Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Growth
2.2. Bacteriophage Isolation and Propagation
2.3. Whole Genome Sequencing and Annotation of Pseudomonas Phages
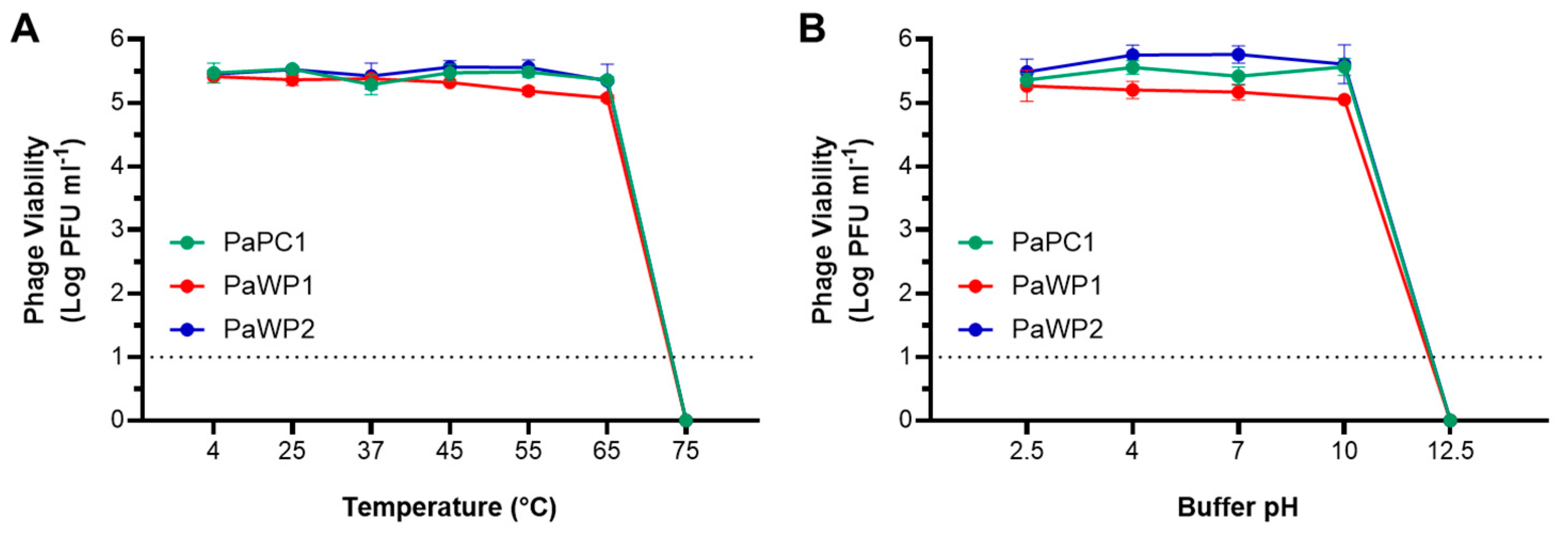
2.4. Temperature and pH Stability of Phages
2.5. Bacterial Lysis Curves
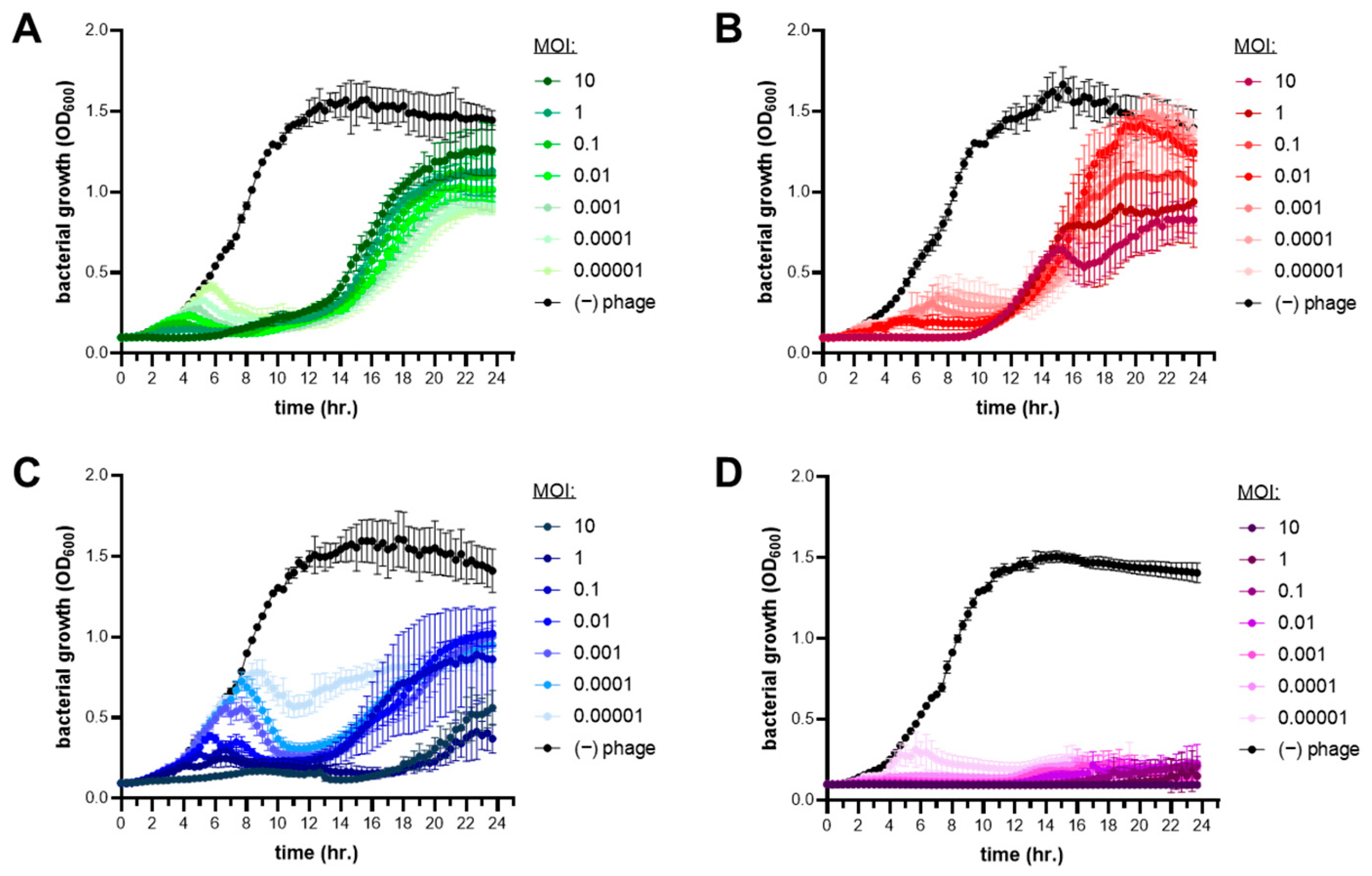
2.6. Phage–Antibiotic Combination Growth Curves
2.7. Biofilm Disruption with PaPC1, PaWP1, and PaWP2
2.8. Data Availability
3. Results
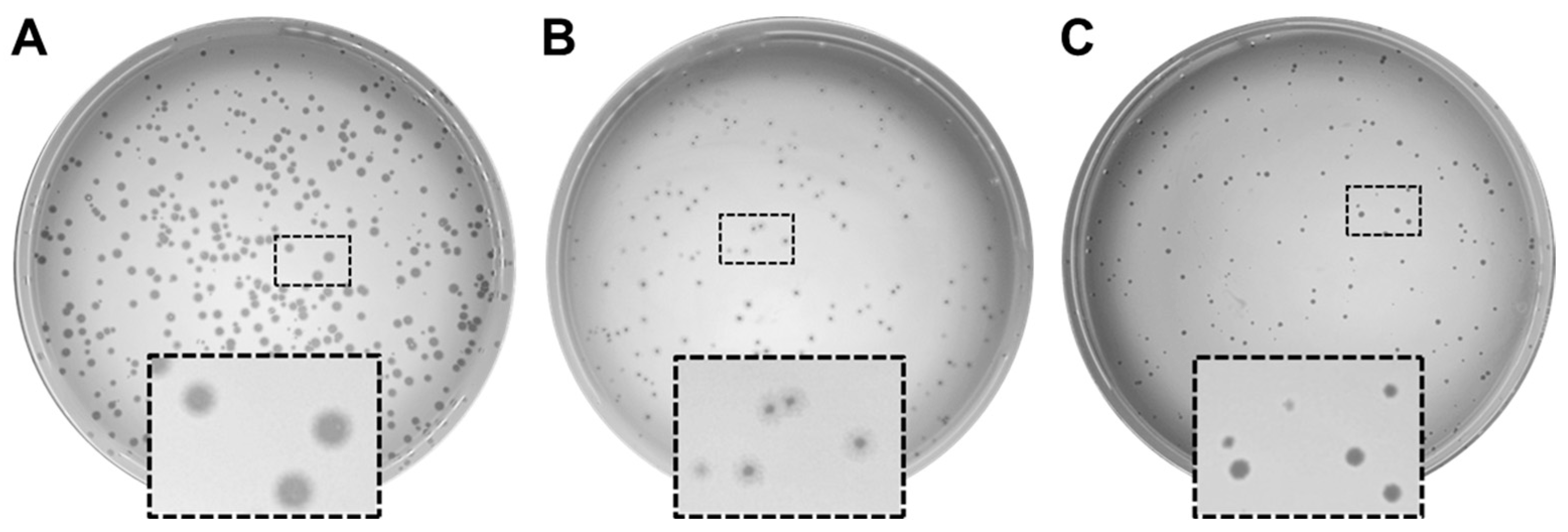
3.1. Isolation and Characterization of Phages Infecting P. aeruginosa PAO1
3.2. Genome Characterization and Genome Similarity of PaPC1, PaWP1, and PaWP2 to Existing Phages
| Features | PaC1Φ | PaWP1Φ | PaWP2Φ |
|---|---|---|---|
| CDS | 90 | 320 | 97 |
| connector | 4 | 1 | 4 |
| DNA, RNA, and nucleotide metabolism | 12 | 11 | 10 |
| head and packaging | 10 | 26 | 10 |
| integration and excision | 0 | 0 | 0 |
| lysis | 3 | 1 | 3 |
| moron, auxiliary metabolic gene and host takeover | 0 | 0 | 1 |
| other | 1 | 5 | 1 |
| Tail | 8 | 27 | 9 |
| transcription regulation | 0 | 0 | 0 |
| unknown function | 52 | 249 | 59 |
| tRNAs | 0 | 0 | 0 |
| CRISPRs | 0 | 0 | 0 |
| tmRNAs | 0 | 0 | 0 |
| † VFDB_Virulence_Factors | 0 | 0 | 0 |
| ‡ CARD_AMR_Genes | 0 | 0 | 0 |
3.3. PaPC1, PaWP1, and PaWP2 Demonstrate Thermostability and pH Tolerance
3.4. Lytic Infection Profile of Phages PaPC1, PaWP1, and PaWP2
3.5. Combinations of Phages with Antibiotics Enhances Carbapenem Sensitivity of P. aeruginosa PAO1
3.6. PaPC1, PaWP1, and PaWP2 Phages Disrupt Biofilms of P. aeruginosa PAO1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Iskandar, K.; Murugaiyan, J.; Hammoudi Halat, D.; Hage, S.E.; Chibabhai, V.; Adukkadukkam, S.; Roques, C.; Molinier, L.; Salameh, P.; Van Dongen, M. Antibiotic Discovery and Resistance: The Chase and the Race. Antibiotics 2022, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. 2020 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019. [Google Scholar]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Khawaldeh, A.; Morales, S.; Dillon, B.; Alavidze, Z.; Ginn, A.N.; Thomas, L.; Chapman, S.J.; Dublanchet, A.; Smithyman, A.; Iredell, J.R. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J. Med. Microbiol. 2011, 60, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Cesta, N.; Pini, M.; Mulas, T.; Materazzi, A.; Ippolito, E.; Wagemans, J.; Kutateladze, M.; Fontana, C.; Sarmati, L.; Tavanti, A.; et al. Application of Phage Therapy in a Case of a Chronic Hip-Prosthetic Joint Infection due to Pseudomonas aeruginosa: An Italian Real-Life Experience and In Vitro Analysis. In Open Forum Infectious Diseases; Oxford University Press: New York, NY, USA, 2023; Volume 10. [Google Scholar] [CrossRef]
- Rezk, N.; Abdelsattar, A.S.; Elzoghby, D.; Agwa, M.M.; Abdelmoteleb, M.; Aly, R.G.; Fayez, M.S.; Essam, K.; Zaki, B.M.; El-Shibiny, A. Bacteriophage as a potential therapy to control antibiotic-resistant Pseudomonas aeruginosa infection through topical application onto a full-thickness wound in a rat model. J. Genet. Eng. Biotechnol. 2022, 20, 133. [Google Scholar] [CrossRef] [PubMed]
- Al-Wrafy, F.; Brzozowska, E.; Górska, S.; Gamian, A. Pathogenic factors of Pseudomonas aeruginosa-the role of biofilm in pathogenicity and as a target for phage therapy. Postępy Hig. Med. Dośw. 2017, 71, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.Y.K.; Das, T.; Manos, J.; Kutter, E.; Morales, S.; Chan, H.-K. Bacteriophage PEV20 and Ciprofloxacin Combination Treatment Enhances Removal of Pseudomonas aeruginosa Biofilm Isolated from Cystic Fibrosis and Wound Patients. AAPS J. 2019, 21, 1–8. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef] [PubMed]
- Bedenić, B.; Plečko, V.; Sardelić, S.; Uzunović, S.; Godič Torkar, K. Carbapenemases in Gram-Negative Bacteria: Laboratory Detection and Clinical Significance. BioMed Res. Int. 2014, 2014, 1–3. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Brown, K.; Kortright, K.E.; Mao, S.; Turner, P.E. Extending the lifetime of antibiotics: How can phage therapy help? Future Microbiol. 2016, 11, 1105–1107. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, A.R.; Fortier, A.; Roush, C.; Lessing, A.J.; Bender, R.G.; Barahman, R.; Grant, R.; Chan, B.K.; Turner, P.E. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 11207–11216. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.; Forsyth, J.H.; Patwa, R.; Kostoulias, X.; Trim, M.; Subedi, D.; Archer, S.K.; Morris, F.C.; Oliveira, C.; Kielty, L.; et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol. 2021, 6, 157–161. [Google Scholar] [CrossRef]
- Hao, G.; Chen, A.I.; Liu, M.; Zhou, H.; Egan, M.; Yang, X.; Kan, B.; Wang, H.; Goulian, M.; Zhu, J. Colistin Resistance-Mediated Bacterial Surface Modification Sensitizes Phage Infection. Antimicrob. Agents Chemother. 2019, 63, e01609-19. [Google Scholar] [CrossRef] [PubMed]
- Kwiatek, M.; Parasion, S.; Rutyna, P.; Mizak, L.; Gryko, R.; Niemcewicz, M.; Olender, A.; Lobocka, M. Isolation of bacteriophages and their application to control Pseudomonas aeruginosa in planktonic and biofilm models. Res. Microbiol. 2017, 168, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Bryers, J.D. Medical biofilms. Biotechnol. Bioeng. 2008, 100, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Akgün, D.; Trampuz, A.; Perka, C.; Renz, N. High failure rates in treatment of streptococcal periprosthetic joint infection: Results from a seven-year retrospective cohort study. Bone Jt. J. 2017, 99, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Campoccia, D.; Montanaro, L.; Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409. [Google Scholar] [CrossRef]
- Amankwah, S.; Adisu, M.; Gorems, K.; Abdella, K.; Kassa, T. Assessment of Phage-Mediated Inhibition and Removal of Multidrug-Resistant Pseudomonas aeruginosa Biofilm on Medical Implants. Infect. Drug Resist. 2022, 15, 2797–2811. [Google Scholar] [CrossRef]
- Rodriguez-Gonzalez, R.A.; Leung, C.Y.; Chan, B.K.; Turner, P.E.; Weitz, J.S. Quantitative Models of Phage-Antibiotic Combination Therapy. mSystems 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, M.; Bouzari, M.; Ghaemi, E.A. Isolation and Characterization of a Lytic Bacteriophage (vB_PmiS-TH) and Its Application in Combination with Ampicillin against Planktonic and Biofilm Forms of Proteus mirabilis Isolated from Urinary Tract Infection. Microb. Physiol. 2018, 28, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Doub, J.B.; Ng, V.Y.; Wilson, E.; Corsini, L.; Chan, B.K. Successful Treatment of a Recalcitrant Staphylococcus epidermidis Prosthetic Knee Infection with Intraoperative Bacteriophage Therapy. Pharmaceuticals 2021, 14, 231. [Google Scholar] [CrossRef] [PubMed]
- Schoeffel, J.; Wang, E.W.; Gill, D.; Frackler, J.; Horne, B.A.; Manson, T.; Doub, J.B. Successful Use of Salvage Bacteriophage Therapy for a Recalcitrant MRSA Knee and Hip Prosthetic Joint Infection. Pharmaceuticals 2022, 15, 177. [Google Scholar] [CrossRef] [PubMed]
- Doub, J.B.; Chan, B.; Johnson, A.J. Salphage: Salvage bacteriophage therapy for a chronic Enterococcus faecalis prosthetic joint infection. IDCases 2023, 33, e01854. [Google Scholar] [CrossRef] [PubMed]
- Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages Using the Small Drop Plaque Assay System. Methods Mol. Biol. 2009, 501, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Pharokka: A fast scalable bacteriophage annotation tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Snesrud, E.; Hall, L.; Mills, E.; Galac, M.; Stam, J.; Ong, A.; Maybank, R.; Kwak, Y.I.; Johnson, S.; et al. A panel of diverse Pseudomonas aeruginosa clinical isolates for research and development. JAC-Antimicrob. Resist. 2021, 3, dlab179. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, e2437. [Google Scholar] [CrossRef]
- Terzian, P.; Ndela, E.P.; Galiez, C.; Lossouarn, J.; Bucio, R.E.P.; Mom, R.; Toussaint, A.; Petit, M.-A.; Enault, F. PHROG: Families of prokaryotic virus proteins clustered using remote homology. NAR Genom. Bioinform. 2021, 3, lqab067. [Google Scholar] [CrossRef] [PubMed]
- Tynecki, P.; Guziński, A.; Kazimierczak, J.; Jadczuk, M.; Dastych, J.; Onisko, A. PhageAI-Bacteriophage Life Cycle Recognition with Machine Learning and Natural Language Processing. BioRxiv 2020. [Google Scholar] [CrossRef]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Adriaenssens, E.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Hughes, K.A.; Sutherland, I.W.; Jones, M.V.; Hughes, K.A.; Sutherland, I.W.; Jones, M.V. Biofilm susceptibility to bacteriophage attack: The role of phage-borne polysaccharide depolymerase. Microbiology 1998, 144, 3039–3047. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Mao, J.; Xie, J.; Yan, J.; Mao, J.; Xie, J. Bacteriophage Polysaccharide Depolymerases and Biomedical Applications. BioDrugs 2013, 28, 265–274. [Google Scholar] [CrossRef]
- Chen, X.; Liu, M.; Zhang, P.; Xu, M.; Yuan, W.; Bian, L.; Liu, Y.; Xia, J.; Leung, S.S.Y. Phage-Derived Depolymerase as an Antibiotic Adjuvant Against Multidrug-Resistant Acinetobacter baumannii. Front. Microbiol. 2022, 13, 845500. [Google Scholar] [CrossRef]
- Shahed-Al-Mahmud, M.; Roy, R.; Sugiokto, F.G.; Islam, M.N.; Lin, M.-D.; Lin, L.-C.; Lin, N.-T. Phage φAB6-Borne Depolymerase Combats Acinetobacter baumannii Biofilm Formation and Infection. Antibiotics 2021, 10, 279. [Google Scholar] [CrossRef]
- Ezemokwe, G.C.; Agwom, F.M.; Okoliegbe, I.; Okonkwo, F.O.; Ngene, A.C.; Gimba, N.; Morenikeji, O.R.; Egwuenu, A.; Okonkwo, C.H.; Aguiyi, J.C.; et al. Complete Genome Sequence of Pseudomonas Phage Zikora. Microbiol. Resour. Announc. 2021, 10, e0048921. [Google Scholar] [CrossRef] [PubMed]
- Atilano, M.L.; Pereira, P.M.; Yates, J.; Reed, P.; Veiga, H.; Pinho, M.G.; Filipe, S.R. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2010, 107, 18991–18996. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, C.J.; Faustoferri, R.C.; Bischer, A.P.; Quivey, R.G. Streptococcus mutans requires mature rhamnose-glucose polysaccharides for proper pathophysiology, morphogenesis and cellular division. Mol. Microbiol. 2019, 112, 944–959. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.; Sousa, J.C.; Silva, A.C.; Melo, L.D.R.; Sillankorva, S. Chestnut Honey and Bacteriophage Application to Control Pseudomonas aeruginosa and Escherichia coli Biofilms: Evaluation in an ex vivo Wound Model. Front. Microbiol. 2018, 9, 392134. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public. Health 2018, 2018, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Law, N.; Logan, C.; Yung, G.; Furr, C.L.; Lehman, S.M.; Morales, S.; Rosas, F.; Gaidamaka, A.; Bilinsky, I.; Grint, P.; et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 2019, 47, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Vashisth, M.; Jaglan, A.B.; Yashveer, S.; Sharma, P.; Bardajatya, P.; Virmani, N.; Bera, B.C.; Vaid, R.K.; Anand, T. Development and Evaluation of Bacteriophage Cocktail to Eradicate Biofilms Formed by an Extensively Drug-Resistant (XDR) Pseudomonas aeruginosa. Viruses 2023, 15, 427. [Google Scholar] [CrossRef]
- Clinical Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 10th ed.; M07-A11; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kovacs, C.J.; Rapp, E.M.; Rankin, W.R.; McKenzie, S.M.; Brasko, B.K.; Hebert, K.E.; Bachert, B.A.; Kick, A.R.; Burpo, F.J.; Barnhill, J.C. Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics. Viruses 2024, 16, 1000. https://doi.org/10.3390/v16071000
Kovacs CJ, Rapp EM, Rankin WR, McKenzie SM, Brasko BK, Hebert KE, Bachert BA, Kick AR, Burpo FJ, Barnhill JC. Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics. Viruses. 2024; 16(7):1000. https://doi.org/10.3390/v16071000
Chicago/Turabian StyleKovacs, Christopher J., Erika M. Rapp, William R. Rankin, Sophia M. McKenzie, Brianna K. Brasko, Katherine E. Hebert, Beth A. Bachert, Andrew R. Kick, F. John Burpo, and Jason C. Barnhill. 2024. "Combinations of Bacteriophage Are Efficacious against Multidrug-Resistant Pseudomonas aeruginosa and Enhance Sensitivity to Carbapenem Antibiotics" Viruses 16, no. 7: 1000. https://doi.org/10.3390/v16071000






