Prevalence, Genotype Diversity, and Distinct Pathogenicity of 205 Gammacoronavirus Infectious Bronchitis Virus Isolates in China during 2019–2023
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Materials, Animals, and Ethics Statement
2.2. Viruses and S1 Gene Sequencing
2.3. Phylogenetic, Recombination, Deletion and Insertion Analysis of S1 Gene
2.4. Growth Characteristics of Isolates in 9-Day-Old SPF Chicken Embryos
2.5. Pathogenicity Evaluation of Isolates
2.6. Statistical Analysis
3. Results
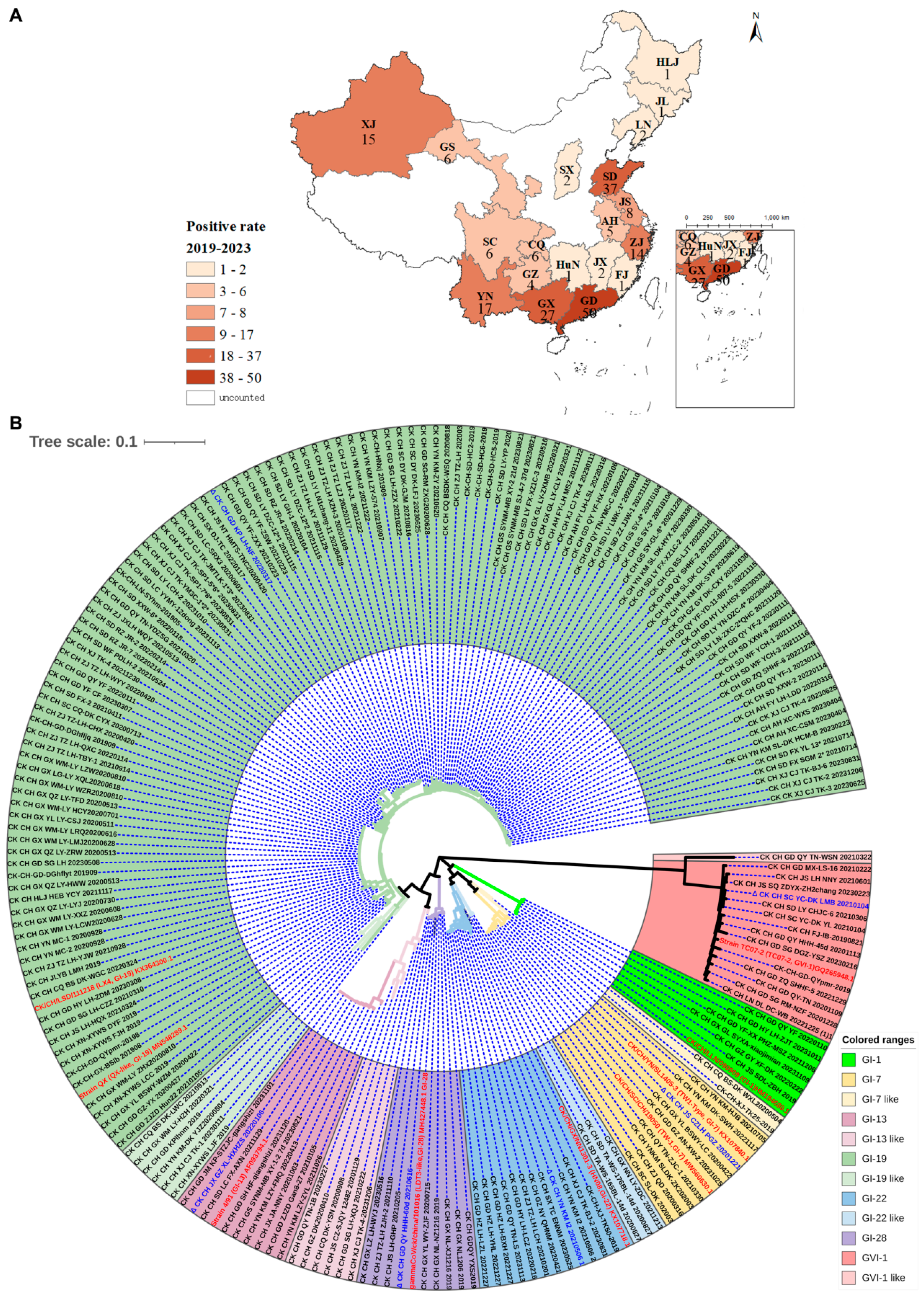
3.1. Isolation, Identification, and Phylogenetic Analysis of Clinical Isolates
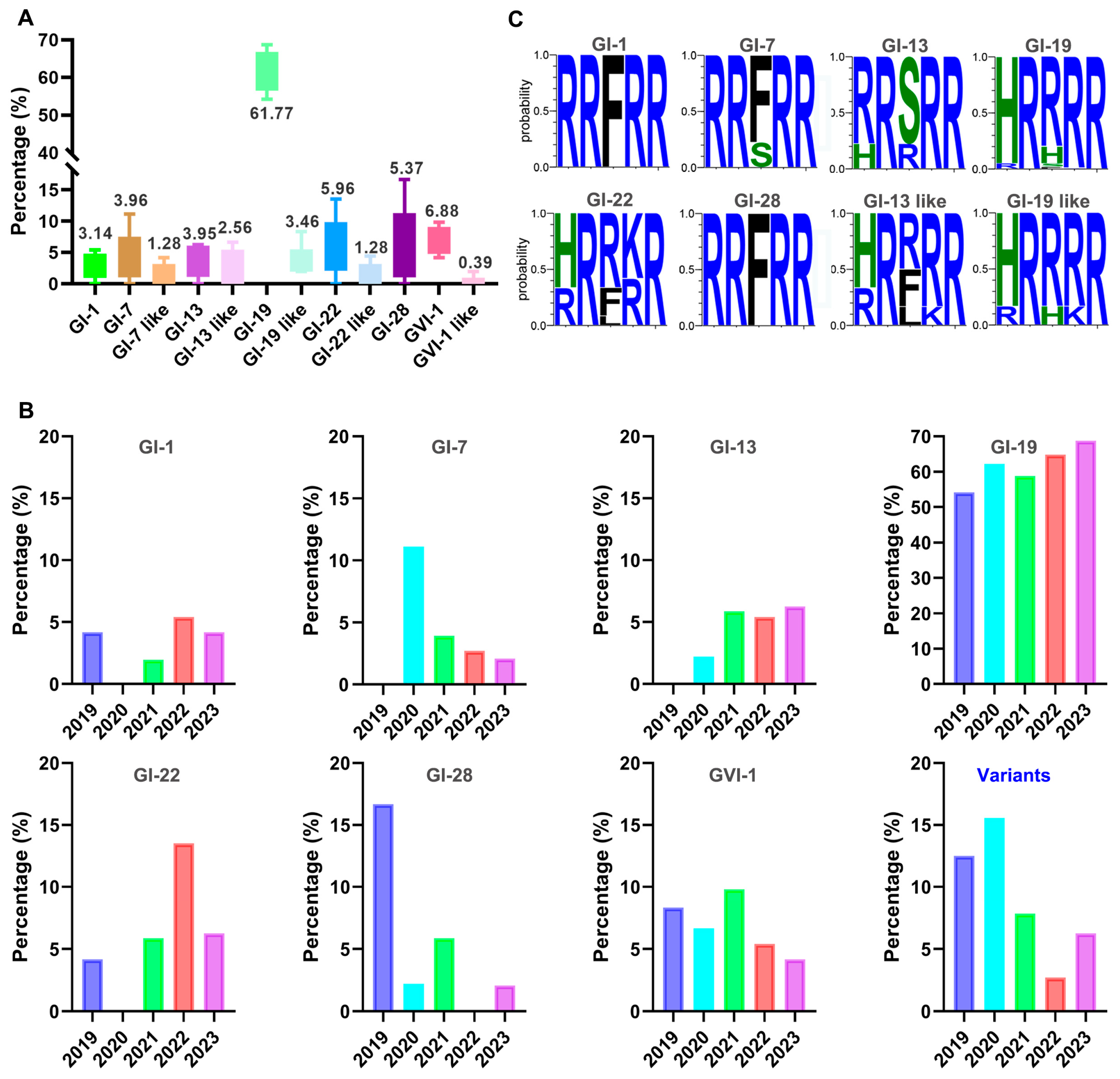
3.2. Prevalence of Different Genotypic Isolates and Sequence Variations in the Furin Motifs of S Genes
3.3. Analysis of Recombination, Deletion, and Insertion on S1 Genes of Different Isolates
3.4. Homology Analysis of Six Representative Isolates with the Vaccine Strain H120 and Their Growth Kinetics and Pathogenicity
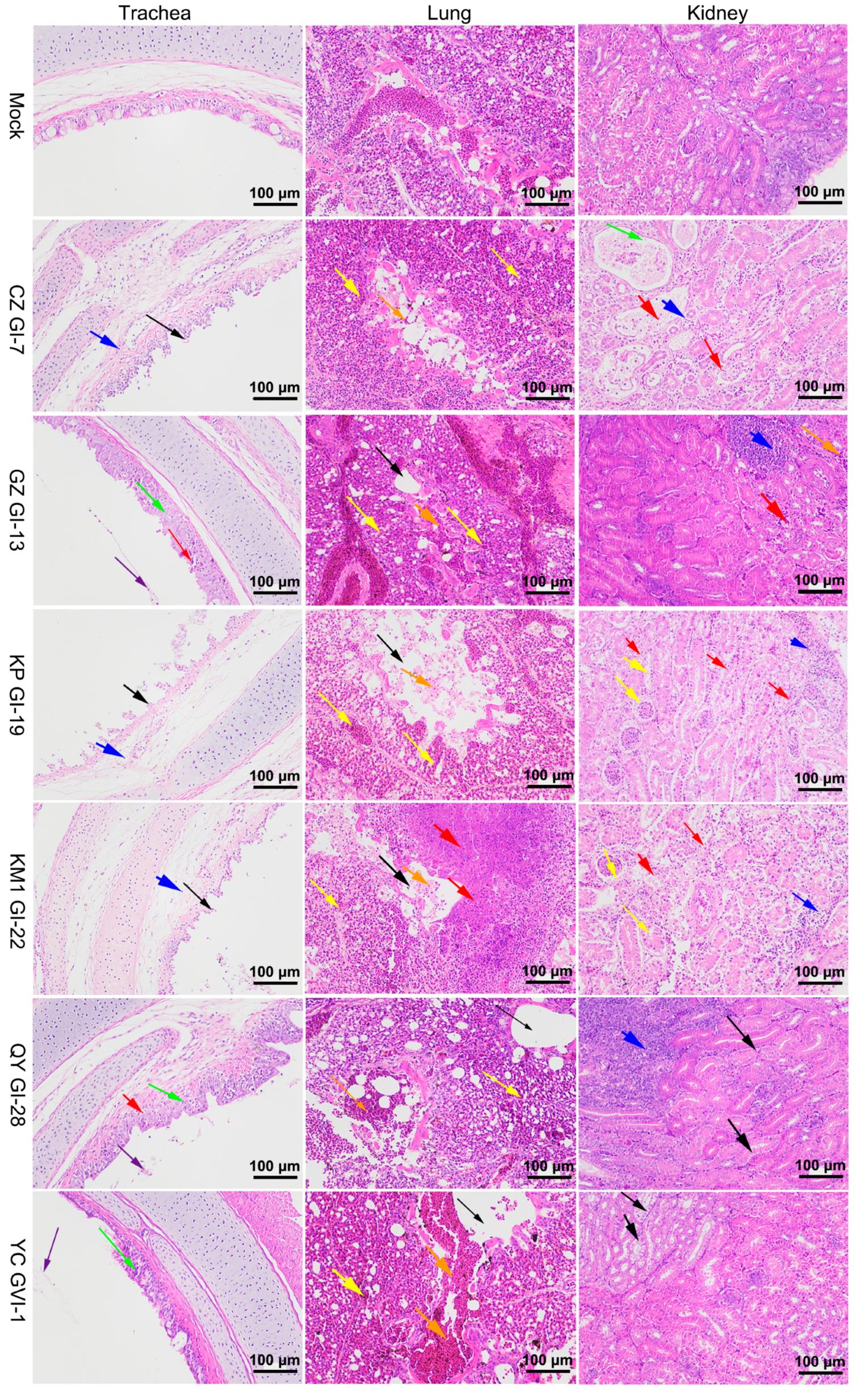
3.5. Histopathological Characterization of Chickens Infected with the Six Representative Isolates
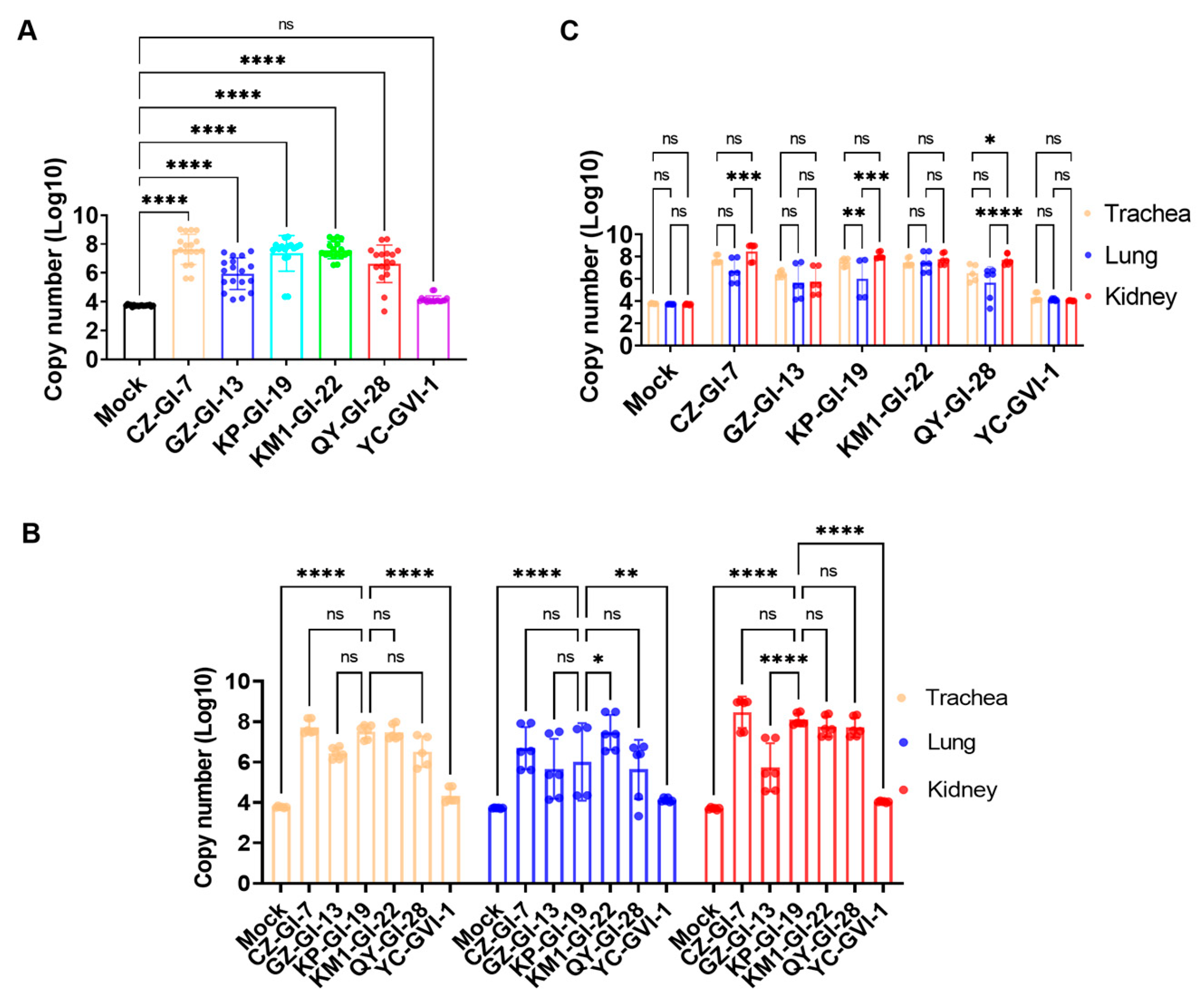
3.6. Determination of Viral Loads in Different Organs in Chickens Infected with the Six Representative Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2020, 2203, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef] [PubMed]
- Hoerr, F.J. The Pathology of Infectious Bronchitis. Avian Dis. 2021, 65, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Xue, Y.; Wang, F.; Chen, F.; Shu, D.; Xie, Q. Analysis of S1 gene of avian infectious bronchitis virus isolated in southern China during 2011–2012. Virus Genes 2014, 49, 292–303. [Google Scholar] [CrossRef]
- Liu, S.; Kong, X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. 2004, 33, 321–327. [Google Scholar] [CrossRef]
- Cavanagh, D.; Davis, P.J.; Mockett, A.P. Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. 1988, 11, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Valastro, V.; Holmes, E.C.; Britton, P.; Fusaro, A.; Jackwood, M.W.; Cattoli, G.; Monne, I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. 2016, 39, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Houta, M.H.; Hassan, K.E.; El-Sawah, A.A.; Elkady, M.F.; Kilany, W.H.; Ali, A.; Abdel-Moneim, A.S. The emergence, evolution and spread of infectious bronchitis virus genotype GI-23. Arch. Virol. 2021, 166, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Xue, Y.; Wang, J.; Chen, W.; Chen, F.; Bi, Y.; Xie, Q. Development and efficacy of a novel live-attenuated QX-like nephropathogenic infectious bronchitis virus vaccine in China. Vaccine 2015, 33, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Sun, Y.; Huang, X.; Jia, W.; Xie, D.; Zhang, G. Molecular characteristics and pathogenicity analysis of QX-like avian infectious bronchitis virus isolated in China in 2017 and 2018. Poult. Sci. 2019, 98, 5336–5341. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Liu, F.; Huang, M.; Li, L.; Shang, H.; Liang, M.; Luo, Q.; Chen, R. Pathogenicity of a QX-like avian infectious bronchitis virus isolated in China. Poult. Sci. 2020, 99, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D. Severe acute respiratory syndrome vaccine development: Experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003, 32, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, I.R.; Villegas, P.; Mossos, N.; Jackwood, M.W. Molecular characterization of avian infectious bronchitis virus strains isolated in Colombia during 2003. Avian Dis. 2005, 49, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Jackwood, M.W.; Hall, D.; Handel, A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012, 12, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Ting, X.; Xiang, C.; Liu, D.X.; Chen, R. Establishment and Cross-Protection Efficacy of a Recombinant Avian Gammacoronavirus Infectious Bronchitis Virus Harboring a Chimeric S1 Subunit. Front. Microbiol. 2022, 13, 897560. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; He, X.; Yao, K.; Du, L.; Liu, P.; Yan, Q.; Wen, Y.; Cao, S.; Han, X.; Huang, Y. Phylogenetic and antigenic analysis of avian infectious bronchitis virus in southwestern China, 2012–2016. Infect. Genet. Evol. 2016, 45, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Ojkic, D.; Elshafiee, E.A.; Shany, S.; El-Safty, M.M.; Shalaby, A.A.; Abdul-Careem, M.F. Genotyping and In Silico Analysis of Delmarva (DMV/1639) Infectious Bronchitis Virus (IBV) Spike 1 (S1) Glycoprotein. Genes 2022, 13, 1617. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Liu, D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009, 83, 8744–8758. [Google Scholar] [CrossRef]
- Ramakrishnan, M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016, 5, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Oumarou, H.H.; Aboudharam, G.; Barbieri, R.; Lepidi, H.; Drancourt, M. Immunohistochemical diagnosis of human infectious diseases: A review. Diagn. Pathol. 2022, 17, 17. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, Y.; Zhang, G. Key Aspects of Coronavirus Avian Infectious Bronchitis Virus. Pathogens 2023, 12, 698. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Wang, F.; Xue, Y.; Zhou, Q.; Chen, F.; Bi, Y.; Xie, Q. Epidemiology and characterization of avian infectious bronchitis virus strains circulating in southern China during the period from 2013–2015. Sci. Rep. 2017, 7, 6576. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Gao, Y.; Chen, Q.; Wu, Q.; Xu, X.; Kan, Y.; Yao, L.; Bi, Y.; Xie, Q. Epidemiological investigation of avian infectious bronchitis and locally determined genotype diversity in central China: A 2016–2018 study. Poult. Sci. 2020, 99, 3001–3008. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xue, C.; Chen, F.; Qin, J.; Xie, Q.; Bi, Y.; Cao, Y. Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004–2008. Vet. Microbiol. 2010, 143, 145–154. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, D.; Bai, Y.; Huang, W.; Gao, S.; Wu, X.; Wang, Y.; Ren, J.; He, J.; Jin, L.; et al. Genetic and pathogenic characterization of new infectious bronchitis virus strains in the GVI-1 and GI-19 lineages isolated in central China. J. Integr. Agric. 2023. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, B.; Hong, Y.; Li, Q.; Du, H.; Lin, Q.; Liao, M.; Ren, T.; Xu, C. Phylogenetic analysis of infectious bronchitis virus circulating in southern China in 2016-2017 and evaluation of an attenuated strain as a vaccine candidate. Arch. Virol. 2021, 166, 73–81. [Google Scholar] [CrossRef]
- Franzo, G.; Massi, P.; Tucciarone, C.M.; Barbieri, I.; Tosi, G.; Fiorentini, L.; Ciccozzi, M.; Lavazza, A.; Cecchinato, M.; Moreno, A. Think globally, act locally: Phylodynamic reconstruction of infectious bronchitis virus (IBV) QX genotype (GI-19 lineage) reveals different population dynamics and spreading patterns when evaluated on different epidemiological scales. PLoS ONE 2017, 12, e184401. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Cheng, Q.; Guo, J.; Li, Z.; Yang, J.; Wang, C.; Liang, Z.; Zhang, X.; Yu, H.; Li, Y.; et al. Detection and genetic characterization of novel infectious bronchitis viruses from recent outbreaks in broiler and layer chicken flocks in southern China, 2021. Poult. Sci. 2022, 101, 102082. [Google Scholar] [CrossRef] [PubMed]
- Sjaak, D.W.J.; Cook, J.K.; van der Heijden, H.M. Infectious bronchitis virus variants: A review of the history, current situation and control measures. Avian Pathol. 2011, 40, 223–235. [Google Scholar] [CrossRef]
- Lin, S.; Chen, H. Infectious Bronchitis Virus Variants: Molecular Analysis and Pathogenicity Investigation. Int. J. Mol. Sci. 2017, 18, 2030. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Ni, R.; Qiu, R.; Wang, F.; Yan, W.; Wang, K.; Li, H.; Fu, X.; Chen, L.; Lei, C.; et al. Evaluation of a novel recombinant strain of infectious bronchitis virus emerged from three attenuated live vaccine strains. Microb. Pathog. 2022, 164, 105437. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zheng, M.; Yang, Y.; Gu, X.; Yang, K.; Li, M.; Liu, Y.; Zhang, Q.; Zhang, P.; Wang, Y.; et al. Furin: A Potential Therapeutic Target for COVID-19. iScience 2020, 23, 101642. [Google Scholar] [CrossRef] [PubMed]
- Tay, F.P.; Huang, M.; Wang, L.; Yamada, Y.; Liu, D.X. Characterization of cellular furin content as a potential factor determining the susceptibility of cultured human and animal cells to coronavirus infectious bronchitis virus infection. Virology 2012, 433, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Tian, S. A 20 Residues Motif Delineates the Furin Cleavage Site and its Physical Properties May Influence Viral Fusion. Biochem. Insights 2009, 2, S2049. [Google Scholar] [CrossRef]
- Chen, H.Y.; Guo, A.Z.; Peng, B.; Zhang, M.F.; Guo, H.Y.; Chen, H.C. Infection of HeLa cells by avian infectious bronchitis virus is dependent on cell status. Avian Pathol. 2007, 36, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Molloy, S.S.; Thomas, L.; Van Slyke, J.K.; Stenberg, P.E.; Thomas, G. Intracellular trafficking and activation of the furin proprotein convertase: Localization to the TGN and recycling from the cell surface. Embo J. 1994, 13, 18–33. [Google Scholar] [CrossRef]
- Bosshart, H.; Humphrey, J.; Deignan, E.; Davidson, J.; Drazba, J.; Yuan, L.; Oorschot, V.; Peters, P.J.; Bonifacino, J.S. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J. Cell Biol. 1994, 126, 1157–1172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhao, Y.; Qin, F. Holocene fire, vegetation, and climate dynamics inferred from charcoal and pollen record in the eastern Tibetan Plateau. J. Asian Earth Sci. 2017, 147, 9–16. [Google Scholar] [CrossRef]
- Yan, W.; Yang, Q.; Huang, S.; Liu, S.; Wang, K.; Tang, Y.; Lei, C.; Wang, H.; Yang, X. Insights on genetic characterization and pathogenesis of a GI-19 (QX-like) infectious bronchitis virus isolated in China. Poult. Sci. 2023, 102, 102719. [Google Scholar] [CrossRef] [PubMed]
- Khataby, K.; Kasmi, Y.; Souiri, A.; Loutfi, C.; Ennaji, M.M. Chapter 33—Avian Coronavirus: Case of Infectious Bronchitis Virus Pathogenesis, Diagnostic Approaches, and Phylogenetic Relationship Among Emerging Strains in Middle East and North Africa Regions. In Emerging and Reemerging Viral Pathogens; Ennaji, M.M., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 729–744. [Google Scholar]
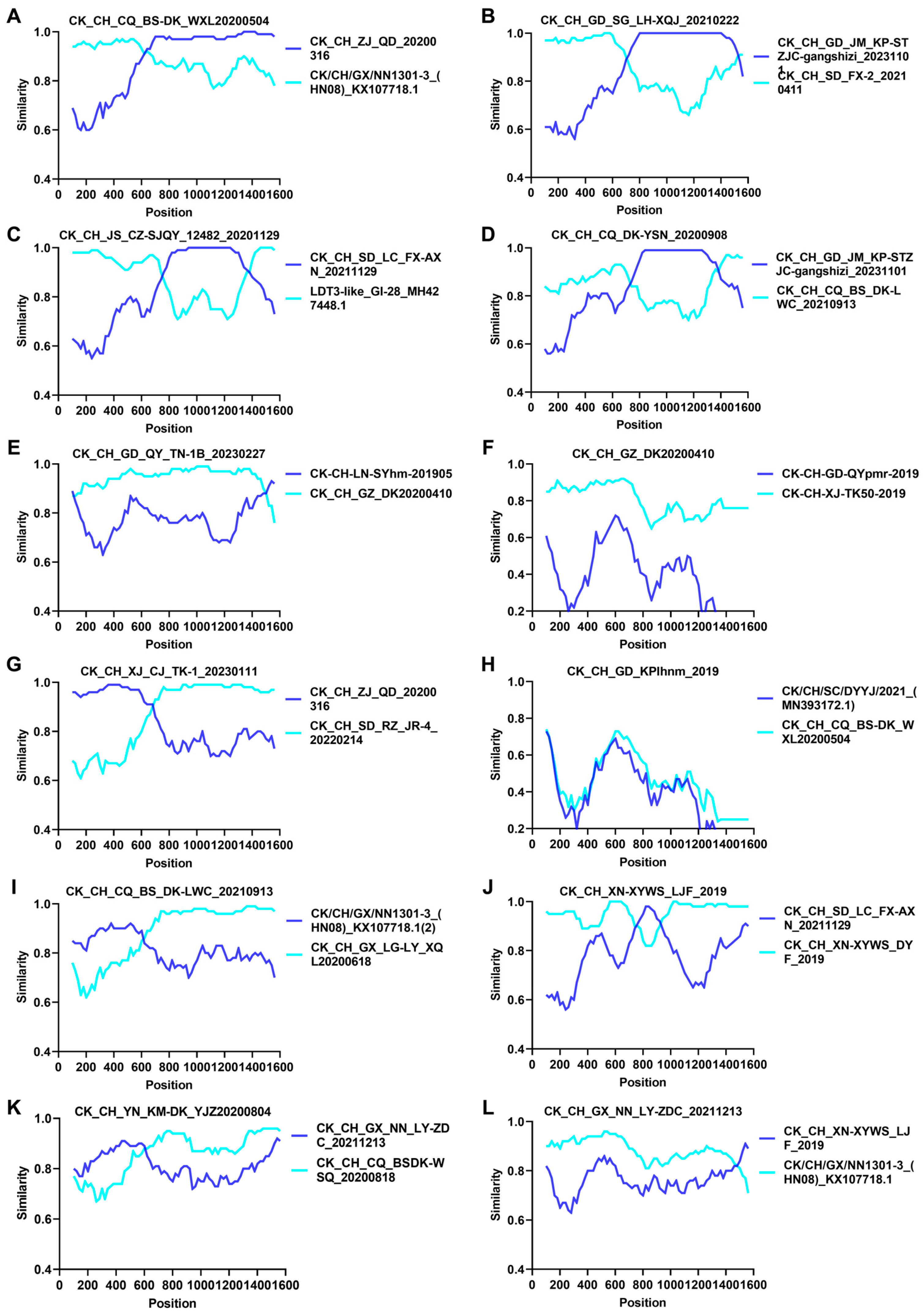



| Recombinant Isolates | Breakpoint Positions | Minor Parent 3 | Major Parent 4 | RDP 6 | |
|---|---|---|---|---|---|
| End | Begin | ||||
| CK CH CQ BS-DK WXL20200504 | 1239 * | 605 | CK CH ZJ QD 20200316 | CK/CH/GX/NN1301-3 (HN08) KX107718.1 | 6.38 × 10−14 |
| CK CH GD SG LH-XQJ 20210222 | 1516 | 666 | CK CH GD JM KP-STZJC-gangshizi 20231101 | CK CH SD FX-2 20210411 | 2.91 × 10−46 |
| CK CH JS CZ-SJQY12482 20201129 | 1358 | 723 | CK CH SD LC FX-AXN 20211129 | LDT3-like_GI-28 MH427448.1 | 4.69 × 10−43 |
| ^CK CH CQ DK-YSN 202009082 | 1370 | 714 | CK CH GD JM KP-STZJC-gangshizi 20231101 | CK CH CQ BS DK-LWC 20210913 | 1.22 × 10−32 |
| ^CK CH GD QY TN-1B 202302272 | 94 | 1475 * | CK-CH-LN-SYhm-201905 | CK CH GZ DK20200410 | 4.65 × 10−13 |
| CK CH GZ DK20200410 | 76 | 18 * | Unknown (CK-CH-GD-QYpmr-2019) 5 | CK-CH-XJ-TK50-2019 | 4.11 × 10−5 |
| CK CH XJ CJ TK-1 20230111 | 666 | 28 | CK CH ZJ QD 20200316 | CK CH SD RZ JR-4 20220214 | 1.33 × 10−32 |
| CK CH GD_KPlhnm_2019 | 1653 | 1040 | CK/CH/SC/DYYJ/2021 (MN393172.1) | CK CH CQ BS-DK WXL20200504 | 5.03 × 10−26 |
| CK CH CQ BS DK-LWC 20210913 | 646 | 46 | CK/CH/GX/NN1301-3 (HN08) KX107718.1 | CK CH GX LG-LY XQL20200618 | 2.94 × 10−12 |
| CK CH XN-XYWS LJF_ 019 | 463 * | 355 | CK CH SD LC FX-AXN 20211129 | CK CH XN-XYWS DYF 2019 | 3.25 × 10−12 |
| CK CH YN KM-DK YJZ20200804 | 666 | 159 * | CK CH GX NN LY-ZDC 20211213 | CK CH CQ BSDK-WSQ 20200818 | 5.28 × 10−4 |
| CK CH GX NN LY-ZDC 20211213 | 36 | 1494 | CK CH XN-XYWS LJF 2019 | CK CH GX NN1301-3 (HN08) KX107718.1 | 6.05 × 10−11 |
| Isolates | Name | Genotype | EID50 |
|---|---|---|---|
| CK CH CZLH PGZ 20201221 | CZ_CI-7 | GI-7 | 106.5 |
| CK CH GD KP LH-NF 20220317 | KP_GI-19 | GI-19 | 105 |
| CK CH YN KM I2 20210506 1 | KM1_GI-22 | GI-22 | 104.5 |
| CK CH GD QY HHHH-60d 20210616 | QY_GI-28 | GI-28 | 105 |
| CK CH SC YC-DK LMB 20210104 | YC_GVI-1 | GVI-1 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, T.; Xie, H.; Li, L.; Liang, S.; Huang, M.; Yu, C.; Zhuang, T.; Liang, X.; Liu, D.; Chen, R. Prevalence, Genotype Diversity, and Distinct Pathogenicity of 205 Gammacoronavirus Infectious Bronchitis Virus Isolates in China during 2019–2023. Viruses 2024, 16, 930. https://doi.org/10.3390/v16060930
Xiong T, Xie H, Li L, Liang S, Huang M, Yu C, Zhuang T, Liang X, Liu D, Chen R. Prevalence, Genotype Diversity, and Distinct Pathogenicity of 205 Gammacoronavirus Infectious Bronchitis Virus Isolates in China during 2019–2023. Viruses. 2024; 16(6):930. https://doi.org/10.3390/v16060930
Chicago/Turabian StyleXiong, Ting, Hangao Xie, Lin Li, Shijin Liang, Meizhen Huang, Chuanzhao Yu, Tingting Zhuang, Xuejing Liang, Dingxiang Liu, and Ruiai Chen. 2024. "Prevalence, Genotype Diversity, and Distinct Pathogenicity of 205 Gammacoronavirus Infectious Bronchitis Virus Isolates in China during 2019–2023" Viruses 16, no. 6: 930. https://doi.org/10.3390/v16060930





