Arg18 Substitutions Reveal the Capacity of the HIV-1 Capsid Protein for Non-Fullerene Assembly
Abstract
1. Introduction
2. Materials and Methods
2.1. Purification and Assembly of HIV-1 CA Arg18 Mutants
2.2. CryoEM Grid Preparation and Image Data Collection
2.3. Structure Determination
2.4. Coordinate Modeling
3. Results
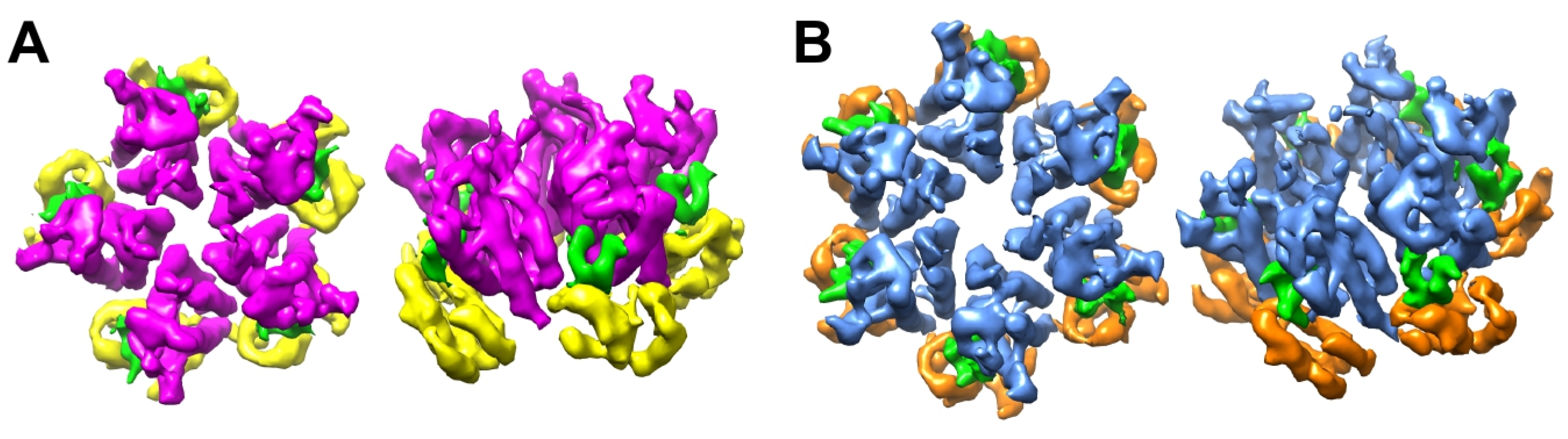
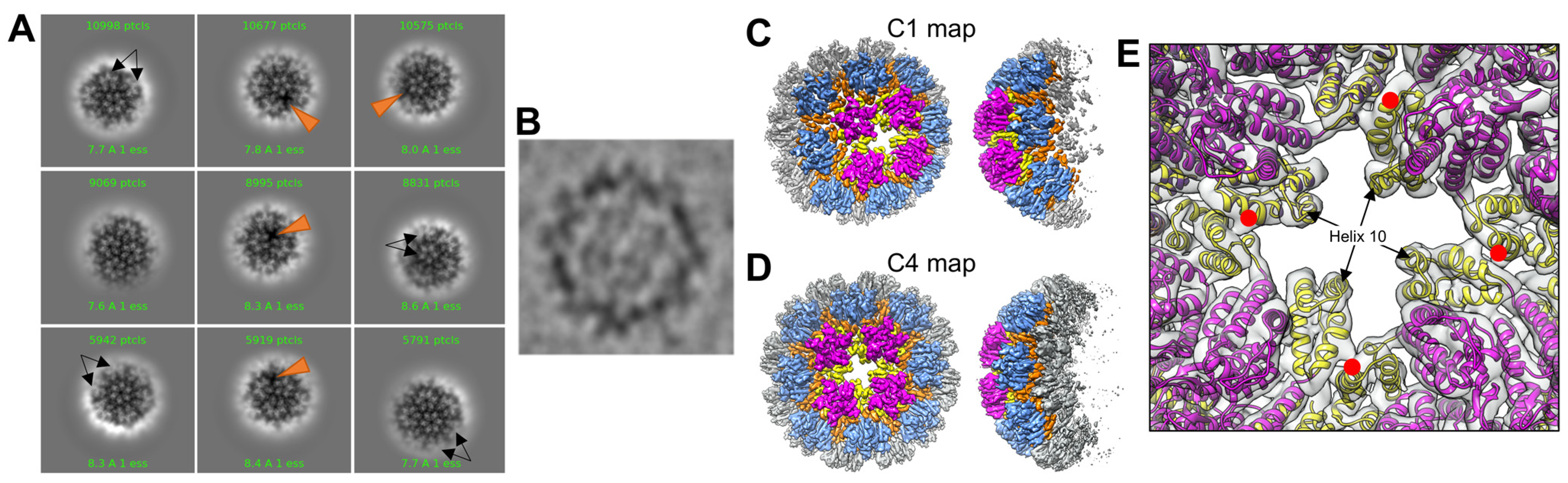
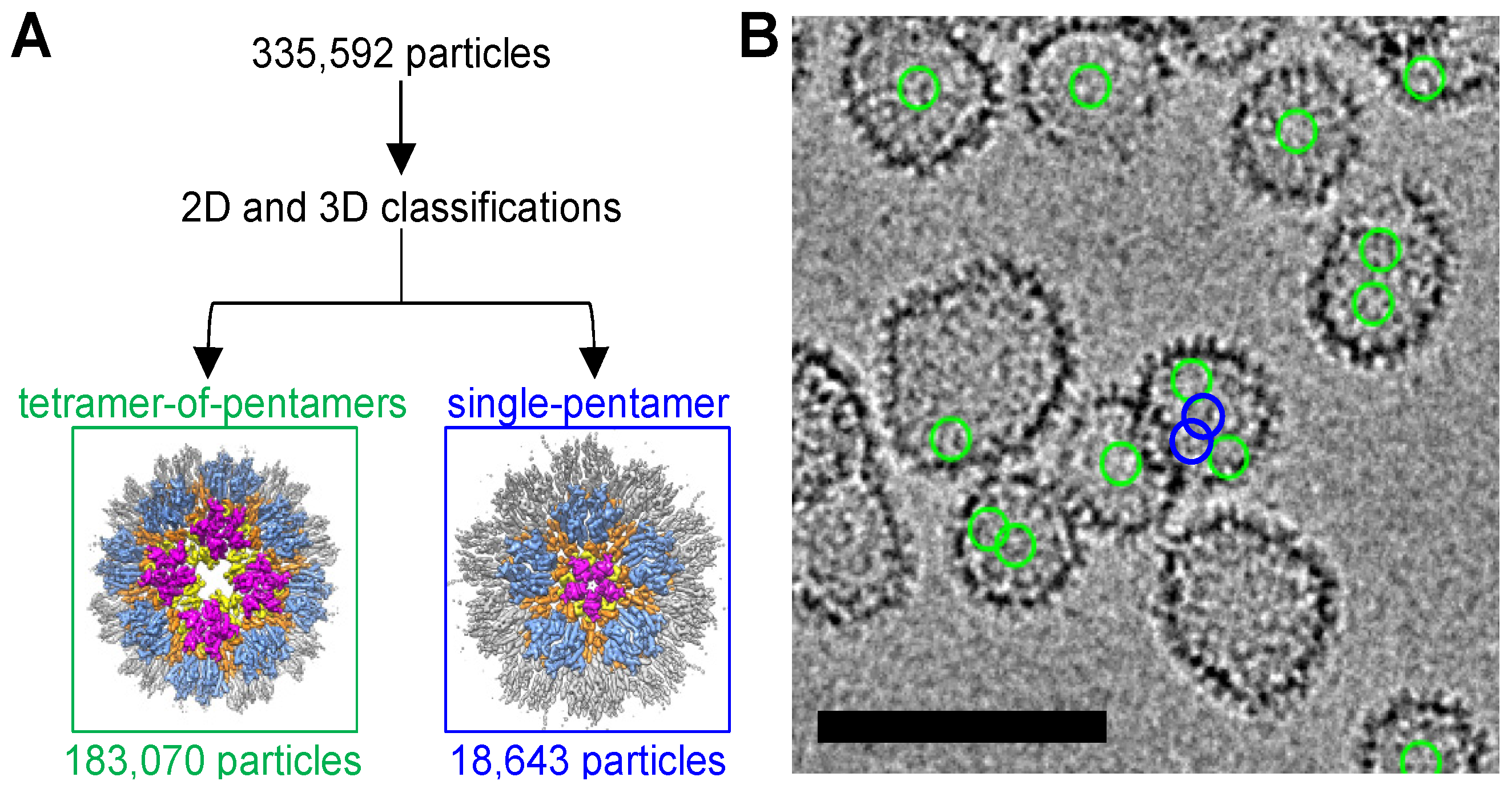
3.1. CryoEM Structures of R18L HIV-1 CA Assemblies
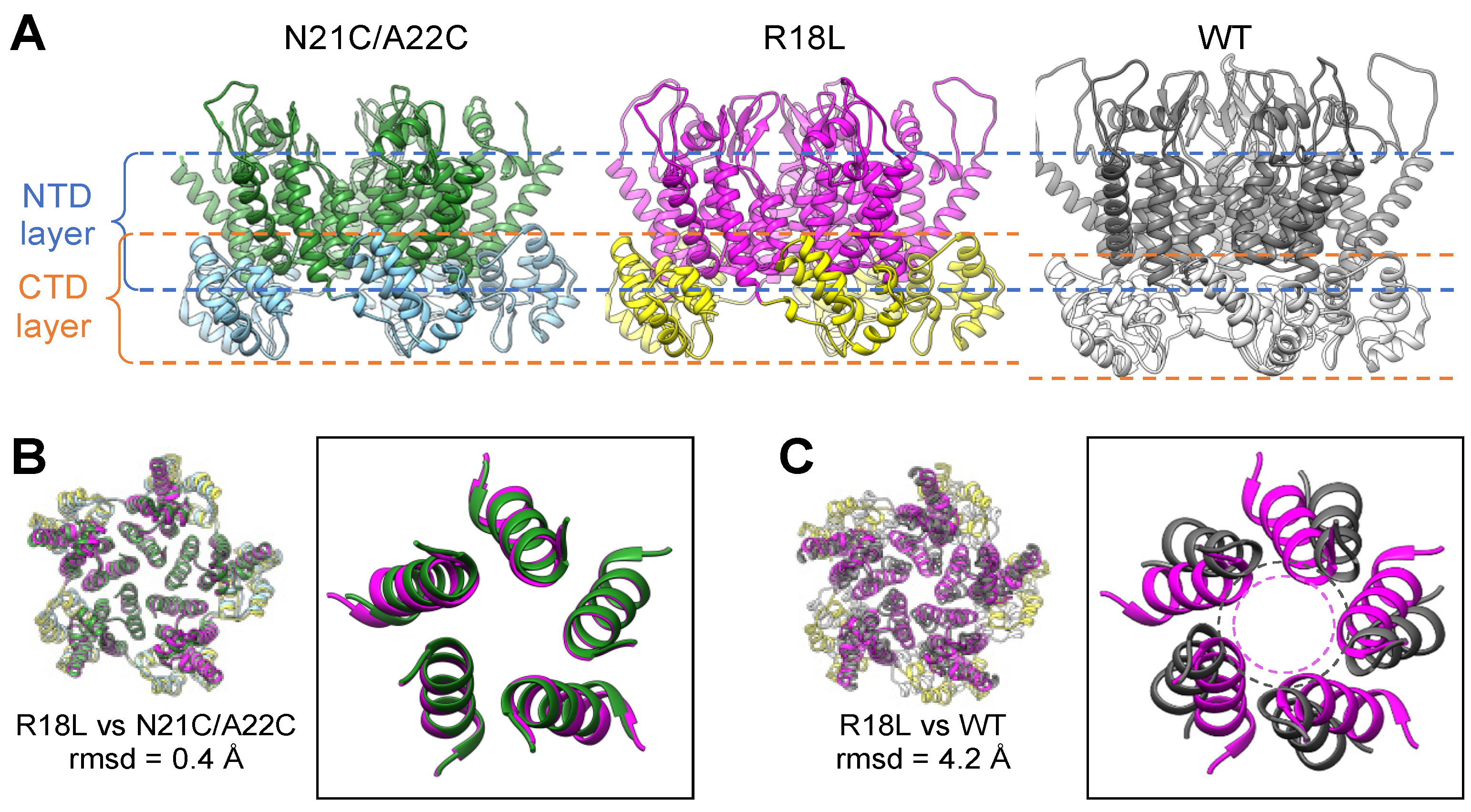
3.2. R18L Allows Pentamer Formation without Triggering the CA Hexamer/Pentamer Switch
3.3. The NTD–CTD Interfaces of the R18L Pentamer and Hexamer Are Functionally Equivalent
3.4. R18L CLPs Combine Icosahedral and Octahedral Geometries
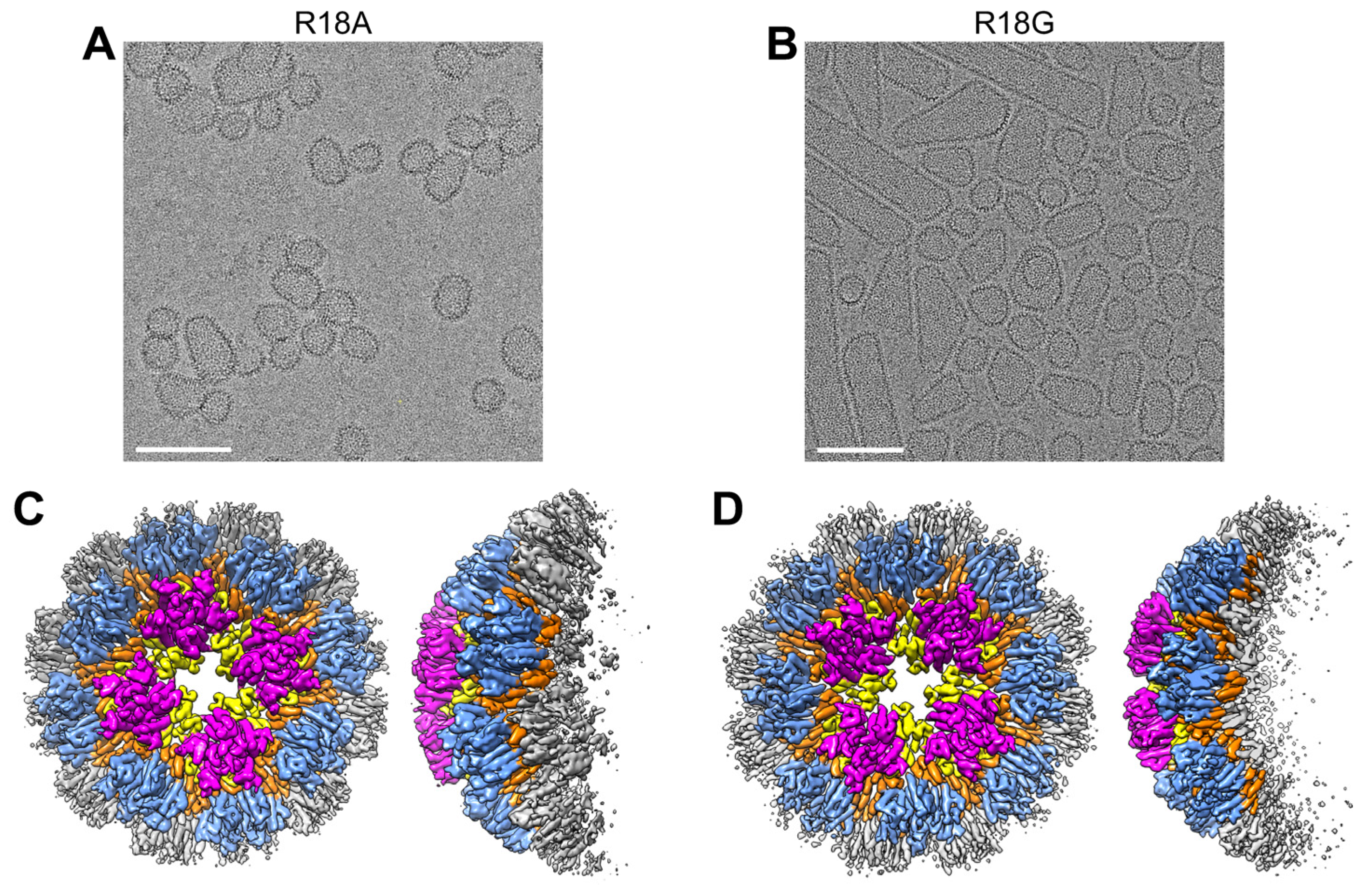
3.5. CA R18A and R18G Form the Same Tetramer-of-Pentamers as R18L
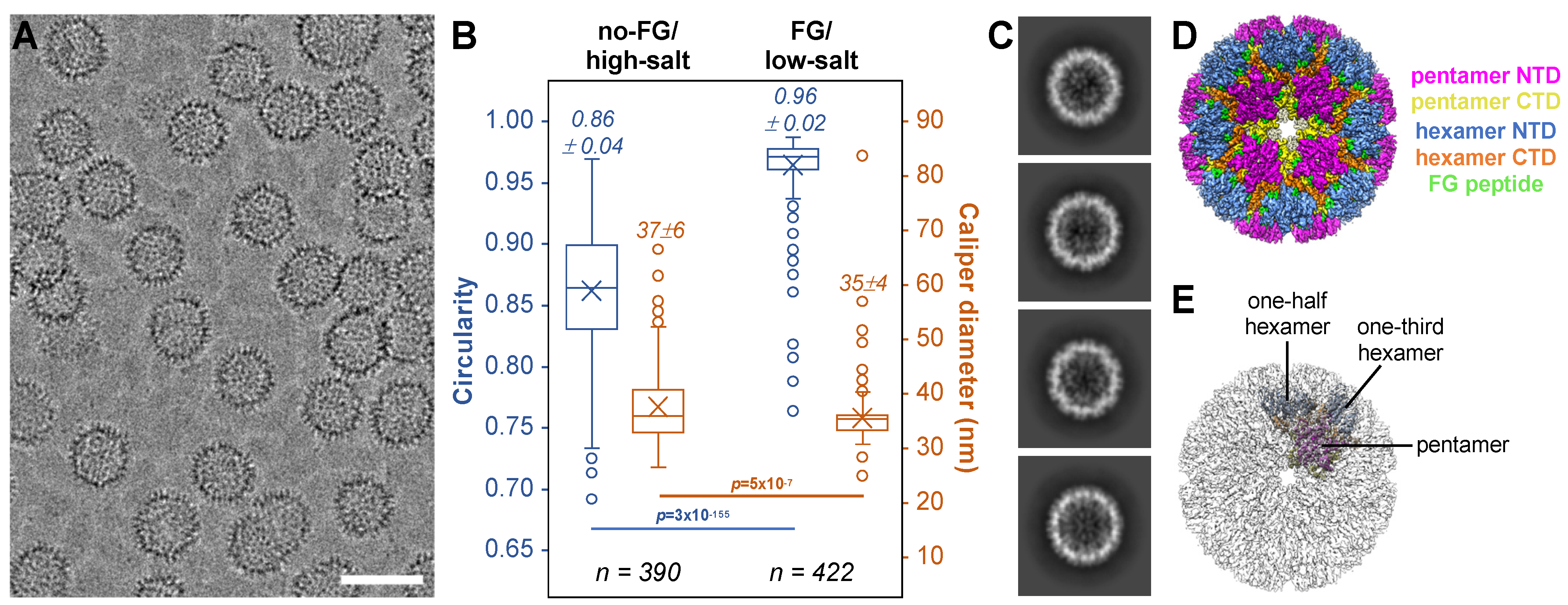
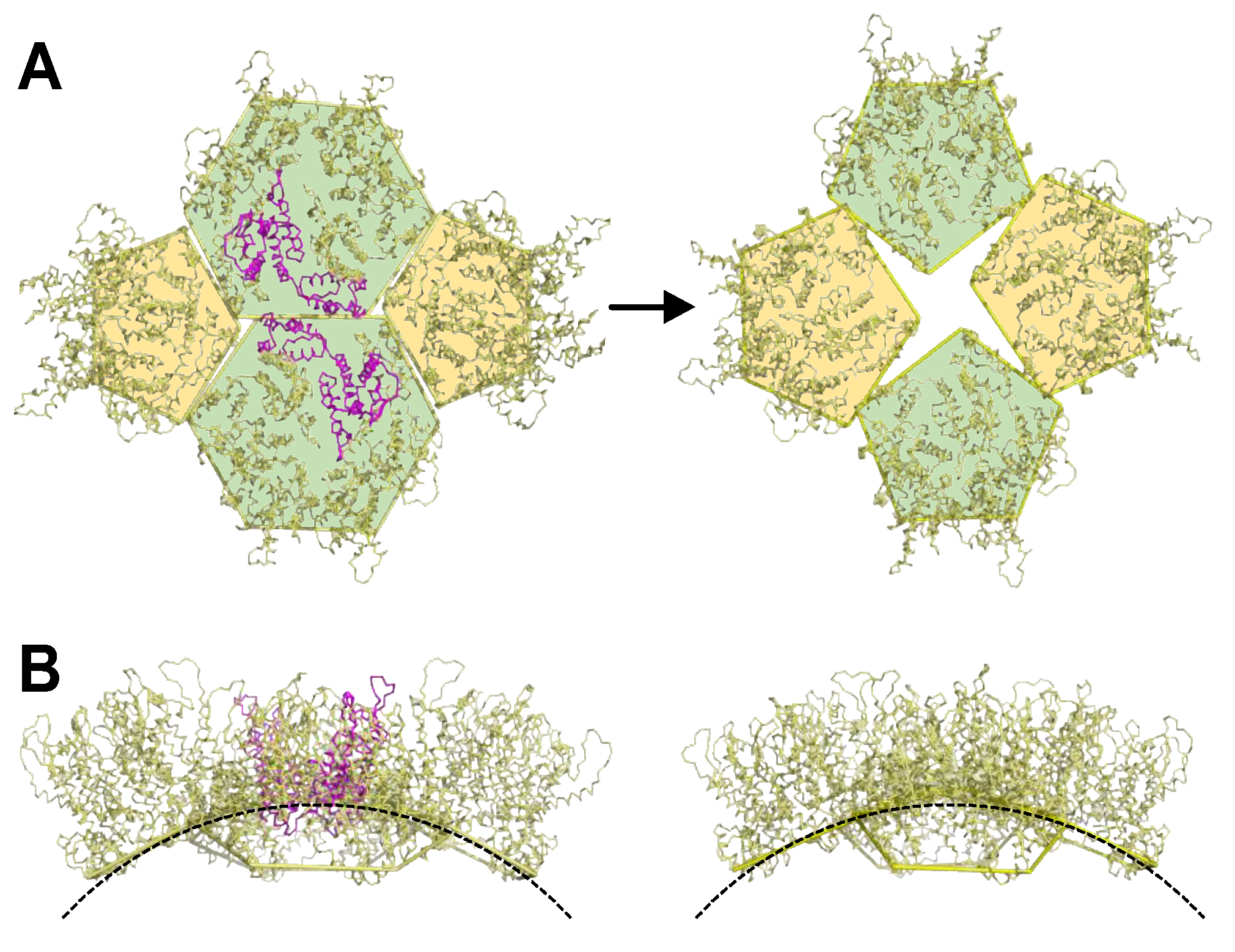
3.6. FG Binding Reinforces Octahedral Symmetry in Arg18 Mutant CA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christensen, D.E.; Ganser-Pornillos, B.K.; Johnson, J.S.; Pornillos, O.; Sundquist, W.I. Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro. Science 2020, 370, eabc8420. [Google Scholar] [CrossRef] [PubMed]
- Jennings, J.; Shi, J.; Varadarajan, J.; Jamieson, P.J.; Aiken, C. The host cell metabolite inositol hexakisphosphate promotes efficient endogenous HIV-1 reverse transcription by stabilizing the viral capsid. mBio 2020, 11, e02820-20. [Google Scholar] [CrossRef] [PubMed]
- Rasaiyaah, J.; Tan, C.P.; Fletcher, A.J.; Price, A.J.; Blondeau, C.; Hilditch, L.; Jacques, D.A.; Selwood, D.L.; James, L.C.; Noursadeghi, M.; et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 2013, 503, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Zila, V.; Müller, T.G.; Müller, B.; Kräusslich, H.G. HIV-1 capsid is the key orchestrator of early viral replication. PLoS Pathog. 2021, 17, e1010109. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.H. HIV-1 capsid exploitation of the host microtubule cytoskeleton during early infection. Retrovirol 2021, 18, 19. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Weiskopf, E.N.; Akkermans, O.; Swanson, N.A.; Cheng, S.; Schwartz, T.U.; Görlich, D. HIV-1 capsids enter the FG phase of nuclear pores like a transport receptor. Nature 2024, 626, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Dickson, C.F.; Hertel, S.; Tuckwell, A.J.; Li, N.; Ruan, J.; Al-Izzi, S.C.; Ariotti, N.; Sierecki, E.; Gambin, Y.; Morris, R.G.; et al. The HIV capsid mimics karyopherin engagement of FG-nucleoporins. Nature 2024, 626, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.M.; Hope, T.J. HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 2015, 13, 471–483. [Google Scholar] [CrossRef]
- Bester, S.M.; Wei, G.; Zhao, H.; Adu-Ampratwum, D.; Iqbal, N.; Courouble, V.V.; Francis, A.C.; Annamalai, A.S.; Singh, P.K.; Shkriabai, N.; et al. Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 2020, 370, 360–364. [Google Scholar] [CrossRef]
- Link, J.O.; Rhee, M.S.; Tse, W.C.; Zheng, J.; Somoza, J.R.; Rowe, W.; Begley, R.; Chiu, A.; Mulato, A.; Hansen, D.; et al. Clinical targeting of HIV capsid protein with a long-acting small molecule. Nature 2020, 584, 614–618. [Google Scholar] [CrossRef]
- Ganser, B.K.; Li, S.; Klishko, V.Y.; Finch, J.T.; Sundquist, W.I. Assembly and analysis of conical models for the HIV-1 core. Science 1999, 283, 80–83. [Google Scholar] [CrossRef]
- Schirra, R.T.; Dos Santos NF, B.; Zadrozny, K.K.; Kucharska, I.; Ganser-Pornillos, B.K.; Pornillos, O. A molecular switch modulates assembly and host factor binding of the HIV-1 capsid. Nat. Struct. Mol. Biol. 2023, 30, 383–390. [Google Scholar] [CrossRef]
- Stacey, J.C.V.; Tan, A.; Lu, J.M.; James, L.C.; Dick, R.A.; Briggs JA, G. Two structural switches in HIV-1 capsid regulate capsid curvature and host factor binding. Proc. Natl. Acad. Sci. USA 2023, 120, e2220557120. [Google Scholar] [CrossRef] [PubMed]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.L.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 2018, 560, 509–512. [Google Scholar] [CrossRef]
- Renner, N.; Mallery, D.L.; Faysal KM, R.; Peng, W.; Jacques, D.A.; Böcking, T.; James, L.C. A lysine ring in HIV capsid pores coordinates IP6 to drive mature capsid assembly. PLoS Pathog. 2021, 17, e1009164. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; von Schwedler, U.K.; Stray, K.M.; Aiken, C.; Sundquist, W.I. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 2004, 78, 2545–2552. [Google Scholar] [CrossRef] [PubMed]
- Ganser-Pornillos, B.K.; Cheng, A.; Yeager, M. Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell 2007, 131, 70–79. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K.; Banumathi, S.; Hua, Y.; Yeager, M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J. Mol. Biol. 2010, 401, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, M.; Bai, S.; Zhang, F.; Jiang, J.; Zheng, E.; Gao, S.; Yan, X.; Li, S.; Gu, Y.; et al. T = 4 icosahedral HIV-1 capsid as an immunogenic vector for HIV-1 V3 loop epitope display. Viruses 2018, 10, 667. [Google Scholar] [CrossRef]
- Wang, J.C.-Y.; Mukhopadhyay, S.; Zlotnick, A. Geometric defects and icosahedral viruses. Viruses 2018, 10, 25. [Google Scholar] [CrossRef]
- Pornillos, O.; Ganser-Pornillos, B.K.; Yeager, M. Atomic-level modelling of the HIV capsid. Nature 2011, 469, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Mattei, S.; Glass, B.; Hagen, W.J.; Kräusslich, H.G.; Briggs, J.A. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef]
- Highland, C.M.; Tan, A.; Ricaña, C.L.; Briggs, J.A.G.; Dick, R.A. Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM. Proc. Natl. Acad. Sci. USA 2023, 120, e2220545120. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Alam, S.L.; Fricke, T.; Zadrozny, K.; Sedzicki, J.; Taylor, A.B.; Demeler, B.; Pornillos, O.; Ganser-Pornillos, B.K.; Diaz-Griffero, F.; et al. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. USA 2014, 111, 18625–18630. [Google Scholar] [CrossRef] [PubMed]
- Price AJJacques, D.A.; McEwan, W.A.; Fletcher, A.J.; Essig, S.; Chin, J.W.; Halambage, U.D.; Aiken, C.; James, L.C. Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS Pathog. 2014, 10, e1004459. [Google Scholar]
- Blair, W.S.; Pickford, C.; Irving, S.L.; Brown, D.G.; Anderson, M.; Bazin, R.; Cao, J.; Ciaramella, G.; Isaacson, J.; Jackson, L.; et al. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 2010, 6, e1001220. [Google Scholar] [CrossRef]
- Yant, S.R.; Mulato, A.; Hansen, D.; Tse, W.C.; Niedziela-Majka, A.; Zhang, J.R.; Stepan, G.J.; Jin, D.; Wong, M.H.; Perreira, J.M.; et al. A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model. Nat. Med. 2019, 25, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.; Teplyakov, A. MOLREP: An automated program for molecular replacement. J. Appl. Cryst. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- von Schwedler, U.K.; Stray, K.M.; Garrus, J.E.; Sundquist, W.I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 2003, 77, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Rihn, S.J.; Wilson, S.J.; Loman, N.J.; Alim, M.; Bakker, S.E.; Bhella, D.; Gifford, R.J.; Rixon, F.J.; Bieniasz, P.D. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog. 2013, 9, e1003461. [Google Scholar] [CrossRef] [PubMed]
- Jacques, D.A.; McEwan, W.A.; Hilditch, L.; Price, A.J.; Towers, G.J.; James, L.C. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 2016, 536, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Caspar, D.L.; Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Salunke, D.M.; Caspar, D.L.; Garcea, R.L. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys. J. 1989, 56, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Mattei, S.; Tan, A.; Glass, B.; Müller, B.; Kräusslich, H.-G.; Briggs, J.A.G. High-resolution structures of HIV-1 Gag cleavage mutants determine structural switch for virus maturation. Proc. Natl. Acad. Sci. USA 2018, 115, E9401–E9410. [Google Scholar] [CrossRef]
- Qu, K.; Glass, B.; Doležal, M.; Schur, F.K.M.; Murciano, B.; Rein, A.; Rumlová, M.; Ruml, T.; Kräusslich, H.-G.; John, A.G.; et al. Structure and architecture of immature and mature murine leukemia virus capsids. Proc. Natl. Acad. Sci. USA 2018, 115, E11751–E11760. [Google Scholar] [CrossRef]
- Renner, N.; Kleinpeter, A.; Mallery, D.L.; Albecka, A.; Rifat Faysal, K.M.; Böcking, T.; Saiardi, A.; Freed, E.O.; James, L.C. HIV-1 is dependent on its immature lattice to recruit IP6 for mature capsid assembly. Nat. Struct. Mol. Biol. 2023, 30, 370–382. [Google Scholar] [CrossRef]
- Guedán, A.; Donaldson, C.D.; Caroe, E.R.; Cosnefroy, O.; Taylor, I.A.; Bishop, K.N. HIV-1 requires capsid remodelling at the nuclear pore for nuclear entry and integration. PLoS Pathog. 2021, 17, e1009484. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.C.; Cereseto, A.; Singh, P.K.; Shi, J.; Poeschla, E.; Engelman, A.N.; Aiken, C.; Melikyan, G.B. Localization and functions of native and eGFP-tagged capsid proteins in HIV-1 particles. PLoS Pathog. 2022, 18, e1010754. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, R.; Ribbeck, K.; Akin, D.; Kent, H.M.; Feldherr, C.M.; Gorlich, D.; Stewart, M. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 1999, 293, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.; Görlich, D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 2007, 130, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Matsui, M.; Handa, H. SV40 virus-like particles as an effective delivery system and its application to a vaccine carrier. Expert. Rev. Vaccines 2013, 12, 199–210. [Google Scholar] [CrossRef]
- Xu, C.; Zhu, W.; Mao, H.; Zhang, W.; Yin, G.-Q.; Zhang, X.-E.; Li, F. Switch from polymorphic to homogenous self-assembly of virus-like particles of simian virus 40 through double-cysteine substitution. Small 2020, 16, e2004484. [Google Scholar] [CrossRef]


| R18L | R18L | R18L | R18L | R18A | R18G | R18L High-Salt + FG | R18L Low-Salt + FG | R18L Low-Salt + FG | |
|---|---|---|---|---|---|---|---|---|---|
| EMDB | EMD-43641 | EMD-43642 | EMD-43632 | EMD-43633 | EMD-43634 | EMD-43635 | EMD-43636 | EMD-43637 | EMD-43638 |
| Focus region | Hexamer | Pentamer | Tetramer of pentamers | Single pentamer | Tetramer of pentamers | Tetramer of pentamers | Tetramer of pentamers | Tetramer of pentamers | Octahed-ron |
| Magnification | 64,000× | 64,000× | 53,000× | 53,000× | 81,000× | 36,000× | 36,000× | 81,000× | 81,000× |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 200 | 200 | 300 | 300 |
| Exposure (e−/Å2) | 50 | 50 | 55 | 55 | 50 | 50 | 45 | 50 | 50 |
| Defocus range (μm) | 1.0 to 2.5 | 1.0 to 2.5 | 1.0 to 2.5 | 1.0 to 2.5 | 1.0 to 2.5 | 1.0 to 2.5 | 1.0 to 2.5 | 1.0 to 2.5 | 1.0 to 2.5 |
| Pixel size (Å) | 0.686 * | 0.686 * | 1.62 | 1.62 | 1.08 | 1.20 | 1.20 | 1.08 | 1.08 |
| Symmetry imposed | C6 | C1 | C4 | C5 | C4 | C4 | C4 | C4 | O |
| Particles | 737,346 | 282,138 | 106,564 | 18,643 | 43,954 | 73,987 | 172,654 | 109,290 | 130,325 |
| Map resolution ** | 3.7 | 4.4 | 5.8 | 6.6 | 6.0 | 7.1 | 5.3 | 5.4 | 4.0 |
| Map resolution range | 2.3–5.4 | 3.0–7.7 | 3.5–75 | 3.9–78 | 4.7–67 | 5.2–64 | 3.2–44 | 3.4–13 | 2.4–8.8 |
| R18L Hexamer | R18L Pentamer | |
|---|---|---|
| EMDB | EMD-43641 | EMD-43642 |
| PDB | 8vxv | 8vxw |
| Model resolution at FSC 0.143/0.5 threshold (Å) * | 3.6/4.1 | 4.3/6.0 |
| Map-sharpening B-factor (Å2) | 250 | 272 |
| No. of nonhydrogen atoms | 1524 | 5020 |
| No. of protein residues | 201 | 1015 |
| Average B-factors (Å2) | 106.4 | 150.4 |
| Rmsd bond lengths (Å) | 0.004 | 0.002 |
| Rmsd bond angles (°) | 0.664 | 0.583 |
| MolProbity score | 1.40 | 0.55 |
| Clash score | 6.97 | 0.14 |
| Poor rotamers (%) | 0 | 0 |
| Ramachandran favored (%) | 98.0 | 99.2 |
| Ramachandran outliers (%) | 0 | 0 |
| Ramachandran Z-score | 1.04 | 2.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schirra, R.T.; dos Santos, N.F.B.; Ganser-Pornillos, B.K.; Pornillos, O. Arg18 Substitutions Reveal the Capacity of the HIV-1 Capsid Protein for Non-Fullerene Assembly. Viruses 2024, 16, 1038. https://doi.org/10.3390/v16071038
Schirra RT, dos Santos NFB, Ganser-Pornillos BK, Pornillos O. Arg18 Substitutions Reveal the Capacity of the HIV-1 Capsid Protein for Non-Fullerene Assembly. Viruses. 2024; 16(7):1038. https://doi.org/10.3390/v16071038
Chicago/Turabian StyleSchirra, Randall T., Nayara F. B. dos Santos, Barbie K. Ganser-Pornillos, and Owen Pornillos. 2024. "Arg18 Substitutions Reveal the Capacity of the HIV-1 Capsid Protein for Non-Fullerene Assembly" Viruses 16, no. 7: 1038. https://doi.org/10.3390/v16071038
APA StyleSchirra, R. T., dos Santos, N. F. B., Ganser-Pornillos, B. K., & Pornillos, O. (2024). Arg18 Substitutions Reveal the Capacity of the HIV-1 Capsid Protein for Non-Fullerene Assembly. Viruses, 16(7), 1038. https://doi.org/10.3390/v16071038






