 6025), and one in the LCR region (nt position 7173) [34]. The latter was strongly associated with an increased mortality hazard rate. These results indicate that nucleotide variations across the HPV16 genome can impact the prognosis of HPV–positive OPC patients [34]. However, HPV infection is a necessary but insufficient condition for cancer development, which is a multifactorial event involving lifestyle, environmental, and genetic host factors.
Moreover, in recent years, evidence suggests that mutations in certain host genomic loci (e.g., HLA) are associated with either cervical cancer or head and neck cancer susceptibility [88,89]. Nevertheless, the precise contribution of these mutations in cancer development remains unknown, and specific studies on this topic need to be addressed [90].
6. Conclusions
To date, the majority of studies aimed at exploring the significance and distribution of HPV variants have been designed and mainly conducted on cervical cancer. Overall, some limitations should be highlighted, including the small sample size analyzed in some studies, the paucity of current studies of HPV variants in HPV–associated cancers other than cervical cancer, the shortage of NGS–based investigations, together with the absence of mechanistic biological studies focusing on specific HPV16 SNPs.
The main findings from the cervical cancer studies could be summarized as follows: the HPV16 sublineages, namely D2, D3, A3, and A4, show an increased risk of cervical cancer development, with the HPV16 sublineages A4, D2, and D3 mainly associated with an increased risk of developing adenocarcinomas. Also, studying the HPV genome variability by NGS-WGS in head and neck cancer specimens has revealed associations between different HPV16 SNPs (e.g., E1, L1, L2, and LCR) and disease prognosis in HPV–related OPC. The potential impact of HPV variants on cancer development at different anatomical sites and their association with disease outcome remains largely unexplored and needs further investigation. Finally, both NGS techniques and the current alphanumeric nomenclature of HPV variants should be used in future epidemiological studies to facilitate a fast and accurate molecular characterization of the HPV16 genome in large-scale studies. In conclusion, with the advent of new molecular techniques such as NGS-WGS, additional studies are warranted to achieve an in-depth and precise characterization of HPV variants in CC and HNSCC, as well as in other HPV–related tumors.
Author Contributions
Conceptualization and original draft preparation: L.G., T.G. and P.D.B.; draft revision and implementation of the bibliography data: P.D.B., T.G., M.V.C. and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are available upon reasonable request.
Acknowledgments
We are grateful to Massimo Tommasino for the helpful discussions. We thank Nicole Suty for her help in the preparation of this manuscript. The authors apologize to those scientists whose important contributions in the topic were accidentally not cited or adequately discussed in this review.
Conflicts of Interest
The authors declare no competing interests. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization and of the Istituto Superiore di Sanità, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization, nor of the Istituto Superiore di Sanità.
References
- Bzhalava, D.; Eklund, C.; Dillner, J. International standardization and classification of human papillomavirus types. Virology 2015, 476, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part B: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- Bernard, H.-U.; Burk, R.D.; Chen, Z.; Van Doorslaer, K.; zur Hausen, H.; de Villiers, E.-M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010, 401, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef]
- Burk, R.D.; Harari, A.; Chen, Z. Human papillomavirus genome variants. Virology 2013, 445, 232–243. [Google Scholar] [CrossRef]
- Bravo, I.G.; Felez-Sanchez, M. Papillomaviruses: Viral evolution, cancer and evolutionary medicine. Evol. Med. Public. Health 2015, 2015, 32–51. [Google Scholar] [CrossRef]
- Simon, V.; Bloch, N.; Landau, N.R. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 2015, 16, 546–553. [Google Scholar] [CrossRef]
- Warren, C.J.; Westrich, J.A.; Van Doorslaer, K.; Pyeon, D. Roles of APOBEC3A and APOBEC3B in Human Papillomavirus Infection and Disease Progression. Viruses 2017, 9, 233. [Google Scholar] [CrossRef]
- Mangeat, B.; Turelli, P.; Caron, G.; Friedli, M.; Perrin, L.; Trono, D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 2003, 424, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Suspène, R.; Aynaud, M.-M.; Koch, S.; Pasdeloup, D.; Labetoulle, M.; Gaertner, B.; Vartanian, J.-P.; Meyerhans, A.; Wain-Hobson, S. Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J. Virol. 2011, 85, 7594–7602. [Google Scholar] [CrossRef]
- Wallace, N.A.; Münger, K. The curious case of APOBEC3 activation by cancer-associated human papillomaviruses. PLoS Pathog. 2018, 14, e1006717. [Google Scholar] [CrossRef] [PubMed]
- Starrett, G.J.; Marcelus, C.; Cantalupo, P.G.; Katz, J.P.; Cheng, J.; Akagi, K.; Thakuria, M.; Rabinowits, G.; Wang, L.C.; Symer, D.E.; et al. Merkel Cell Polyomavirus Exhibits Dominant Control of the Tumor Genome and Transcriptome in Virus-Associated Merkel Cell Carcinoma. mBio 2017, 8, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Santiago, M.L.; Pyeon, D. APOBEC3: Friend or Foe in Human Papillomavirus Infection and Oncogenesis? Annu. Rev. Virol. 2022, 9, 375–395. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Van Doorslaer, K.; Pandey, A.; Espinosa, J.M.; Pyeon, D. Role of the host restriction factor APOBEC3 on papil-lomavirus evolution. Virus Evol. 2015, 1, vev015. [Google Scholar] [CrossRef] [PubMed]
- Tommasino, M. The human papillomavirus family and its role in carcinogenesis. Semin. Cancer Biol. 2014, 26, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. Papillomavirus life cycle organization and biomarker selection. Dis. Markers 2007, 23, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Egawa, N.; Egawa, K.; Griffin, H.; Doorbar, J. Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses 2015, 7, 3863–3890. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine 2012, 30 (Suppl. S5), F55–F70. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. The papillomavirus life cycle. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2005, 32 (Suppl. S1), S7–S15. [Google Scholar] [CrossRef]
- Yamada, T.; Wheeler, C.M.; Halpern, A.L.; Stewart, A.C.; Hildesheim, A.; Jenison, S.A. Human papillomavirus type 16 variant lineages in United States populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J. Virol. 1995, 69, 7743–7753. [Google Scholar] [CrossRef]
- Smith, J.S.; Lindsay, L.; Hoots, B.; Keys, J.; Franceschi, S.; Winer, R.; Clifford, G.M. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: A meta-analysis update. Int. J. Cancer 2007, 121, 621–632. [Google Scholar] [CrossRef]
- Chen, Z.; DeSalle, R.; Schiffman, M.; Herrero, R.; Wood, C.E.; Ruiz, J.C.; Clifford, G.M.; Chan, P.K.S.; Burk, R.D. Niche adaptation and viral transmission of human papillomaviruses from archaic hominins to modern humans. PLoS Pathog. 2018, 14, e1007352. [Google Scholar] [CrossRef] [PubMed]
- Cornet, I.; Gheit, T.; Franceschi, S.; Vignat, J.; Burk, R.D.; Sylla, B.S.; Tommasino, M.; Clifford, G.M.; IARC HPV Variant Study Group. Human papillomavirus type 16 genetic variants: Phylogeny and classification based on E6 and LCR. J. Virol. 2012, 86, 6855–6861. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Onuki, M.; Tenjimbayashi, Y.; Yamaguchi-Naka, M.; Mori, S.; Tasaka, N.; Satoh, T.; Morisada, T.; Iwata, T.; Kiyono, T.; et al. Whole-Genome Analysis of Human Papillomavirus Type 16 Prevalent in Japanese Women with or without Cervical Lesions. Viruses 2019, 11, 350. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Clarke, M.A.; Nelson, C.W.; Dean, M.; Wentzensen, N.; Yeager, M.; Cullen, M.; Boland, J.F.; Workshop, N.H.; Schiffman, M.; et al. The Intersection of HPV Epidemiology, Genomics and Mechanistic Studies of HPV-Mediated Carcinogenesis. Viruses 2018, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Bletsa, G.; Zagouri, F.; Amoutzias, G.D.; Nikolaidis, M.; Zografos, E.; Markoulatos, P.; Tsakogiannis, D. Genetic variability of the HPV16 early genes and LCR. Present and future perspectives. Expert. Rev. Mol. Med. 2021, 23, e19. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Koch, W.M.; Capone, R.B.; Spafford, M.; Westra, W.H.; Wu, L.; Zahurak, M.L.; Daniel, R.W.; Viglione, M.; Symer, D.E.; et al. Evidence for a Causal Association Between Human Papillomavirus and a Subset of Head and Neck Cancers. JNCI J. Natl. Cancer Inst. 2000, 92, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Badaracco, G.; Rizzo, C.; Mafera, B.; Pichi, B.; Giannarelli, D.; Rahimi, S.S.; Vigili, M.G.; Venuti, A. Molecular analyses and prognostic relevance of HPV in head and neck tumours. Oncol. Rep. 2007, 17, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, Y.; Koch, W.M.; Xiao, W.; Westra, W.H.; Trivett, A.L.; Symer, D.E.; Gillison, M.L. Oral human papillomavirus infection before and after treatment for human papillomavirus 16-positive and human papillomavirus 16-negative head and neck squamous cell carcinoma. Clin. Cancer Res. 2008, 14, 7143–7150. [Google Scholar] [CrossRef]
- Barbieri, D.; Nebiaj, A.; Strammiello, R.; Agosti, R.; Sciascia, S.; Gallinella, G.; Landini, M.P.; Caliceti, U.; Venturoli, S. Detection of HPV16 African variants and quantitative analysis of viral DNA methylation in oropharyngeal squamous cell carcinomas. J. Clin. Virol. 2014, 60, 243–249. [Google Scholar] [CrossRef]
- Betiol, J.C.; Sichero, L.; Costa, H.O.d.O.; de Matos, L.L.; Andreoli, M.A.; Ferreira, S.; Faraj, S.F.; de Mello, E.S.; Sobrinho, J.S.; Brandão, L.G.; et al. Prevalence of human papillomavirus types and variants and p16(INK4a) expression in head and neck squamous cells carcinomas in São Paulo, Brazil. Infect. Agents Cancer 2016, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- LeConte, B.A.; Szaniszlo, P.; Fennewald, S.M.; Lou, D.I.; Qiu, S.; Chen, N.-W.; Lee, J.H.; Resto, V.A. Differences in the viral genome between HPV-positive cervical and oropharyngeal cancer. PLoS ONE 2018, 13, e0203403. [Google Scholar] [CrossRef]
- Lewis, J.S., Jr.; Mirabello, L.; Liu, P.; Wang, X.; Dupont, W.D.; Plummer, W.D.; Pinheiro, M.; Yeager, M.; Boland, J.F.; Cullen, M.; et al. Oropharyngeal Squamous Cell Car-cinoma Morphology and Subtypes by Human Papillomavirus Type and by 16 Lineages and Sublineages. Head. Neck Pathol. 2021, 15, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Kuhs, K.L.; Faden, D.; Chen, L.; Smith, D.; Pinheiro, M.; Wood, C.; Davis, S.; Yeager, M.; Boland, J.; Cullen, M.; et al. Genetic variation within the human papillomavirus type 16 genome is associated with oropharyngeal cancer prognosis. Ann. Oncol. 2022, 33, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Kottaridi, C.; Resta, P.; Leventakou, D.; Gioti, K.; Zygouras, I.; Gouloumi, A.R.; Sakagiannis, G.; Alzahrani, K.J.; Venetikou, M.S.; Anthouli-Anagnostopoulou, F.; et al. The T350G Variation of Human Papillomavirus 16 E6 Gene Prevails in Oro-pharyngeal Cancer from a Small Cohort of Greek Patients. Viruses 2022, 14, 1724. [Google Scholar] [CrossRef] [PubMed]
- Gameiro, S.F.; Salnikov, M.Y.; Zeng, P.Y.F.; Barrett, J.W.; Nichols, A.C.; Mymryk, J.S. HPV16 Intratypic Variants in Head and Neck Cancers: A North American Perspective. Viruses 2023, 15, 2411. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Lohrey, C.; Hunziker, A.; Kahn, T.; Schwarz, E. Human papillomavirus type 16 E6 and E7 genotypes in head-and-neck carcinomas. Oral. Oncol. 2004, 40, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Da Mosto, M.C.; Fuson, R.; Frayle-Salamanca, H.; Trevisan, R.; Del Mistro, A. HPV-16 E6 L83V variant in squamous cell carcinomas of the upper aerodigestive tract. J. Cancer Res. Clin. Oncol. 2008, 135, 559–566. [Google Scholar] [CrossRef]
- Du, J.; Nordfors, C.; Näsman, A.; Sobkowiak, M.; Romanitan, M.; Dalianis, T.; Ramqvist, T. Human papillomavirus (HPV) 16 E6 variants in tonsillar cancer in comparison to those in cervical cancer in Stockholm, Sweden. PLoS ONE 2012, 7, e36239. [Google Scholar] [CrossRef]
- Hassani, S.; Castillo, A.; Ohori, J.; Higashi, M.; Kurono, Y.; Akiba, S.; Koriyama, C. Molecular Pathogenesis of Human Papil-lomavirus Type 16 in Tonsillar Squamous Cell Carcinoma. Anticancer. Res. 2015, 35, 6633–6638. [Google Scholar]
- de Sanjose, S.; Quint, W.G.V.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.-R.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef] [PubMed]
- de Sanjosé, S.; Serrano, B.; Tous, S.; Alejo, M.; Lloveras, B.; Quirós, B.; Clavero, O.; Vidal, A.; Ferrándiz-Pulido, C.; Pavón, M.; et al. Burden of Human Pap-illomavirus (HPV)-Related Cancers Attributable to HPVs 6/11/16/18/31/33/45/52 and 58. JNCI Cancer Spectr. 2018, 2, pky045. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.M.; Smith, J.S.; Plummer, M.; Muñoz, N.; Franceschi, S. Human papillomavirus types in invasive cervical cancer worldwide: A meta-analysis. Br. J. Cancer 2003, 88, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, M.L.; Duraturo, M.L.; Salatiello, I.; Buonaguro, L.; Losito, S.; Botti, G.; Stellato, G.; Greggi, S.; Piccoli, R.; Pilotti, S.; et al. Analysis of human papillomavirus type-16 variants in Italian women with cervical intraepithelial neoplasia and cervical cancer. J. Med. Virol. 2004, 74, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.D.; Terai, M.; Gravitt, P.E.; Brinton, L.A.; Kurman, R.J.; Barnes, W.A.; Greenberg, M.D.; Hadjimichael, O.C.; Fu, L.; McGowan, L.; et al. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer Res. 2003, 63, 7215–7220. [Google Scholar] [PubMed]
- Hildesheim, A.; Schiffman, M.; Bromley, C.; Wacholder, S.; Herrero, R.; Rodriguez, A.C.; Bratti, M.C.; Sherman, M.E.; Scarpidis, U.; Lin, Q.-Q.; et al. Human Papillomavirus Type 16 Variants and Risk of Cervical Cancer. JNCI J. Natl. Cancer Inst. 2001, 93, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Nicolás-Párraga, S.; Gandini, C.; Pimenoff, V.N.; Alemany, L.; de Sanjosé, S.; Bosch, F.X.; Bravo, I.G. HPV16 variants distribution in invasive cancers of the cervix, vulva, vagina, penis, and anus. Cancer Med. 2016, 5, 2909–2919. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.M.; Tenet, V.; Georges, D.; Alemany, L.; Pavón, M.A.; Chen, Z.; Yeager, M.; Cullen, M.; Boland, J.F.; Bass, S.; et al. Human papillo-mavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: Whole viral genome sequences from 7116 HPV16-positive women. Papillomavirus Res. 2019, 7, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, M.; Rodriguez, A.C.; Chen, Z.; Wacholder, S.; Herrero, R.; Hildesheim, A.; Desalle, R.; Befano, B.; Yu, K.; Safaeian, M.; et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010, 70, 3159–3169. [Google Scholar] [CrossRef]
- Villa, L.L.; Sichero, L.; Rahal, P.; Caballero, O.; Ferenczy, A.; Rohan, T.; Franco, E.L. Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J. Gen. Virol. 2000, 81, 2959–2968. [Google Scholar] [CrossRef]
- Sichero, L.; Ferreira, S.; Trottier, H.; Duarte-Franco, E.; Ferenczy, A.; Franco, E.L.; Villa, L.L. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int. J. Cancer 2007, 120, 1763–1768. [Google Scholar] [CrossRef]
- Berumen, J.; Ordoñez, R.M.; Lazcano, E.; Salmeron, J.; Galvan, S.C.; Estrada, R.A.; Yunes, E.; Garcia-Carranca, A.; Gonzalez-Lira, G.; la Campa, A.M.-D. Asian-American Variants of Human Papillomavirus 16 and Risk for Cervical Cancer: A Case-Control Study. JNCI J. Natl. Cancer Inst. 2001, 93, 1325–1330. [Google Scholar] [CrossRef]
- van der Weele, P.; Meijer, C.; King, A.J. Whole-Genome Sequencing and Variant Analysis of Human Papillomavirus 16 Infec-tions. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Totaro, M.E.; Gili, J.A.; Liotta, D.J.; Schurr, T.G.; Picconi, M.A.; Badano, I. Genetic variation in the E6 and E7 genes of human papillomavirus type 16 in northeastern Argentina. J. Med. Virol. 2021, 94, 745–751. [Google Scholar] [CrossRef]
- Togtema, M.; Jackson, R.; Richard, C.; Niccoli, S.; Zehbe, I. The human papillomavirus 16 European-T350G E6 variant can immortalize but not transform keratinocytes in the absence of E7. Virology 2015, 485, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Zehbe, I.; Lichtig, H.; Westerback, A.; Lambert, P.F.; Tommasino, M.; Sherman, L. Rare human papillomavirus 16 E6 variants reveal significant oncogenic potential. Mol. Cancer 2011, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Lichtig, H.; Algrisi, M.; Botzer, L.E.; Abadi, T.; Verbitzky, Y.; Jackman, A.; Tommasino, M.; Zehbe, I.; Sherman, L. HPV16 E6 natural variants exhibit different activities in functional assays relevant to the carcinogenic potential of E6. Virology 2006, 350, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Hochmann, J.; Sobrinho, J.S.; Villa, L.L.; Sichero, L. The Asian-American variant of human papillomavirus type 16 exhibits higher activation of MAPK and PI3K/AKT signaling pathways, transformation, migration and invasion of primary human keratinocytes. Virology 2016, 492, 145–154. [Google Scholar] [CrossRef]
- Sichero, L.; Sobrinho, J.S.; Villa, L.L. Oncogenic potential diverge among human papillomavirus type 16 natural variants. Virology 2012, 432, 127–132. [Google Scholar] [CrossRef]
- Zehbe, I.; Voglino, G.; Delius, H.; Wilander, E.; Tommasino, M. Risk of cervical cancer and geographical variations of human papillomavirus 16 E6 polymorphisms. Lancet 1998, 352, 1441–1442. [Google Scholar] [CrossRef]
- Xi, L.F.; Koutsky, L.A.; Hildesheim, A.; Galloway, D.A.; Wheeler, C.M.; Winer, R.L.; Ho, J.; Kiviat, N.B. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiology Biomarkers Prev. 2007, 16, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.F.; Kiviat, N.B.; Hildesheim, A.; Galloway, D.A.; Wheeler, C.M.; Ho, J.; Koutsky, L.A. Human papillomavirus type 16 and 18 variants: Race-related distribution and persistence. JNCI J. Natl. Cancer Inst. 2006, 98, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Yeager, M.; Cullen, M.; Boland, J.F.; Chen, Z.; Wentzensen, N.; Zhang, X.; Yu, K.; Yang, Q.; Mitchell, J.; et al. HPV16 Sublineage Asso-ciations With Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J. Natl. Cancer Inst. 2016, 108, djw100. [Google Scholar] [CrossRef] [PubMed]
- Cornet, I.; Gheit, T.; Clifford, G.M.; Combes, J.-D.; Dalstein, V.; Franceschi, S.; Tommasino, M.; Clavel, C. Human papillomavirus type 16 E6 variants in France and risk of viral persistence. Infect. Agents Cancer 2013, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Gheit, T.; Cornet, I.; Clifford, G.M.; Iftner, T.; Munk, C.; Tommasino, M.; Kjaer, S.K. Risks for persistence and progression by human papillomavirus type 16 variant lineages among a population-based sample of Danish women. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1315–1321. [Google Scholar] [CrossRef]
- Ai, W.; Wu, C.; Jia, L.; Xiao, X.; Xu, X.; Ren, M.; Xue, T.; Zhou, X.; Wang, Y.; Gao, C. Deep Sequencing of HPV16 E6 Region Reveals Unique Mutation Pattern of HPV16 and Predicts Cervical Cancer. Microbiol. Spectr. 2022, 10, e0140122. [Google Scholar] [CrossRef] [PubMed]
- Cullen, M.; Boland, J.F.; Schiffman, M.; Zhang, X.; Wentzensen, N.; Yang, Q.; Chen, Z.; Yu, K.; Mitchell, J.; Roberson, D.; et al. Deep sequencing of HPV16 genomes: A new high-throughput tool for exploring the carcinogenicity and natural history of HPV16 infection. Papillomavirus Res. 2015, 1, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Yeager, M.; Yu, K.; Clifford, G.M.; Xiao, Y.; Zhu, B.; Cullen, M.; Boland, J.F.; Wentzensen, N.; Nelson, C.W.; et al. HPV16 E7 Genetic Conservation Is Critical to Carcinogenesis. Cell 2017, 170, 1164–1174.e6. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Boland, J.F.; Torres-Gonzalez, E.; Albanez, A.; Zhou, W.; Steinberg, M.K.; Diaw, L.; Mitchell, J.; Roberson, D.; Cullen, M.; et al. The D2 and D3 Sublineages of Human Papilloma Virus 16-Positive Cervical Cancer in Guatemala Differ in Integration Rate and Age of Diagnosis. Cancer Res. 2020, 80, 3803–3809. [Google Scholar] [CrossRef]
- Freitas, L.B.; Chen, Z.; Muqui, E.F.; Boldrini, N.A.T.; Miranda, A.E.; Spano, L.C.; Burk, R.D. Human papillomavirus 16 non-European variants are preferentially associated with high-grade cervical lesions. PLoS ONE 2014, 9, e100746. [Google Scholar] [CrossRef]
- Lizano, M.; De la Cruz-Hernández, E.; Carrillo-García, A.; García-Carrancá, A.; de Leon-Rosales, S.P.; Dueñas-González, A.; Hernández-Hernández, D.M.; Mohar, A. Distribution of HPV16 and 18 intratypic variants in normal cytology, intraepithelial lesions, and cervical cancer in a Mexican population. Gynecol. Oncol. 2006, 102, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Schlecht, N.F.; Burk, R.D.; Palefsky, J.M.; Minkoff, H.; Xue, X.; Massad, L.S.; Bacon, M.; Levine, A.M.; Anastos, K.; Gange, S.J.; et al. Variants of human papillomaviruses 16 and 18 and their natural history in human immunodeficiency virus-positive women. J. Gen. Virol. 2005, 86, 2709–2720. [Google Scholar] [CrossRef]
- Rajeevan, M.S.; Swan, D.C.; Nisenbaum, R.; Lee, D.R.; Vernon, S.D.; Ruffin, M.T.; Horowitz, I.R.; Flowers, L.C.; Kmak, D.; Tadros, T.; et al. Epidemiologic and viral factors associated with cervical neoplasia in HPV-16-positive women. Int. J. Cancer 2005, 115, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Zuna, R.E.; Moore, W.E.; Shanesmith, R.P.; Dunn, S.T.; Wang, S.S.; Schiffman, M.; Blakey, G.L.; Teel, T. Association of HPV16 E6 variants with diagnostic severity in cervical cytology samples of 354 women in a US population. Int. J. Cancer 2009, 125, 2609–2613. [Google Scholar] [CrossRef]
- Gonçalves, M.G.; Ferreira, M.T.; López, R.V.M.; Ferreira, S.; Sirak, B.; Baggio, M.L.; Lazcano-Ponce, E.; Nyitray, A.G.; Giuliano, A.R.; Villa, L.L.; et al. Prevalence and persistence of HPV-16 molecular variants in the anal canal of men: The HIM study. J. Clin. Virol. 2022, 149, 105128. [Google Scholar] [CrossRef] [PubMed]
- Nicolás-Párraga, S.; Alemany, L.; de Sanjosé, S.; Bosch, F.; Bravo, I. Differential HPV16 variant distribution in squamous cell carcinoma, adenocarcinoma and adenosquamous cell carcinoma. Int. J. Cancer 2017, 140, 2092–2100. [Google Scholar] [CrossRef] [PubMed]
- Quint, K.D.; de Koning, M.N.; van Doorn, L.-J.; Quint, W.G.; Pirog, E.C. HPV genotyping and HPV16 variant analysis in glandular and squamous neoplastic lesions of the uterine cervix. Gynecol. Oncol. 2010, 117, 297–301. [Google Scholar] [CrossRef]
- De Boer, M.A.; Peters, L.A.; Aziz, M.F.; Siregar, B.; Cornain, S.; Vrede, M.A.; Jordanova, E.S.; Fleuren, G.J. Human papil-lomavirus type 18 variants: Histopathology and E6/E7 polymorphisms in three countries. Int. J. Cancer 2005, 114, 422–425. [Google Scholar] [CrossRef]
- Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Combes, J.D.; Chen, A.A.; Franceschi, S. Prevalence of human papillomavirus in cancer of the oropharynx by gender. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 2954–2958. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Chen, X.; Tang, C.; Xue, P.; Jiang, Y.; Qiao, Y. Artificial intelligence for HPV status prediction based on disease-specific images in head and neck cancer: A systematic review and meta-analysis. J. Med. Virol. 2023, 95, e29080. [Google Scholar] [CrossRef] [PubMed]
- Castellsagué, X.; Alemany, L.; Quer, M.; Halec, G.; Quirós, B.; Tous, S.; Clavero, O.; Alòs, L.; Biegner, T.; Szafarowski, T.; et al. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J. Natl. Cancer Inst. 2016, 108, djv403. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [PubMed]
- Galati, L.; Chiocca, S.; Duca, D.; Tagliabue, M.; Simoens, C.; Gheit, T.; Arbyn, M.; Tommasino, M. HPV and head and neck cancers: Towards early diagnosis and prevention. Tumour Virus Res. 2022, 14, 200245. [Google Scholar] [CrossRef] [PubMed]
- Cochicho, D.; Gil da Costa, R.; Felix, A. Exploring the roles of HPV16 variants in head and neck squamous cell carcinoma: Current challenges and opportunities. Virol. J. 2021, 18, 217. [Google Scholar] [CrossRef] [PubMed]
- Schrank, T.P.; Landess, L.; Stepp, W.H.; Rehmani, H.; Weir, W.H.; Lenze, N.; Lal, A.; Wu, D.; Kothari, A.; Hackman, T.G.; et al. Comprehensive Viral Genotyping Reveals Prognostic Viral Phylogenetic Groups in HPV16-Associated Squamous Cell Carcinoma of the Oropharynx. Mol. Cancer Res. 2022, 20, 1489–1501. [Google Scholar] [CrossRef]
- Ramachandran, D.; Schürmann, P.; Mao, Q.; Wang, Y.; Bretschneider, L.; Speith, L.; Hülse, F.; Enßen, J.; Bousset, K.; Jentschke, M.; et al. Association of genomic variants at the human leukocyte antigen locus with cervical cancer risk, HPV status and gene expression levels. Int. J. Cancer 2020, 147, 2458–2468. [Google Scholar] [CrossRef]
- Lindström, S.; Wang, L.; Feng, H.; Majumdar, A.; Huo, S.; Macdonald, J.; Harrison, T.; Turman, C.; Chen, H.; Mancuso, N.; et al. Genome-wide analyses characterize shared heritability among cancers and identify novel cancer susceptibility regions. JNCI J. Natl. Cancer Inst. 2023, 115, 712–732. [Google Scholar] [CrossRef]
- Ramachandran, D.; Dörk, T. Genomic Risk Factors for Cervical Cancer. Cancers 2021, 13, 5137. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
