Interleukin 27, Similar to Interferons, Modulates Gene Expression of Tripartite Motif (TRIM) Family Members and Interferes with Mayaro Virus Replication in Human Macrophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
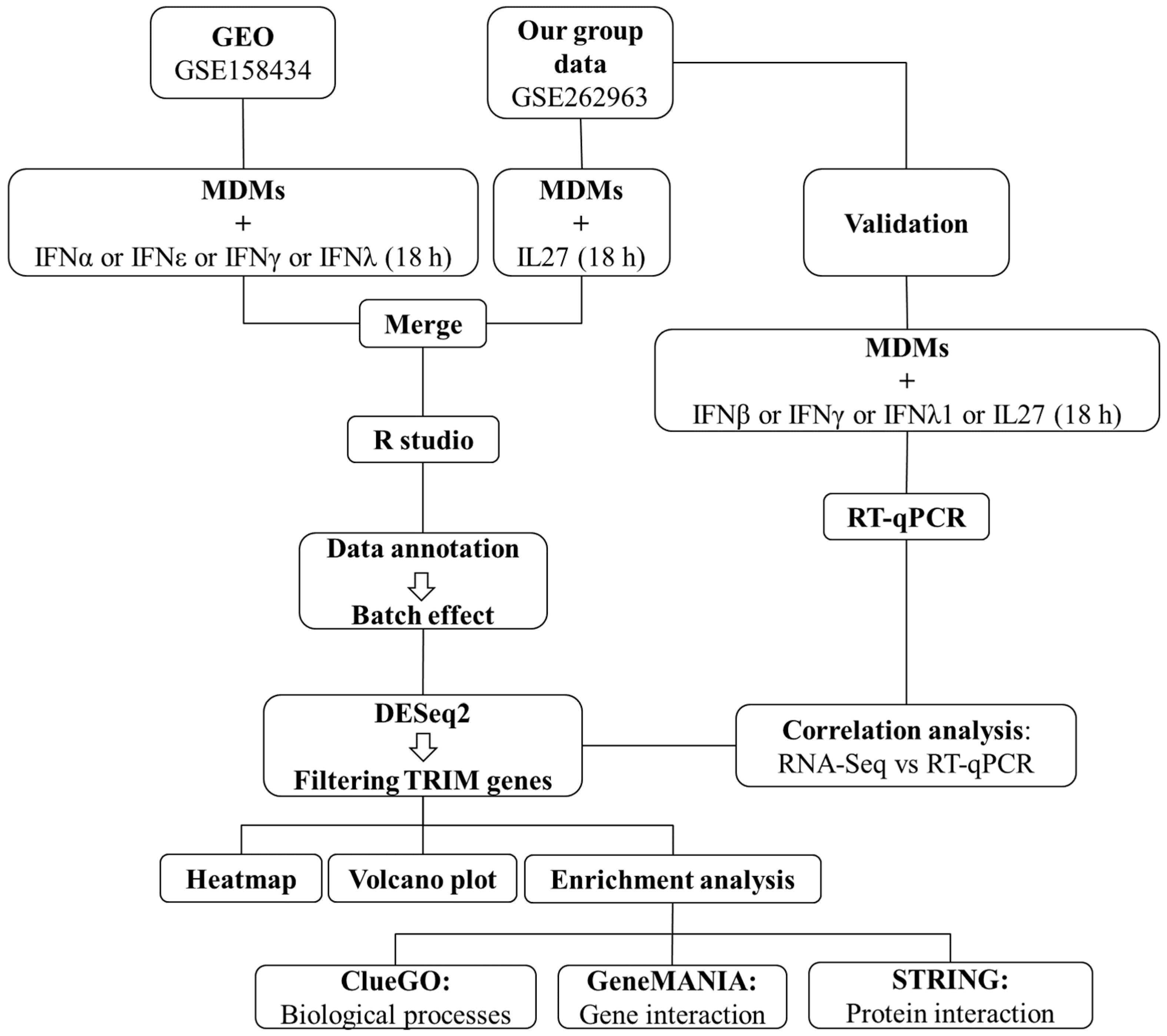
2.2. Transcriptomic Analysis of Previously Published RNA-Seq Studies and Prediction of TRIM Expression in MDMs
2.3. Data Annotation and Batch Effect Correction
2.4. Analysis of TRIM Gene Expression
2.5. Gene Set Enrichment Analysis
2.6. Culture of Primary Human Monocytes and Differentiation into Monocyte-Derived Macrophages
2.7. Treatment of Monocyte-Derived Macrophages with IL27, IFNβ, IFNγ, or IFNλ1
2.8. Cell Lines and Mayaro Virus Stocks
2.9. In Vitro MAYV Infection of MDM and In Vitro Antiviral Assay
2.10. RNA Extraction, cDNA Synthesis, and Real-Time RT-qPCR
3. Results
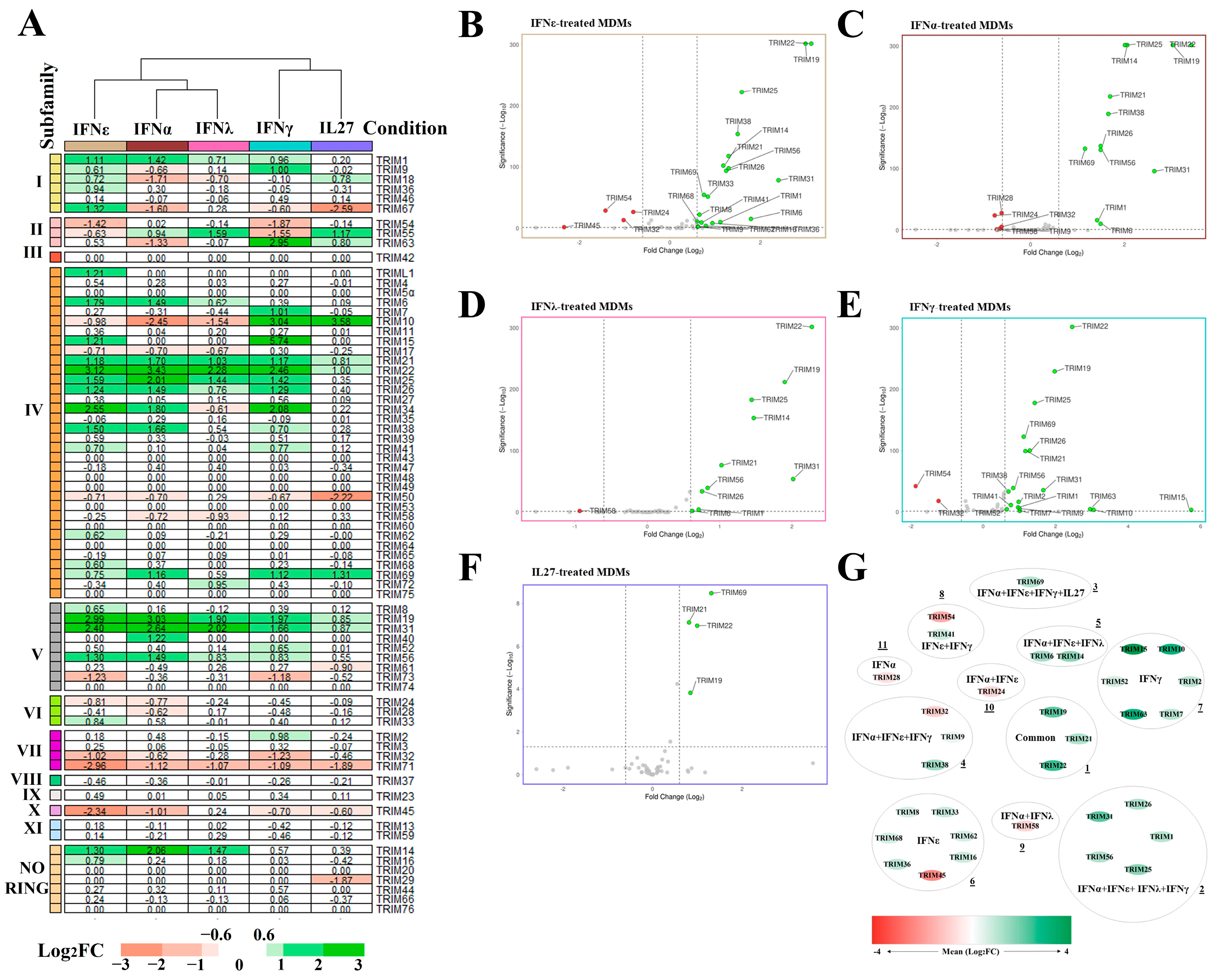
3.1. Identification of TRIM mRNA Expression in MDMs Treated with IL27 or IFNs
3.2. Differentially Expressed TRIM Genes in MDMs Treated with IL27 or IFNs
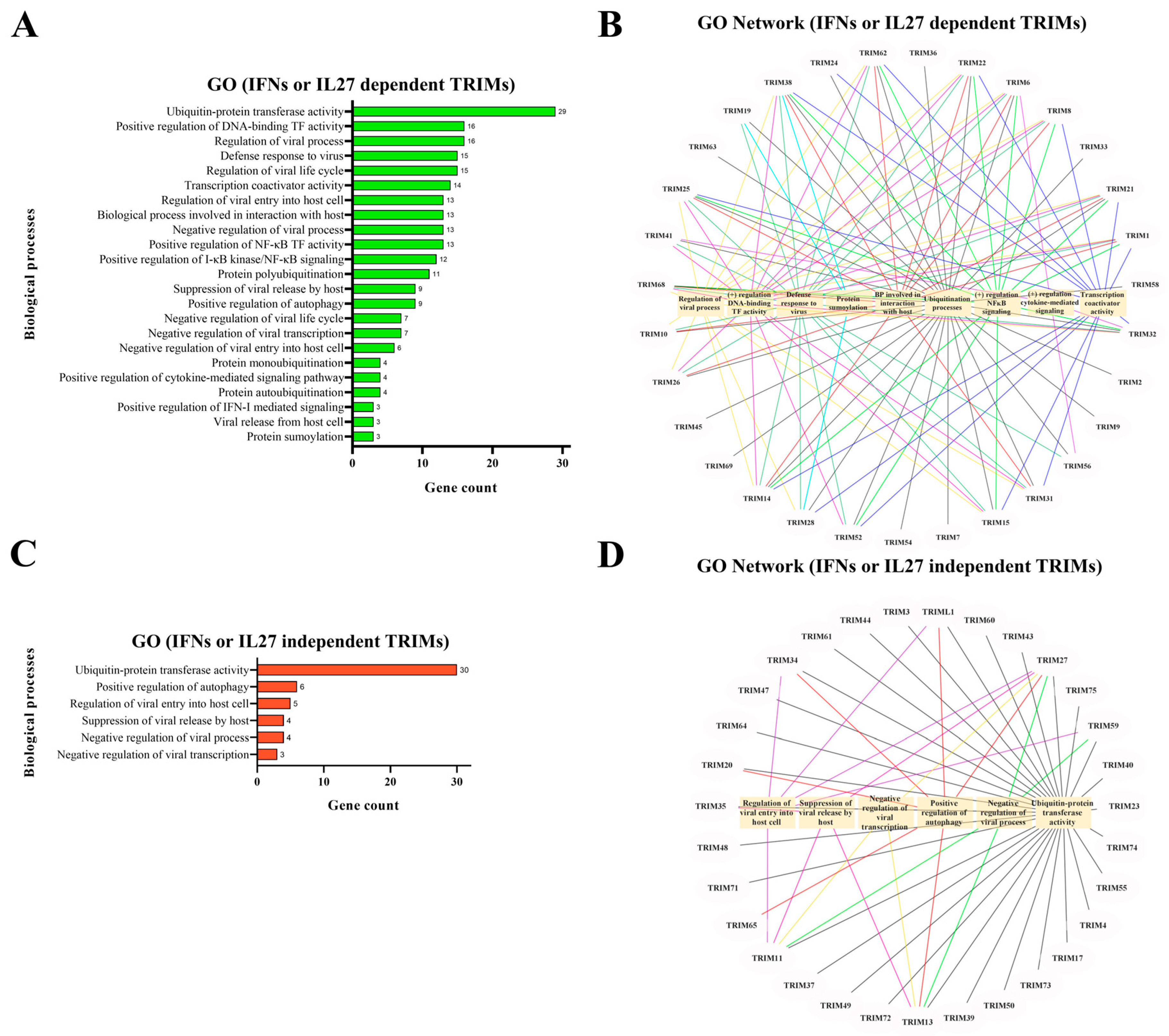
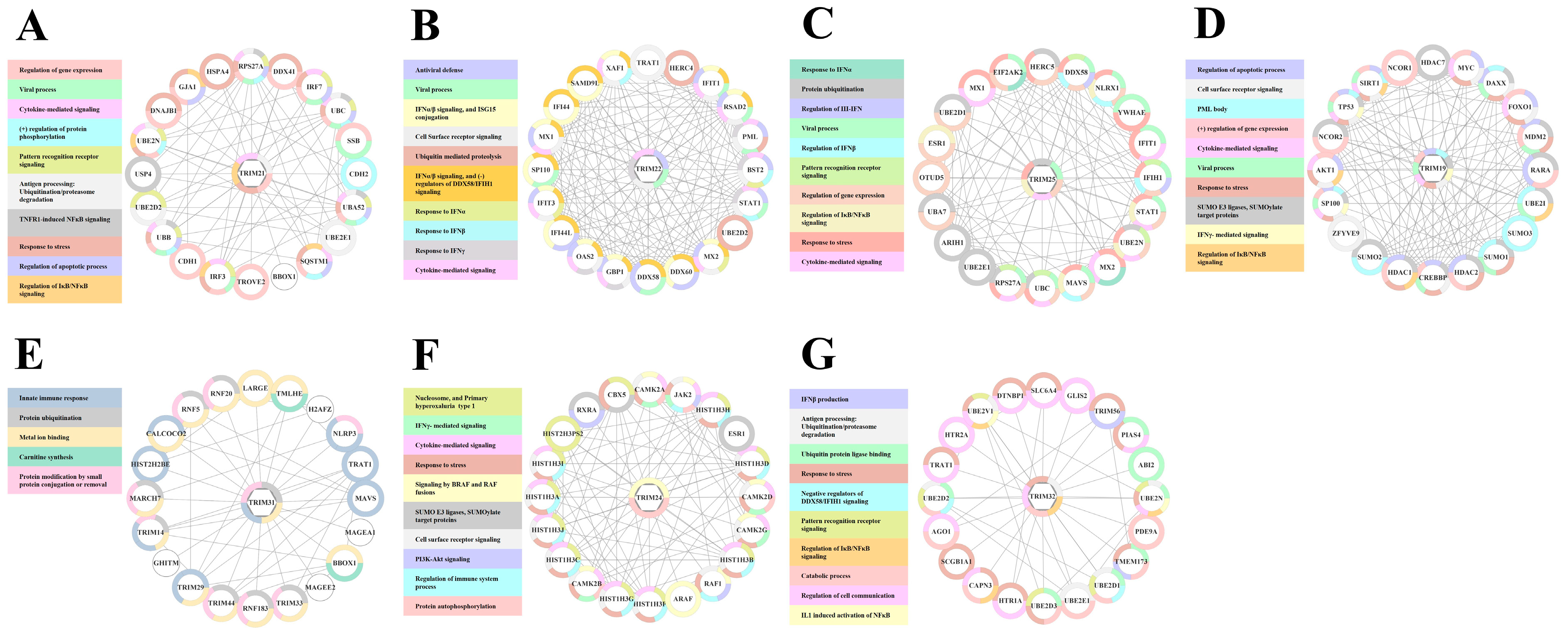
3.3. Functional Response of TRIM Genes in MDMs Treated with IL27 or IFNs
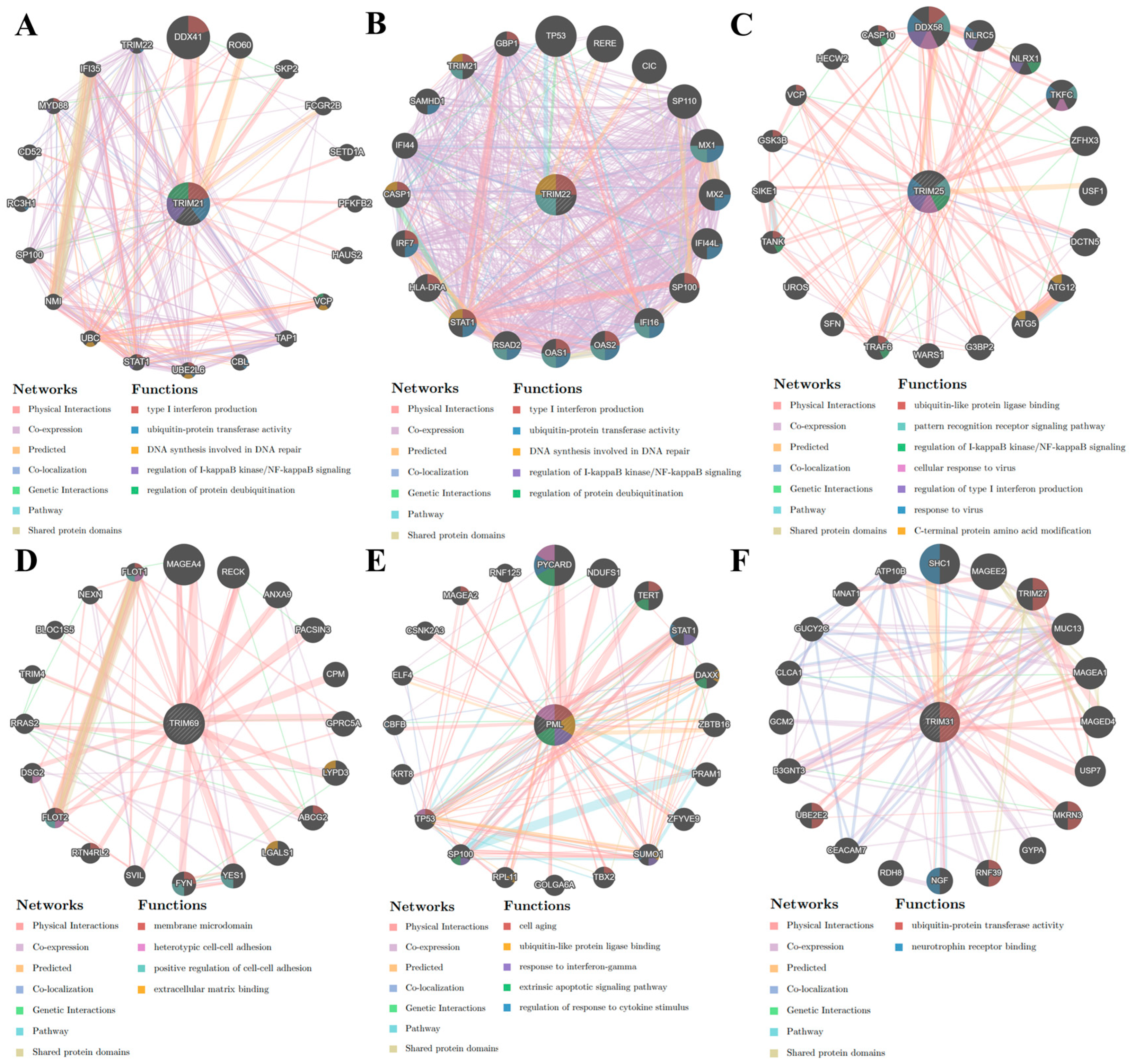
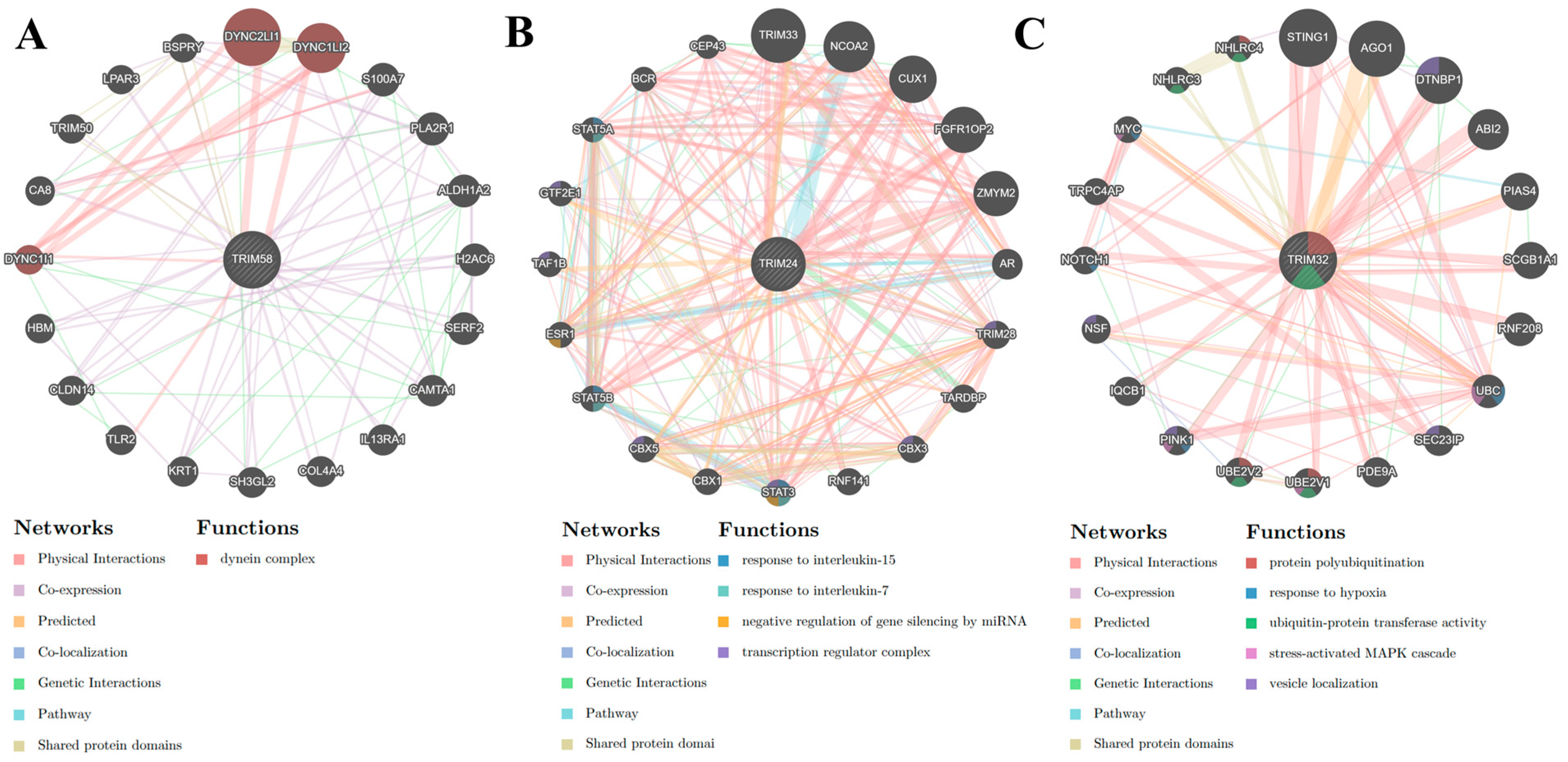
3.4. Gene-Gene Interaction Network of TRIMs Modulated by IL27 and/or IFNs
3.5. Protein-Protein Interaction Network of TRIMs Modulated by Il27 and/or IFNs
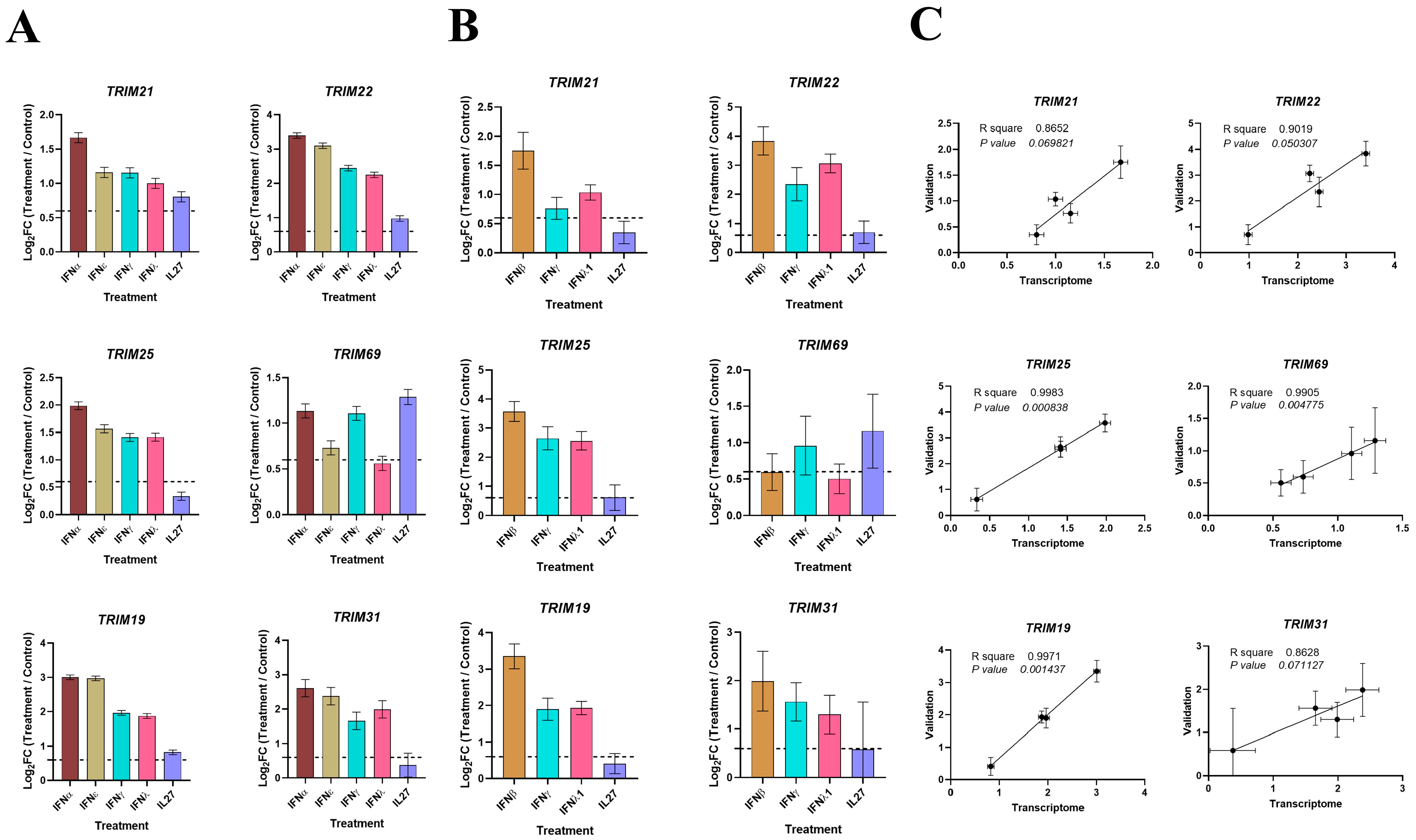
3.6. Validation of Key TRIMs by RT-qPCR
3.7. IL27, Similar to IFNs, Promotes the Expression of TRIMs and Interferes with the Replication of MAYV in MDMs
4. Discussion
5. Limitations of the Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoneyama, M.; Fujita, T. Recognition of Viral Nucleic Acids in Innate Immunity. Rev. Med. Virol. 2010, 20, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Walker, F.C.; Sridhar, P.R.; Baldridge, M.T. Differential Roles of Interferons in Innate Responses to Mucosal Viral Infections. Trends Immunol. 2021, 42, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Hardy, M.P.; Owczarek, C.M.; Jermiin, L.S.; Ejdebäck, M.; Hertzog, P.J. Characterization of the Type I Interferon Locus and Identification of Novel Genes. Genomics 2004, 84, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Gray, P.W.; Goeddel, D.V. Structure of the Human Immune Interferon Gene. Nature 1982, 298, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.Y.; Solanki, H.S.; Advani, J.; Khan, A.A.; Keshava Prasad, T.S.; Gowda, H.; Thiyagarajan, S.; Chatterjee, A. Comprehensive Network Map of Interferon Gamma Signaling. J. Cell Commun. Signal. 2018, 12, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Antiviral Actions of Interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef]
- Mesev, E.V.; LeDesma, R.A.; Ploss, A. Decoding Type I and III Interferon Signalling during Viral Infection. Nat. Microbiol. 2019, 4, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Vilcek, J. Novel Interferons. Nat. Immunol. 2003, 4, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Kotenko, S.V.; Gallagher, G.; Baurin, V.V.; Lewis-Antes, A.; Shen, M.; Shah, N.K.; Langer, J.A.; Sheikh, F.; Dickensheets, H.; Donnelly, R.P. IFN-Λs Mediate Antiviral Protection through a Distinct Class II Cytokine Receptor Complex. Nat. Immunol. 2003, 4, 69–77. [Google Scholar] [CrossRef]
- Stetson, D.B.; Medzhitov, R. Type I Interferons in Host Defense. Immunity 2006, 25, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Darnell, J.E. Studies of IFN-Induced Transcriptional Activation Uncover the Jak-Stat Pathway. J. Interf. Cytokine Res. 1998, 18, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Goodbourn, S.; Didcock, L.; Randall, R.E. Interferons: Cell Signalling, Immune Modulation, Antiviral Response and Virus Countermeasures. J. Gen. Virol. 2000, 81, 2341–2364. [Google Scholar] [CrossRef]
- Blaszczyk, K.; Olejnik, A.; Nowicka, H.; Ozgyin, L.; Chen, Y.-L.; Chmielewski, S.; Kostyrko, K.; Wesoly, J.; Balint, B.L.; Lee, C.-K.; et al. STAT2/IRF9 Directs a Prolonged ISGF3-like Transcriptional Response and Antiviral Activity in the Absence of STAT1. Biochem. J. 2015, 466, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Borden, E.C.; Sen, G.C.; Uze, G.; Silverman, R.H.; Ransohoff, R.M.; Foster, G.R.; Stark, G.R. Interferons at Age 50: Past, Current and Future Impact on Biomedicine. Nat. Rev. Drug Discov. 2007, 6, 975–990. [Google Scholar] [CrossRef]
- Versteeg, G.A.; García-Sastre, A. Viral Tricks to Grid-Lock the Type I Interferon System. Curr. Opin. Microbiol. 2010, 13, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Darnell, J.E. The JAK-STAT Pathway at Twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Qing, Y.; Stark, G.R. Alternative Activation of STAT1 and STAT3 in Response to Interferon-γ. J. Biol. Chem. 2004, 279, 41679–41685. [Google Scholar] [CrossRef] [PubMed]
- Decker, T.; Kovarik, P.; Meinke, A. GAS Elements: A Few Nucleotides with a Major Impact on Cytokine-Induced Gene Expression. J. Interf. Cytokine Res. 1997, 17, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Pflanz, S.; Timans, J.C.; Cheung, J.; Rosales, R.; Kanzler, H.; Gilbert, J.; Hibbert, L.; Churakova, T.; Travis, M.; Vaisberg, E.; et al. IL-27, a Heterodimeric Cytokine Composed of EBI3 and P28 Protein, Induces Proliferation of Naive CD4+ T Cells. Immunity 2002, 16, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, F.; Basset, L.; Froger, J.; Dinguirard, N.; Chevalier, S.; Gascan, H. IL-27 Structural Analysis Demonstrates Similarities with Ciliary Neurotrophic Factor (CNTF) and Leads to the Identi Fi Cation of Antagonistic Variants. Proc. Natl. Acad. Sci. USA 2010, 107, 19420–19425. [Google Scholar] [CrossRef] [PubMed]
- Valdés-López, J.F.; Fernandez, G.J.; Urcuqui-Inchima, S. Synergistic Effects of Toll-Like Receptor 1/2 and Toll-Like Receptor 3 Signaling Triggering Interleukin 27 Gene Expression in Chikungunya Virus-Infected Macrophages. Front. Cell Dev. Biol. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Pflanz, S.; Hibbert, L.; Mattson, J.; Rosales, R.; Vaisberg, E.; Bazan, J.F.; Phillips, J.H.; McClanahan, T.K.; de Waal Malefyt, R.; Kastelein, R.A. WSX-1 and Glycoprotein 130 Constitute a Signal-Transducing Receptor for IL-27. J. Immunol. 2004, 172, 2225–2231. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Steinwald, V.; Guralnik, A.; Brustle, A.; Kleemann, P.; Rosenplanter, C.; Decker, T.; Lohoff, M. IL-27 Inhibits the Development of Regulatory T Cells via STAT3. Int. Immunol. 2008, 20, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Bender, H.; Wiesinger, M.Y.; Nordhoff, C.; Schoenherr, C.; Haan, C.; Ludwig, S.; Weiskirchen, R.; Kato, N.; Heinrich, P.C.; Haan, S. Interleukin-27 Displays Interferon-γ-like Functions in Human Hepatoma Cells and Hepatocytes. Hepatology 2009, 50, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Kwock, J.T.; Handfield, C.; Suwanpradid, J.; Hoang, P.; McFadden, M.J.; Labagnara, K.F.; Floyd, L.; Shannon, J.; Uppala, R.; Sarkar, M.K.; et al. IL-27 Signaling Activates Skin Cells to Induce Innate Antiviral Proteins and Protects against Zika Virus Infection. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Hibbert, L.; Pflanz, S.; de Waal Malefyt, R.; Kastelein, R.A. IL-27 and IFN- α Signal via Stat1 and Stat3 and Induce T-Bet and IL-12R β 2 in Naive T Cells. J. Interf. Cytokine Res. 2003, 23, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Hamano, S.; Yamanaka, A.; Hanada, T.; Ishibashi, T.; Mak, T.W.; Yoshimura, A.; Yoshida, H. Cutting Edge: Role of IL-27/WSX-1 Signaling for Induction of T-Bet Through Activation of STAT1 During Initial Th1 Commitment. J. Immunol. 2003, 170, 4886–4890. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.A. New IL-12-Family Members: IL-23 and IL-27, Cytokines with Divergent Functions. Nat. Rev. Immunol. 2005, 5, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.; Ghilardi, N.; Li, J.; de Sauvage, F.J. IL-27 Regulates IL-12 Responsiveness of Naïve CD4+ T Cells through Stat1-Dependent and -Independent Mechanisms. Proc. Natl. Acad. Sci. 2003, 100, 15047–15052. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, T.; Yoshimoto, T.; Yasuda, K.; Mizuguchi, J.; Nakanishi, K. IL-27 Suppresses Th2 Cell Development and Th2 Cytokines Production from Polarized Th2 Cells: A Novel Therapeutic Way for Th2-Mediated Allergic Inflammation. J. Immunol. 2007, 179, 4415–4423. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Rohowsky-Kochan, C. Interleukin-27-Mediated Suppression of Human Th17 Cells Is Associated with Activation of STAT1 and Suppressor of Cytokine Signaling Protein 1. J. Interf. Cytokine Res. 2011, 31, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Diveu, C.; McGeachy, M.J.; Boniface, K.; Stumhofer, J.S.; Sathe, M.; Joyce-Shaikh, B.; Chen, Y.; Tato, C.M.; McClanahan, T.K.; de Waal Malefyt, R.; et al. IL-27 Blocks RORc Expression to Inhibit Lineage Commitment of Th17 Cells. J. Immunol. 2009, 182, 5748–5756. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Yoshiyuki, M. Regulation of Immune Responses by Interleukin-27. Immunol. Rev. 2008, 226, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Guzzo, C.; Che Mat, N.F.; Gee, K. Interleukin-27 Induces a STAT1/3- and NF-ΚB-Dependent Proinflammatory Cytokine Profile in Human Monocytes. J. Biol. Chem. 2010, 285, 24404–24411. [Google Scholar] [CrossRef] [PubMed]
- Valdés-López, J.F.; Hernández-Sarmiento, L.J.; Tamayo-Molina, Y.S.; Velilla-Hernández, P.A.; Rodenhuis-Zybert, I.A.; Urcuqui-Inchima, S. Interleukin 27, like Interferons, Activates JAK-STAT Signaling and Promotes pro-Inflammatory and Antiviral States That Interfere with Dengue and Chikungunya Viruses Replication in Human Macrophages. Front. Immunol. 2024, 15, 1385473. [Google Scholar] [CrossRef]
- Frank, A.C.; Zhang, X.; Katsounas, A.; Bharucha, J.P.; Kottilil, S.; Imamichi, T. Interleukin-27, an Anti-HIV-1 Cytokine, Inhibits Replication of Hepatitis C Virus. J. Interf. Cytokine Res. 2010, 30, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhang, R.; Zhang, W.; Zhu, C.; Yu, Y.; Song, Y.; Wang, Q.; Bai, L.; Liu, Y.; Wu, K.; et al. IL-27, a Cytokine, and IFN-Λ1, a Type III IFN, Are Coordinated To Regulate Virus Replication through Type I IFN. J. Immunol. 2014, 192, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.D.M.; Kenngott, E.E.; Schröter, M.F.; Kühl, A.; Jennrich, S.; Watzlawick, R.; Hoffmann, U.; Wolff, T.; Norley, S.; Scheffold, A.; et al. Timed Action of IL-27 Protects from Immunopathology While Preserving Defense in Influenza. PLoS Pathog. 2014, 10, e1004110. [Google Scholar] [CrossRef]
- Hernández-Sarmiento, L.J.; Valdés-López, J.F.; Urcuqui-Inchima, S. American-Asian- and African Lineages of Zika Virus Induce Differential pro-Inflammatory and Interleukin 27-Dependent Antiviral Responses in Human Monocytes. Virus Res. 2023, 325, 199040. [Google Scholar] [CrossRef] [PubMed]
- Valdés-López, J.F.; Fernandez, G.J.; Urcuqui-Inchima, S. Interleukin 27 as an Inducer of Antiviral Response against Chikungunya Virus Infection in Human Macrophages. Cell. Immunol. 2021, 367, 104411. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, H.; Wu, W.; Zhuo, W.; Liu, W.; Zhang, Y.; Cheng, M.; Chen, Y.-G.; Gao, N.; Yu, H.; et al. Structural Insights into the TRIM Family of Ubiquitin E3 Ligases. Cell Res. 2014, 24, 762–765. [Google Scholar] [CrossRef] [PubMed]
- McNab, F.W.; Rajsbaum, R.; Stoye, J.P.; O’Garra, A. Tripartite-Motif Proteins and Innate Immune Regulation. Curr. Opin. Immunol. 2011, 23, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Short, K.M.; Cox, T.C. Subclassification of the RBCC/TRIM Superfamily Reveals a Novel Motif Necessary for Microtubule Binding. J. Biol. Chem. 2006, 281, 8970–8980. [Google Scholar] [CrossRef] [PubMed]
- Meroni, G.; Diez-Roux, G. TRIM/RBCC, a Novel Class of ‘Single Protein RING Finger’ E3 Ubiquitin Ligases. BioEssays 2005, 27, 1147–1157. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Giraldo, M.I.; Hage, A.; van Tol, S.; Rajsbaum, R. TRIM Proteins in Host Defense and Viral Pathogenesis. Curr. Clin. Microbiol. Reports 2020, 7, 101–114. [Google Scholar] [CrossRef]
- Hage, A.; Rajsbaum, R. To TRIM or Not to TRIM: The Balance of Host–Virus Interactions Mediated by the Ubiquitin System. J. Gen. Virol. 2019, 100, 1641–1662. [Google Scholar] [CrossRef] [PubMed]
- van Tol, S.; Hage, A.; Giraldo, M.; Bharaj, P.; Rajsbaum, R. The TRIMendous Role of TRIMs in Virus–Host Interactions. Vaccines 2017, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- D’Cruz, A.A.; Kershaw, N.J.; Chiang, J.J.; Wang, M.K.; Nicola, N.A.; Babon, J.J.; Gack, M.U.; Nicholson, S.E. Crystal Structure of the TRIM25 B30.2 (PRYSPRY) Domain: A Key Component of Antiviral Signalling. Biochem. J. 2013, 456, 231–240. [Google Scholar] [CrossRef]
- Versteeg, G.A.; Rajsbaum, R.; Sánchez-Aparicio, M.T.; Maestre, A.M.; Valdiviezo, J.; Shi, M.; Inn, K.-S.; Fernandez-Sesma, A.; Jung, J.; García-Sastre, A. The E3-Ligase TRIM Family of Proteins Regulates Signaling Pathways Triggered by Innate Immune Pattern-Recognition Receptors. Immunity 2013, 38, 384–398. [Google Scholar] [CrossRef]
- Roy, M.; Singh, R. TRIMs: Selective Recruitment at Different Steps of the NF-ΚB Pathway—Determinant of Activation or Resolution of Inflammation. Cell. Mol. Life Sci. 2021, 78, 6069–6086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, X.; Hu, W.; Li, C. Emerging Role of TRIM Family Proteins in Cardiovascular Disease. Cardiology 2020, 145, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Uchil, P.D.; Hinz, A.; Siegel, S.; Coenen-Stass, A.; Pertel, T.; Luban, J.; Mothes, W. TRIM Protein-Mediated Regulation of Inflammatory and Innate Immune Signaling and Its Association with Antiretroviral Activity. J. Virol. 2013, 87, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhao, X.; Sun, D.; Yang, L.; Chong, C.; Pan, Y.; Chi, X.; Gao, Y.; Wang, M.; Shi, X.; et al. Interferon Alpha (IFNα)-Induced TRIM22 Interrupts HCV Replication by Ubiquitinating NS5A. Cell. Mol. Immunol. 2016, 13, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Nigos, L.R.; Scott, N.E.; Brooks, A.G.; Ait-Goughoulte, M.; Londrigan, S.L.; Reading, P.C.; Farrukee, R. TRIM16 Overexpression in HEK293T Cells Results in Cell Line-Specific Antiviral Activity. Pathogens 2023, 12, 852. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Dong, H.; Lai, X.; Ou, G.; Cao, J.; Shi, J.; Xiang, C.; Wang, L.; Zhang, X.; Zhang, K.; et al. TRIM56 Impairs HBV Infection and Replication by Inhibiting HBV Core Promoter Activity. Antivir. Res. 2022, 207, 105406. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Wang, L.; Ding, H.; Schwamborn, J.C.; Li, S.; Dorf, M.E. TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase. PLoS Pathog. 2015, 11, e1004960. [Google Scholar] [CrossRef] [PubMed]
- Carthagena, L.; Bergamaschi, A.; Luna, J.M.; David, A.; Uchil, P.D.; Margottin-Goguet, F.; Mothes, W.; Hazan, U.; Transy, C.; Pancino, G.; et al. Human TRIM Gene Expression in Response to Interferons. PLoS ONE 2009, 4, e4894. [Google Scholar] [CrossRef] [PubMed]
- Rajsbaum, R.; Stoye, J.P.; O’Garra, A. Type I Interferon-dependent and -independent Expression of Tripartite Motif Proteins in Immune Cells. Eur. J. Immunol. 2008, 38, 619–630. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. 2022. Available online: https://www.R-project.org (accessed on 10 June 2024).
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-Seq: Batch Effect Adjustment for RNA-Seq Count Data. NAR Genom. Bioinforma 2020, 2, lqaa078. [Google Scholar] [CrossRef]
- Van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef] [PubMed]
- Ozato, K.; Shin, D.M.; Chang, T.H.; Morse, H.C. TRIM Family Proteins and Their Emerging Roles in Innate Immunity. Nat. Rev. Immunol. 2008, 8, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulou, A.-I.; Xanthopoulos, C.; Piperi, C.; Kostareli, E. Emerging Roles of TRIM Family Proteins in Gliomas Pathogenesis. Cancers 2022, 14, 4536. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8—A Global View on Proteins and Their Functional Interactions in 630 Organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef] [PubMed]
- Tamayo-Molina, Y.S.; Velilla, P.A.; Hernández-Sarmiento, L.J.; Urcuqui-Inchima, S. Multitranscript Analysis Reveals an Effect of 2-Deoxy-d-Glucose on Gene Expression Linked to Unfolded Protein Response and Integrated Stress Response in Primary Human Monocytes and Monocyte-Derived Macrophages. Biochim. Biophys. Acta Gen. Subj. 2023, 1867, 130397. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Sarmiento, L.J.; Tamayo-Molina, Y.S.; Valdés-López, J.F.; Urcuqui-Inchima, S. Mayaro Virus Infection Elicits a Robust Pro-Inflammatory and Antiviral Response in Human Macrophages. Acta Trop. 2024, 252, 107146. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Khan, A.; Ali, A.; Idrees, M. The Interplay between Viruses and TRIM Family Proteins. Rev. Med. Virol. 2019, 29, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.-X.; Hong, X.; Liao, B.-B.; Shi, S.-Z.; Lai, X.-F.; Zheng, H.-Y.; Xie, L.; Wang, Y.; Wang, X.-L.; Xin, H.-B.; et al. Expression Profiling of TRIM Protein Family in THP1-Derived Macrophages Following TLR Stimulation. Sci. Rep. 2017, 7, 42781. [Google Scholar] [CrossRef]
- Rhodes, D.A.; de Bono, B.; Trowsdale, J. Relationship between SPRY and B30.2 Protein Domains. Evolution of a Component of Immune Defence? Immunology 2005, 116, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Nisole, S.; Stoye, J.P.; Saïb, A. TRIM Family Proteins: Retroviral Restriction and Antiviral Defence. Nat. Rev. Microbiol. 2005, 3, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.W.; Stoye, J.P. TRIM Proteins and the Innate Immune Response to Viruses. Adv. Exp. Med. Biol. 2012, 770, 93–104. [Google Scholar] [PubMed]
- Martinez, F.O.; Gordon, S.; Locati, M.; Mantovani, A. Transcriptional Profiling of the Human Monocyte-to-Macrophage Differentiation and Polarization: New Molecules and Patterns of Gene Expression. J. Immunol. 2006, 177, 7303–7311. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Liu, D.; Li, R.; Qian, H.; Qiu, W.; Ye, Q.; Kong, F. Comprehensive Analysis of TRIM Family Genes in Hepatitis Virus B-Related Hepatoma Carcinoma. Front. Genet. 2022, 13, 913743. [Google Scholar] [CrossRef] [PubMed]
- Der, S.D.; Zhou, A.; Williams, B.R.G.; Silverman, R.H. Identification of Genes Differentially Regulated by Interferon α, β, or γ Using Oligonucleotide Arrays. Proc. Natl. Acad. Sci. USA 1998, 95, 15623–15628. [Google Scholar] [CrossRef] [PubMed]
- Temerozo, J.R.; Ferreira, P.L.C.; Linhares-Lacerda, L.; Vieira, R.C.; Cister-Alves, B.; Gobbo, L.; Ribeiro-Alves, M.; Menna-Barreto, R.F.S.; Bou-Habib, D.C. Interleukin-27 Promotes Divergent Effects on HIV-1 Infection in Peripheral Blood Mononuclear Cells through BST-2/Tetherin. J. Virol. 2023, 97, e0175222. [Google Scholar] [CrossRef] [PubMed]
- Koepke, L.; Gack, M.U.; Sparrer, K.M. The Antiviral Activities of TRIM Proteins. Curr. Opin. Microbiol. 2021, 59, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Z.; Li, F.H.; Wang, X.J. Regulation of Tripartite Motif-Containing Proteins on Immune Response and Viral Evasion. Front. Microbiol. 2021, 12, 794882. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Shin, Y.C.; Joo, C.-H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-Finger E3 Ubiquitin Ligase Is Essential for RIG-I-Mediated Antiviral Activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Zha, J.; Han, K.-J.; Xu, L.-G.; He, W.; Zhou, Q.; Chen, D.; Zhai, Z.; Shu, H.-B. The Ret Finger Protein Inhibits Signaling Mediated by the Noncanonical and Canonical IκB Kinase Family Members. J. Immunol. 2006, 176, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liu, T.-T.; Lin, H.; Zhang, M.; Wei, J.; Luo, W.-W.; Hu, Y.-H.; Zhong, B.; Hu, M.-M.; Shu, H.-B. TRIM32-TAX1BP1-Dependent Selective Autophagic Degradation of TRIF Negatively Regulates TLR3/4-Mediated Innate Immune Responses. PLoS Pathog. 2017, 13, e1006600. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Li, N.L.; Wang, J.; Liu, B.; Lester, S.; Li, K. TRIM56 Is an Essential Component of the TLR3 Antiviral Signaling Pathway. J. Biol. Chem. 2012, 287, 36404–36413. [Google Scholar] [CrossRef]
- Fang, R.; Wang, C.; Jiang, Q.; Lv, M.; Gao, P.; Yu, X.; Mu, P.; Zhang, R.; Bi, S.; Feng, J.-M.; et al. NEMO–IKKβ Are Essential for IRF3 and NF-ΚB Activation in the CGAS–STING Pathway. J. Immunol. 2017, 199, 3222–3233. [Google Scholar] [CrossRef] [PubMed]
- Foss, S.; Bottermann, M.; Jonsson, A.; Sandlie, I.; James, L.C.; Andersen, J.T. TRIM21—From Intracellular Immunity to Therapy. Front. Immunol. 2019, 10, 2049. [Google Scholar] [CrossRef] [PubMed]
- Mu, T.; Zhao, X.; Zhu, Y.; Fan, H.; Tang, H. The E3 Ubiquitin Ligase TRIM21 Promotes HBV DNA Polymerase Degradation. Viruses 2020, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.; Kong, N.; Jiao, Y.; Sun, D.; Dong, S.; Qin, W.; Zhai, H.; Yu, L.; Zheng, H.; et al. TRIM21 Inhibits Porcine Epidemic Diarrhea Virus Proliferation by Proteasomal Degradation of the Nucleocapsid Protein. Arch. Virol. 2021, 166, 1903–1911. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wu, X.; Xu, Y.; Zhu, J.; Li, J.; Zou, Z.; Chen, L.; Zhang, B.; Hua, C.; Rui, H.; et al. HPV E7 Inhibits Cell Pyroptosis by Promoting TRIM21-Mediated Degradation and Ubiquitination of the IFI16 Inflammasome. Int. J. Biol. Sci. 2020, 16, 2924–2937. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Cao, W.; Chen, S.; Tian, R.; Xue, B.; Wang, L.; Liu, Q.; Deng, R.; Wang, X.; Wang, Z.; et al. TRIM21 Regulates Virus-Induced Cell Pyroptosis through Polyubiquitination of ISG12a. J. Immunol. 2022, 209, 1987–1998. [Google Scholar] [CrossRef]
- Tan, T.; Xia, L. TRIM21 Aggravates Herpes Simplex Virus Epithelial Keratitis by Attenuating STING-IRF3-Mediated Type I Interferon Signaling. Front. Microbiol. 2020, 11, 703. [Google Scholar] [CrossRef] [PubMed]
- Sanda, C.; Weitzel, P.; Tsukahara, T.; Schaley, J.; Edenberg, H.J.; Stephens, M.A.; McClintick, J.N.; Blatt, L.M.; Li, L.; Brodsky, L.; et al. Differential Gene Induction by Type I and Type II Interferons and Their Combination. J. Interf. Cytokine Res. 2006, 26, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Barr, S.D.; Smiley, J.R.; Bushman, F.D. The Interferon Response Inhibits HIV Particle Production by Induction of TRIM22. PLoS Pathog. 2008, 4, e1000007. [Google Scholar] [CrossRef] [PubMed]
- Zu, S.; Li, C.; Li, L.; Deng, Y.-Q.; Chen, X.; Luo, D.; Ye, Q.; Huang, Y.-J.; Li, X.-F.; Zhang, R.-R.; et al. TRIM22 Suppresses Zika Virus Replication by Targeting NS1 and NS3 for Proteasomal Degradation. Cell Biosci. 2022, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Tosi, G.; Turrini, F.; Poli, G.; Vicenzi, E.; Accolla, R.S. Tripartite Motif-Containing Protein 22 Interacts with Class II Transactivator and Orchestrates Its Recruitment in Nuclear Bodies Containing TRIM19/PML and Cyclin T1. Front. Immunol. 2017, 8, 564. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, A.; Kajaste-Rudnitski, A.; Oteiza, A.; Nicora, L.; Towers, G.J.; Mechti, N.; Vicenzi, E. TRIM22 Inhibits Influenza A Virus Infection by Targeting the Viral Nucleoprotein for Degradation. J. Virol. 2013, 87, 4523–4533. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Duan, Z.; Xu, W.; Xiong, S. Tripartite Motif-Containing 22 Inhibits the Activity of Hepatitis B Virus Core Promoter, Which Is Dependent on Nuclear-Located RING Domain. Hepatology 2009, 50, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, R.; Rahimi, P.; Hamidi-Fard, M.; Eybpoosh, S.; Doroud, D.; Sadeghi, S.A.; Zaheri Birgani, M.; Aghasadeghi, M.; Fateh, A. Impact of TRIM5α and TRIM22 Genes Expression on the Clinical Course of Coronavirus Disease 2019. Arch. Med. Res. 2023, 54, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Xiao, Q.; Song, H.; Ma, F.; Xu, F.; Peng, D.; Li, N.; Wang, X.; Niu, J.; Gao, P.; et al. Type I IFN Augments IL-27-Dependent TRIM25 Expression to Inhibit HBV Replication. Cell. Mol. Immunol. 2018, 15, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Yi, Z.; Song, H.; Xu, F.; Li, F.; Aliyari, R.; Zhang, H.; Du, P.; Ding, Y.; Niu, J.; et al. Type-I-IFN-Stimulated Gene TRIM5γ Inhibits HBV Replication by Promoting HBx Degradation. Cell Rep. 2019, 29, 3551–3563.e3. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Xiao, Q.; Xu, F.; Wei, Q.; Wang, F.; Tan, G. TRIM25 Inhibits HBV Replication by Promoting HBx Degradation and the RIG-I-Mediated PgRNA Recognition. Chin. Med. J. 2023, 136, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, N.R.; Zhou, L.; Guo, Y.R.; Zhao, C.; Tao, Y.J.; Krug, R.M.; Sawyer, S.L. Nuclear TRIM25 Specifically Targets Influenza Virus Ribonucleoproteins to Block the Onset of RNA Chain Elongation. Cell Host Microbe 2017, 22, 627–638.e7. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zou, C.; Wang, X.; Huang, C.; Feng, T.; Pan, W.; Wu, Q.; Wang, P.; Dai, J. Interferon-Stimulated TRIM69 Interrupts Dengue Virus Replication by Ubiquitinating Viral Nonstructural Protein 3. PLoS Pathog. 2018, 14, e1007287. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, N.L.; Wang, J.; Shi, P.-Y.; Wang, T.; Miller, M.A.; Li, K. Overlapping and Distinct Molecular Determinants Dictating the Antiviral Activities of TRIM56 against Flaviviruses and Coronavirus. J. Virol. 2014, 88, 13821–13835. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Li, N.L.; Wei, D.; Liu, B.; Guo, F.; Elbahesh, H.; Zhang, Y.; Zhou, Z.; Chen, G.-Y.; Li, K. The E3 Ligase TRIM56 Is a Host Restriction Factor of Zika Virus and Depends on Its RNA-Binding Activity but Not MiRNA Regulation, for Antiviral Function. PLoS Negl. Trop. Dis. 2019, 13, e0007537. [Google Scholar] [CrossRef] [PubMed]
- Heidary, F.; Gharebaghi, R. Systematic Review of the Antiviral Properties of TRIM56: A Potential Therapeutic Intervention for COVID-19. Expert Rev. Clin. Immunol. 2020, 16, 973–984. [Google Scholar] [CrossRef] [PubMed]

| Tripartite Motif (TRIM) Family Members Expression in Macrophages | ||||||||||
| Sp. | Homo sapiens (Human) | Mus musculus (C57BL/6mice) | Homo sapiens (Human) | Homo sapiens (Human) | Homo sapiens (Human) | Homo sapiens (Human) | Homo sapiens (Human) | Homo sapiens (Human) | Homo sapiens (Human) | |
| Cell type | MDMs FCS + M-CSF, 7 d.d. | BMDM M-CSF | MDMs h-AB serum, 7 to 11 d.d. | MDMs h-AB serum, 7 to 11 d.d. | MDMs h-AB serum, 7 d.d. | MDMs h-AB serum, 7 d.d. | MDMs h-AB serum, 7 d.d. | MDMs h-AB serum, 7 d.d. | MDMs FBS, 7 d.d. | |
| Stimulus | 100 ng/ml LPS + 20 ng/ml IFNγ 18 h | Influenza virus (IFNβ-dependent antiviral response), 24 hpi. | 1000 UI/ml universal IFN-I (recombinant IFNαA-E), 8 h | 1000 UI/ml IFN-II (IFNγ), 8 h | 25 gr/mL IFN-I (IFNα), 18 h | 25 ng/mL IFN-I (IFNε), 18 h | 25 ng/mL IFN-II (IFNγ), 18 h | 25 ng/mL IFN-III (IFNλ), 18 h | 25 ng/mL Interleukin 27, 18 h | |
| Technique | Microarray | RT-qPCR | RT-PCR Array | RT-PCR Array | Bulk RNA-Seq. and/or RT-qPCR | Bulk RNA-Seq. and/or RT-qPCR | Bulk RNA-Seq. and/or RT-qPCR | Bulk RNA-Seq. and/or RT-qPCR | Bulk RNA-Seq. and/or RT-qPCR | |
| Reference. | [77] | [78] | [73] | [73] | Our study | Our study | Our study | Our study | Our study | |
| SubFamily | TRIM Gene | mRNA Expression in Macrophages | ||||||||
| I | TRIM1 | N.C. | N.C. | N.C. | N.C. | Up-regulated | Up-regulated | Up-regulated | Up-regulated | N.C. |
| TRIM9 | N.R. | N.C. | N.C. | Down-regulated | Down-regulated | Up-regulated | Up-regulated | N.C. | N.C. | |
| TRIM18 | Up-regulated | Up-regulated | N.D. | N.D. | No significant, Down-regulated | No significant, Up-regulated | N.C. | No significant, Down-regulated | No significant, Up-regulated | |
| TRIM36 | N.R | N.D. | N.D. | N.D. | N.C. | Up-regulated | N.C. | N.C. | N.C. | |
| TRIM46 | Up-regulated | Up-regulated | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM67 | N.R. | N.R. | N.D. | N.D. | No significant, Down-regulated | No significant, Up-regulated | No significant, Down-regulated | N.C. | No significant, Down-regulated | |
| II | TRIM54 | N.R. | N.R. | Down-regulated | Down-regulated | N.C. | Down-regulated | Down-regulated | N.C. | N.C. |
| TRIM55 | N.R. | N.R. | N.D. | N.D. | No significant, Up-regulated | No significant, Down-regulated | No significant, Down-regulated | No significant, Up-regulated | No significant, Up-regulated | |
| TRIM63 | N.R. | N.R. | N.D. | N.D. | No significant, Down-regulated | N.C. | No significant, Up-regulated | N.C. | No significant, Up-regulated | |
| III | TRIM42 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. |
| IV | TRIML1 | N.R. | N.R. | N.D. | N.D. | N.C. | No significant, Up-regulated | N.C. | N.C. | N.C. |
| TRIM4 | N.R. | N.R. | N.C. | Down-regulated | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM5α | Up-regulated | No homologue in mouse | Up-regulated | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM6 | Up-regulated | Up-regulated | N.D. | N.D. | Up-regulated | Up-regulated | N.C. | Up-regulated | N.C. | |
| TRIM7 | N.R | N.R. | N.D. | N.D. | N.C. | N.C. | Up-regulated | N.C. | N.C. | |
| TRIM10 | Up-regulated | N.D. | N.D. | N.D. | No significant, Down-regulated | No significant, Down-regulated | Up-regulated | No significant, Down-regulated | No significant, Up-regulated | |
| TRIM11 | N.R. | N.R. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM15 | N.C. | N.D. | N.D. | N.D. | N.C. | No significant, Up-regulated | Up-regulated | N.C. | N.C. | |
| TRIM17 | Up-regulated | N.D. | N.D. | N.D. | No significant, Down-regulated | No significant, Down-regulated | N.C. | No significant, Down-regulated | N.C. | |
| TRIM21 | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | |
| TRIM22 | Up-regulated | No homologue in mouse | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | |
| TRIM25 | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | N.C. | |
| TRIM26 | Up-regulated | Up-regulated | Up-regulated | N.C. | Up-regulated | Up-regulated | Up-regulated | Up-regulated | N.C. | |
| TRIM27 | Down-regulated | N.C. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM34 | Up-regulated | Up-regulated | Up-regulated | N.C. | No significant, Up-regulated | No significant, Up-regulated | No significant, Up-regulated | No significant, Down-regulated | N.C. | |
| TRIM35 | Up-regulated | Up-regulated | Up-regulated | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM38 | Up-regulated | N.C. | Up-regulated | N.C. | Up-regulated | Up-regulated | Up-regulated | N.C. | N.C. | |
| TRIM39 | N.C. | N.C. | N.C. | N.C. | N.C. | Up-regulated | N.C. | N.C. | N.C. | |
| TRIM41 | N.R. | N.R. | N.D. | N.D. | N.C. | No significant, Up-regulated | Up-regulated | N.C. | N.C. | |
| TRIM43 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM47 | N.R. | N.R. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM48 | Up-regulated | No homologue in mouse | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM49 | N.C. | No homologue in mouse | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM50 | N.R. | N.R. | N.D. | N.D. | No significant, Down-regulated | No significant, Down-regulated | No significant, Down-regulated | N.C. | No significant, Down-regulated | |
| TRIM53 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM58 | N.C. | N.D. | Up-regulated | N.C. | Down-regulated | N.C. | N.C. | Down-regulated | N.C. | |
| TRIM60 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM62 | Up-regulated | N.D. | N.C. | N.C. | N.C. | Up-regulated | N.C. | N.C. | N.C. | |
| TRIM64 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM65 | N.R. | N.R. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM68 | Up-regulated | N.C. | N.C. | N.C. | N.C. | Up-regulated | N.C. | N.C. | N.C. | |
| TRIM69 | N.R. | N.R. | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | N.C. | Up-regulated | |
| TRIM72 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | No significant, Up-regulated | N.C. | |
| TRIM75 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| V | TRIM8 | Up-regulated | Up-regulated | N.C. | N.C. | N.C. | Up-regulated | N.C. | N.C. | N.C. |
| TRIM19 | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | Up-regulated | |
| TRIM31 | Up-regulated | N.D. | Up-regulated | N.D. | Up-regulated | Up-regulated | Up-regulated | Up-regulated | No significant, Up-regulated | |
| TRIM40 | N.R. | N.R. | N.D. | N.D. | No significant, Up-regulated | N.C. | N.C. | N.C. | N.C. | |
| TRIM52 | Down-regulated | N.D. | N.C. | N.C. | N.C. | N.C. | Up-regulated | N.C. | N.C. | |
| TRIM56 | N.R. | N.R. | Up-regulated | Up-regulated | Up-regulated | Up-regulated | No significant, Up-regulated | Up-regulated | N.C. | |
| TRIM61 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | No significant, Down-regulated | |
| TRIM73 | N.R. | N.R. | N.C. | N.C. | N.C. | No significant, Down-regulated | No significant, Down-regulated | N.C. | N.C. | |
| TRIM74 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| VI | TRIM24 | N.C. | N.C. | N.C. | N.C. | Down-regulated | Down-regulated | N.C. | N.C. | N.C. |
| TRIM28 | N.C. | N.C. | Down-regulated | Down-regulated | Down-regulated | N.C. | N.C. | N.C. | N.C. | |
| TRIM33 | Up-regulated | N.D. | N.C. | N.C. | N.C. | Up-regulated | N.C. | N.C. | N.C. | |
| VII | TRIM2 | N.R. | N.R. | N.C. | Down-regulated | N.C. | N.C. | No significant, Up-regulated | N.C. | N.C. |
| TRIM3 | Up-regulated | Up-regulated | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM32 | Down-regulated | N.D. | N.C. | Down-regulated | Down-regulated | Down-regulated | Down-regulated | N.C. | N.C. | |
| TRIM71 | N.R. | N.R. | N.D. | N.D. | No significant, Down-regulated | No significant, Down-regulated | No significant, Down-regulated | No significant, Up-regulated | No significant, Down-regulated | |
| VIII | TRIM37 | N.C. | N.C. | Down-regulated | Down-regulated | N.C. | N.C. | N.C. | N.C. | N.C. |
| IX | TRIM23 | Down-regulated | Up-regulated | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. |
| X | TRIM45 | Up-regulated | Up-regulated | N.D. | N.D. | No significant, Down-regulated | Down-regulated | No significant, Down-regulated | N.C. | No significant, Down-regulated |
| XI | TRIM13 | Up-regulated | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. |
| TRIM59 | N.C. | N.C. | Down-regulated | Down-regulated | N.C. | N.C. | N.C. | N.C. | N.C. | |
| NO RING | TRIM14 | Up-regulated | Up-regulated | Up-regulated | N.C. | Up-regulated | Up-regulated | N.C. | Up-regulated | N.C. |
| TRIM16 | Down-regulated | Down-regulated | N.C. | Down-regulated | N.C. | Up-regulated | N.C. | N.C. | N.C. | |
| TRIM20 | N.C. | Up-regulated | Up-regulated | Up-regulated | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM29 | Up-regulated | N.D. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | No significant, Down-regulated | |
| TRIM44 | Down-regulated | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM66 | Down-regulated | N.D. | Down-regulated | Down-regulated | N.C. | N.C. | N.C. | N.C. | N.C. | |
| TRIM76 | N.R. | N.R. | N.D. | N.D. | N.C. | N.C. | N.C. | N.C. | N.C. | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Sarmiento, L.J.; Tamayo-Molina, Y.S.; Valdés-López, J.F.; Urcuqui-Inchima, S. Interleukin 27, Similar to Interferons, Modulates Gene Expression of Tripartite Motif (TRIM) Family Members and Interferes with Mayaro Virus Replication in Human Macrophages. Viruses 2024, 16, 996. https://doi.org/10.3390/v16060996
Hernández-Sarmiento LJ, Tamayo-Molina YS, Valdés-López JF, Urcuqui-Inchima S. Interleukin 27, Similar to Interferons, Modulates Gene Expression of Tripartite Motif (TRIM) Family Members and Interferes with Mayaro Virus Replication in Human Macrophages. Viruses. 2024; 16(6):996. https://doi.org/10.3390/v16060996
Chicago/Turabian StyleHernández-Sarmiento, Lady Johana, Y. S. Tamayo-Molina, Juan Felipe Valdés-López, and Silvio Urcuqui-Inchima. 2024. "Interleukin 27, Similar to Interferons, Modulates Gene Expression of Tripartite Motif (TRIM) Family Members and Interferes with Mayaro Virus Replication in Human Macrophages" Viruses 16, no. 6: 996. https://doi.org/10.3390/v16060996






