Viromic and Metagenomic Analyses of Commercial Spirulina Fermentations Reveal Remarkable Microbial Diversity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Processing
2.2. Metagenome DNA Extraction and Analysis
2.3. Virome DNA Extraction and Analysis
3. Results and Discussion
3.1. Metagenome Sequencing
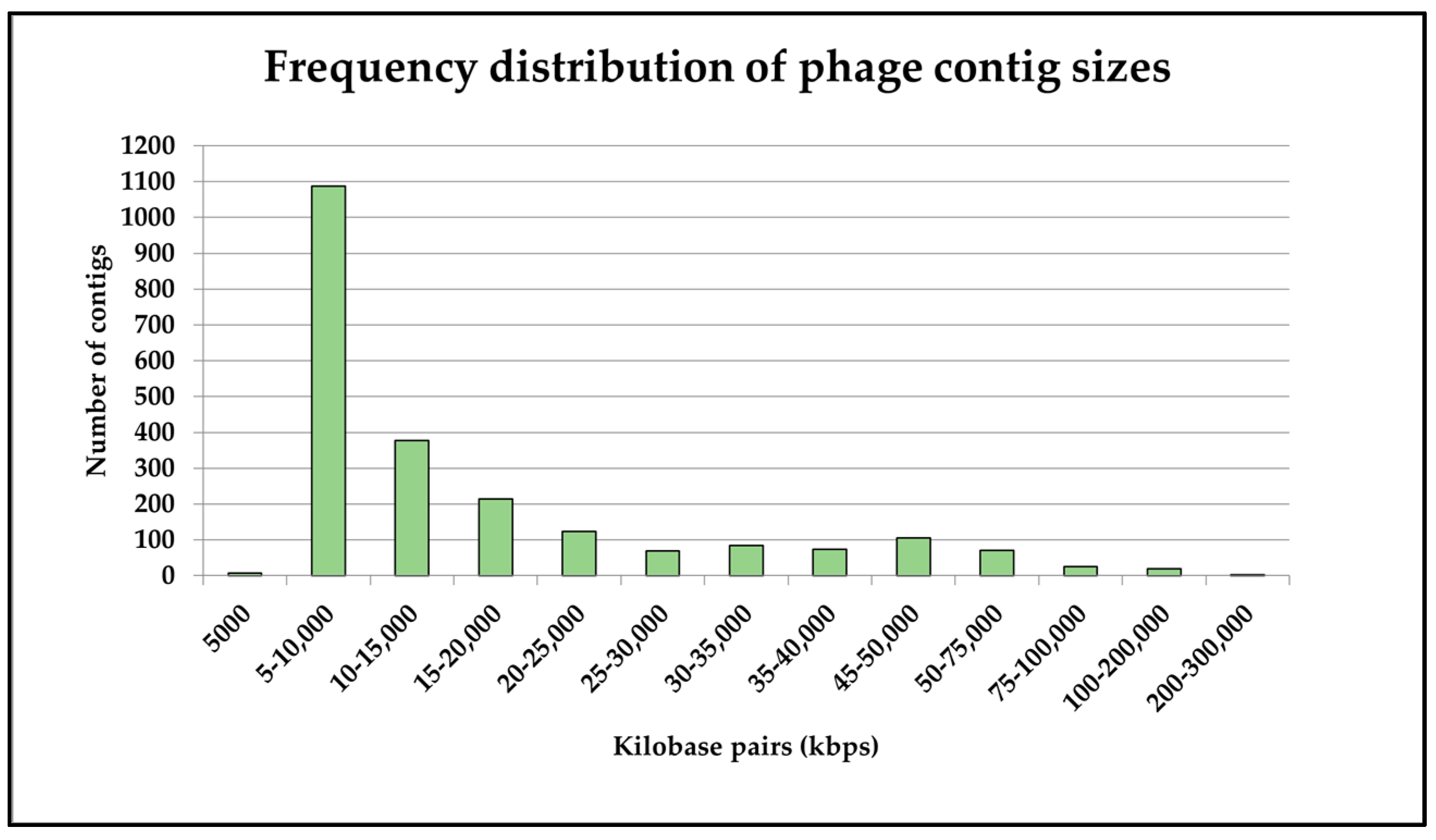
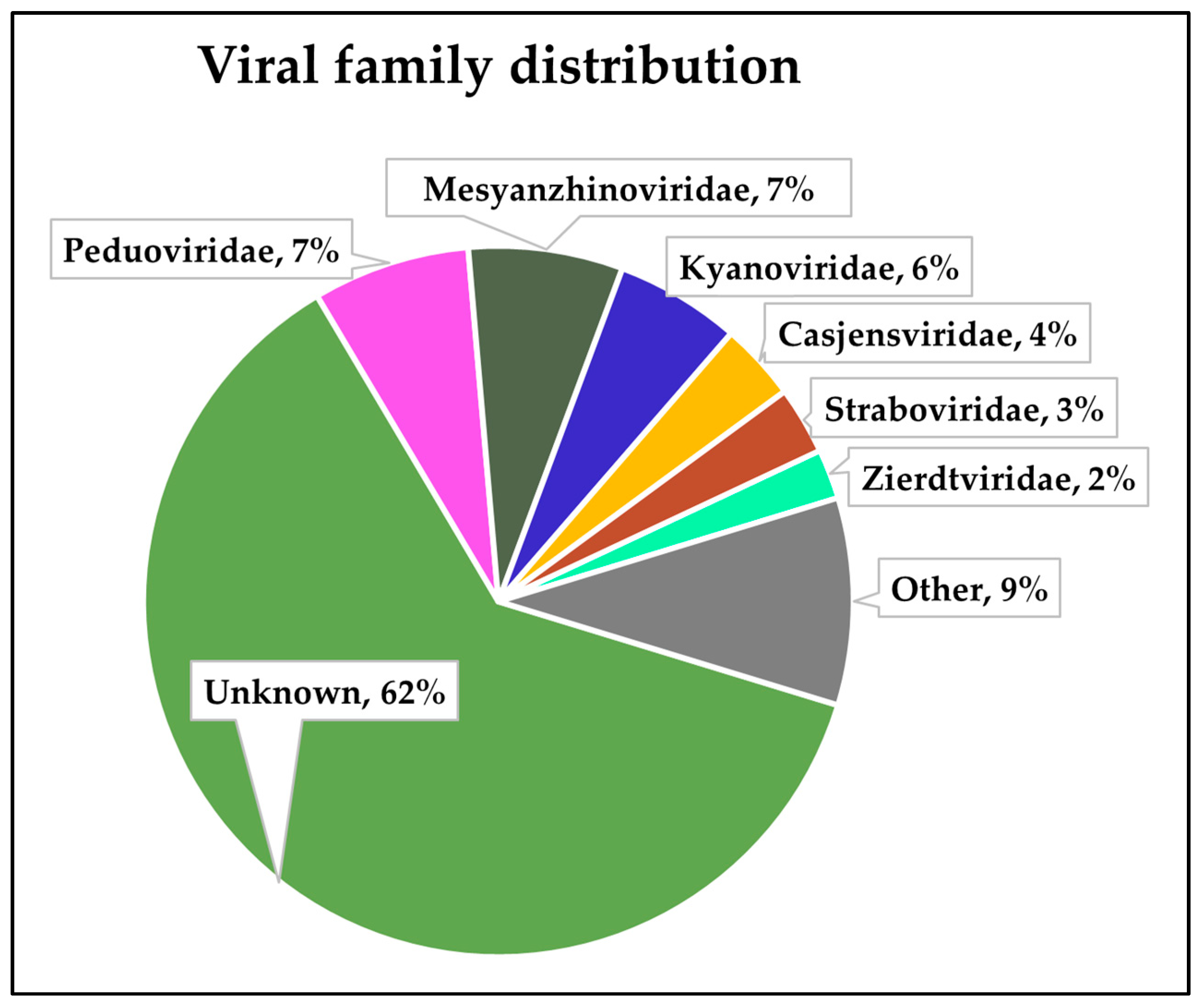
3.2. Virome Sequencing
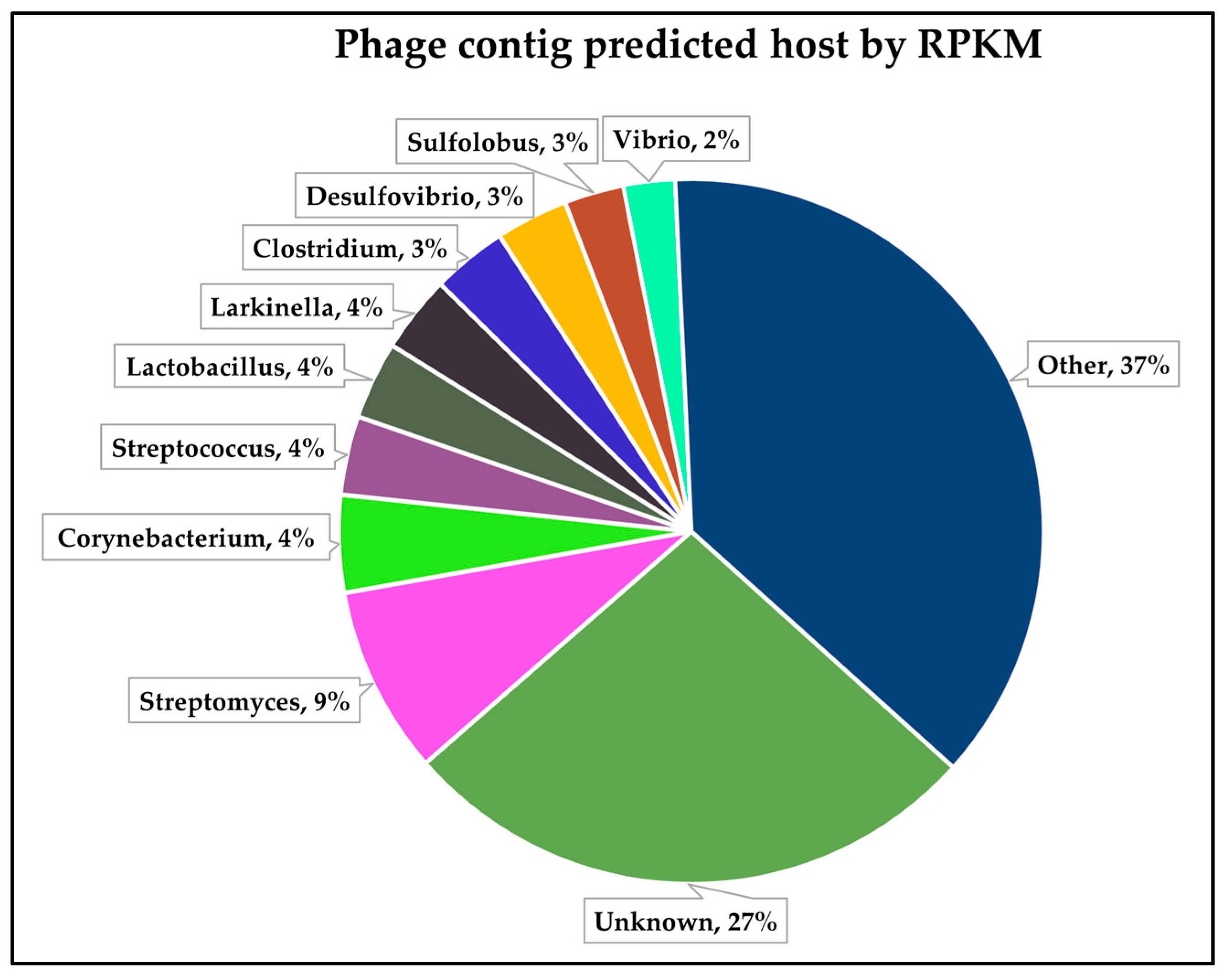
3.3. Identification and Analysis of Putative Limnospira-Infecting Phages
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ciferri, O. Spirulina, the Edible Microorganism. Microbiol. Rev. 1983, 47, 551–578. [Google Scholar] [CrossRef] [PubMed]
- Soni, R.A.; Sudhakar, K.; Rana, R.S. Spirulina—From Growth to Nutritional Product: A Review. Trends Food Sci. Technol. 2017, 69, 157–171. [Google Scholar] [CrossRef]
- Nedeva, R.; Jordanova, G.; Kistanova, E.; Shumkov, K.; Georgiev, B.; Abadgieva, D.; Kacheva, D.; Shimkus, A.; Shimkine, A. Effect of the Addition of Spirulina Platensis on the Productivity and Some Blood Parameters on Growing Pigs. Bulg. J. Agric. Sci. 2014, 20, 680–684. [Google Scholar]
- Grosshagauer, S.; Kraemer, K.; Somoza, V. The True Value of Spirulina. J. Agric. Food Chem. 2020, 68, 4109–4115. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Bhadouria, P.; Bisen, P.S. Nutritional and Therapeutic Potential of Spirulina. Curr. Pharm. Biotechnol. 2005, 6, 373–379. [Google Scholar] [CrossRef] [PubMed]
- AlFadhly, N.K.Z.; Alhelfi, N.; Altemimi, A.B.; Verma, D.K.; Cacciola, F. Tendencies Affecting the Growth and Cultivation of Genus Spirulina: An Investigative Review on Current Trends. Plants 2022, 11, 3063. [Google Scholar] [CrossRef]
- Vardaka, E.; Kormas, K.A.; Katsiapi, M.; Genitsaris, S.; Moustaka-Gouni, M. Molecular Diversity of Bacteria in Commercially Available “Spirulina” Food Supplements. PeerJ 2016, 4, e1610. [Google Scholar] [CrossRef] [PubMed]
- Toyomizu, M.; Sato, K.; Taroda, H.; Kato, T.; Akiba, Y. Effects of Dietary Spirulina on Meat Colour in Muscle of Broiler Chickens. Br. Poult. Sci. 2001, 42, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Ross, E.; Dominy, W. The Nutritional Value of Dehydrated, Blue-Green Algae (Spirulina plantensis) for Poultry. Poult. Sci. 1990, 69, 794–800. [Google Scholar] [CrossRef]
- Peiretti, P.G.; Meineri, G. Effects of Diets with Increasing Levels of Spirulina platensis on the Performance and Apparent Digestibility in Growing Rabbits. Livest. Sci. 2008, 118, 173–177. [Google Scholar] [CrossRef]
- Holman, B.W.B.; Malau-Aduli, A.E.O. Spirulina as a Livestock Supplement and Animal Feed. J. Anim. Physiol. Anim. Nutr. 2013, 97, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Santiago, C.B.; Pantastico, J.B.; Baldia, S.F.; Reyes, O.S. Milkfish (Chanos chanos) Fingerling Production in Freshwater Ponds with the Use of Natural and Artificial Feeds. Aquaculture 1989, 77, 307–318. [Google Scholar] [CrossRef]
- Tayag, C.M.; Lin, Y.-C.; Li, C.-C.; Liou, C.-H.; Chen, J.-C. Administration of the Hot-Water Extract of Spirulina Platensis Enhanced the Immune Response of White Shrimp Litopenaeus Vannamei and Its Resistance against Vibrio Alginolyticus. Fish Shellfish. Immunol. 2010, 28, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, M.G.; Umino, T.; Nakagawa, H. The Effect of Spirulina Feeding on Muscle Protein Deposition in Red Sea Bream, Pagrus major. J. Appl. Ichthyol. 1994, 10, 141–145. [Google Scholar] [CrossRef]
- Ragaza, J.A.; Hossain, M.S.; Meiler, K.A.; Velasquez, S.F.; Kumar, V. A Review on Spirulina: Alternative Media for Cultivation and Nutritive Value as an Aquafeed. Rev. Aquac. 2020, 12, 2371–2395. [Google Scholar] [CrossRef]
- DiNicolantonio, J.J.; Bhat, A.G.; OKeefe, J. Effects of Spirulina on Weight Loss and Blood Lipids: A Review. Open Heart 2020, 7, e001003. [Google Scholar] [CrossRef] [PubMed]
- McHenry, M.S.; Dixit, A.; Vreeman, R.C. A Systematic Review of Nutritional Supplementation in HIV-Infected Children in Resource-Limited Settings. J. Int. Assoc. Provid. AIDS Care (JIAPAC) 2014, 14, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Grobler, L.; Siegfried, N.; Visser, M.E.; Mahlungulu, S.S.N.; Volmink, J. Nutritional Interventions for Reducing Morbidity and Mortality in People with HIV. Cochrane Database Syst. Rev. 2013, 2, CD004536. [Google Scholar] [CrossRef] [PubMed]
- Tallec, J.; Vandermies, M.; Coene, C.; Lamaze-Lefebvre, B.; Demey, D.; Frappart, M.; Couallier, E. Implementation of an Automated Process for Limnospira Indica Harvesting and Culture Medium Recycling for Space Applications. Front. Astron. Space Sci. 2023, 10, 1229043. [Google Scholar] [CrossRef]
- Carcea, M.; Sorto, M.; Batello, C.; Narducci, V.; Aguzzi, A.; Azzini, E.; Fantauzzi, P.; Finotti, E.; Gabrielli, P.; Galli, V.; et al. Nutritional Characterization of Traditional and Improved Dihé, Alimentary Blue-Green Algae from the Lake Chad Region in Africa. LWT-Food Sci. Technol. 2015, 62, 753–763. [Google Scholar] [CrossRef]
- Lu, Y.-M.; Xiang, W.-Z.; Wen, Y.-H. Spirulina (Arthrospira) Industry in Inner Mongolia of China: Current Status and Prospects. J. Appl. Phycol. 2011, 23, 265–269. [Google Scholar] [CrossRef]
- Shimamatsu, H. Mass Production of Spirulina, an Edible Microalga. Hydrobiologia 2004, 512, 39–44. [Google Scholar] [CrossRef]
- Peduzzi, P.; Gruber, M.; Gruber, M.; Schagerl, M. The Virus’s Tooth: Cyanophages Affect an African Flamingo Population in a Bottom-up Cascade. ISME J. 2014, 8, 1346–1351. [Google Scholar] [CrossRef]
- Watanabe, F.; Katsura, H.; Takenaka, S.; Fujita, T.; Abe, K.; Tamura, Y.; Nakatsuka, T.; Nakano, Y. Pseudovitamin B12 Is the Predominant Cobamide of an Algal Health Food, Spirulina Tablets. J. Agric. Food Chem. 1999, 47, 4736–4741. [Google Scholar] [CrossRef] [PubMed]
- Muys, M.; Sui, Y.; Schwaiger, B.; Lesueur, C.; Vandenheuvel, D.; Vermeir, P.; Vlaeminck, S.E. High Variability in Nutritional Value and Safety of Commercially Available Chlorella and Spirulina Biomass Indicates the Need for Smart Production Strategies. Bioresour. Technol. 2019, 275, 247–257. [Google Scholar] [CrossRef]
- Gilroy, D.J.; Kauffman, K.W.; Hall, R.A.; Huang, X.; Chu, F.S. Assessing Potential Health Risks from Microcystin Toxins in Blue-Green Algae Dietary Supplements. Environ. Health Perspect. 2000, 108, 435–439. [Google Scholar] [CrossRef]
- Heussner, A.H.; Mazija, L.; Fastner, J.; Dietrich, D.R. Toxin Content and Cytotoxicity of Algal Dietary Supplements. Toxicol. Appl. Pharmacol. 2012, 265, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Nowicka-Krawczyk, P.; Mühlsteinová, R.; Hauer, T. Detailed Characterization of the Arthrospira Type Species Separating Commercially Grown Taxa into the New Genus Limnospira (Cyanobacteria). Sci. Rep. 2019, 9, 694. [Google Scholar] [CrossRef] [PubMed]
- Maghembe, R.; Michael, A.; Harish, A.; Nyandoro, S.S.; Lyantagaye, S.L.; Hati-Kaul, R. Draft Genome Sequence of Limnospira sp. Strain BM01, Isolated from a Hypersaline Lake of the Momela Ecosystem in Tanzania. Microbiol. Resour. Announc. 2021, 10, e00132-21. [Google Scholar] [CrossRef]
- Hicks, M.; Tran-Dao, T.-K.; Mulroney, L.; Bernick, D.L. De-Novo Assembly of Limnospira Fusiformis Using Ultra-Long Reads. Front. Microbiol. 2021, 12, 657995. [Google Scholar] [CrossRef]
- Misztak, A.E.; Waleron, M.; Furmaniak, M.; Waleron, M.M.; Bazhenova, O.; Daroch, M.; Waleron, K.F. Comparative Genomics and Physiological Investigation of a New Arthrospira/Limnospira Strain O9.13F Isolated from an Alkaline, Winter Freezing, Siberian Lake. Cells 2021, 10, 3411. [Google Scholar] [CrossRef] [PubMed]
- Roussel, T.; Halary, S.; Duval, C.; Piquet, B.; Cadoret, J.-P.; Vernès, L.; Bernard, C.; Marie, B. Monospecific Renaming within the Cyanobacterial Genus Limnospira (Spirulina) and Consequences for Food Authorization. J. Appl. Microbiol. 2023, 134, lxad159. [Google Scholar] [CrossRef] [PubMed]
- de S. Santos, K.R.; Hentschke, G.S.; Ferrari, G.; Andreote, A.P.D.; de F. Fiore, M.; Vasconcelos, V.; Sant’Anna, C.L. Molecular, Morphological and Ecological Studies of Limnospira Platensis (Cyanobacteria), from Saline and Alkaline Lakes, Pantanal Biome, Braz. Front. Environ. Sci. 2023, 11, 1204787. [Google Scholar] [CrossRef]
- Jacquet, S.; Zhong, X.; Parvathi, A.; Ram, A.S.P. First Description of a Cyanophage Infecting the Cyanobacterium Arthrospira Platensis (Spirulina). J. Appl. Phycol. 2013, 25, 195–203. [Google Scholar] [CrossRef]
- Zekri, M.A.; Schagerl, M.; Schweichhart, J.; Lang, I. Confocal Microscopy Reveals Alterations of Thylakoids in Limnospira Fusiformis during Prophage Induction. Protoplasma 2021, 258, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Castillo, M.; Guevara, G.; Baldanta, S.; Rodríguez, P.S.; Agudo, L.; Nogales, J.; Carrasco, A.D.; Arribas-Aguilar, F.; Pérez-Pérez, J.; García, J.L.; et al. Characterization of Limnospira Platensis PCC 9108 R-M and CRISPR-Cas Systems. Microbiol. Res. 2024, 279, 127572. [Google Scholar] [CrossRef]
- Sturino, J.M.; Klaenhammer, T.R. Engineered Bacteriophage-Defence Systems in Bioprocessing. Nat. Rev. Microbiol. 2006, 4, 395–404. [Google Scholar] [CrossRef]
- Milani, C.; Lugli, G.A.; Fontana, F.; Mancabelli, L.; Alessandri, G.; Longhi, G.; Anzalone, R.; Viappiani, A.; Turroni, F.; van Sinderen, D. METAnnotatorX2: A Comprehensive Tool for Deep and Shallow Metagenomic Data Set Analyses. Msystems 2021, 6, e00583-21. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Kleiner, M.; Hooper, L.V.; Duerkop, B.A. Evaluation of Methods to Purify Virus-like Particles for Metagenomic Sequencing of Intestinal Viromes. BMC Genom. 2015, 16, 7. [Google Scholar] [CrossRef]
- Milani, C.; Casey, E.; Lugli, G.A.; Moore, R.; Kaczorowska, J.; Feehily, C.; Mangifesta, M.; Mancabelli, L.; Duranti, S.; Turroni, F.; et al. Tracing Mother-Infant Transmission of Bacteriophages by Means of a Novel Analytical Tool for Shotgun Metagenomic Datasets: METAnnotatorX. Microbiome 2018, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1989; ISBN 0-87969-309-6. [Google Scholar]
- Shang, J.; Peng, C.; Liao, H.; Tang, X.; Sun, Y. PhaBOX: A Web Server for Identifying and Characterizing Phage Contigs in Metagenomic Data. Bioinform. Adv. 2023, 3, vbad101. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Jiang, J.; Sun, Y. Bacteriophage Classification for Assembled Contigs Using Graph Convolutional Network. Bioinformatics 2021, 37, i25–i33. [Google Scholar] [CrossRef]
- Shang, J.; Sun, Y. CHERRY: A Computational metHod for accuratE pRediction of Virus–pRokarYotic Interactions Using a Graph Encoder–Decoder Model. Brief. Bioinform. 2022, 23, bbac182. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Tang, X.; Sun, Y. PhaTYP: Predicting the Lifestyle for Bacteriophages Using BERT. Brief. Bioinform. 2023, 24, bbac487. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-P.; Chang, H.-Y.; Chen, J.-S.; Jean, W.D.; Shieh, W.Y. Aliidiomarina taiwanensis gen. nov., sp. nov., Isolated from Shallow Coastal Water. Int. J. Syst. Evol. Microbiol. 2012, 62, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xue, Y.; Grant, W.D.; Collins, N.C.; Duckworth, A.W.; van Steenbergen, R.P.; Jones, B.E. Alkalimonas amylolytica gen. nov., sp. nov., and Aliidiomarina taiwanensis gen. nov., sp. nov., Novel Alkaliphilic Bacteria from Soda Lakes in China and East Africa. Extremophiles 2004, 8, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Takano, T.; Liu, S. Isolation and Characterization of Novel Bacterial Taxa from Extreme Alkali-Saline Soil. World J. Microbiol. Biotechnol. 2012, 28, 2147–2157. [Google Scholar] [CrossRef]
- Xu, L.; Sun, J.-Q.; Wang, L.-J.; Liu, X.-Z.; Ji, Y.-Y.; Shao, Z.-Q.; Wu, X.-L. Aliidiomarina soli sp. nov., Isolated from Saline–Alkaline Soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 724–728. [Google Scholar] [CrossRef]
- Borsodi, A.K.; Knáb, M.; Czeibert, K.; Márialigeti, K.; Vörös, L.; Somogyi, B. Planktonic Bacterial Community Composition of an Extremely Shallow Soda Pond during a Phytoplankton Bloom Revealed by Cultivation and Molecular Cloning. Extremophiles 2013, 17, 575–584. [Google Scholar] [CrossRef]
- Yang, M.; Xue, Q.; Zuo, Z.; Zhou, J.; Zhang, S.; Li, M.; Zhou, H.; Zhang, M.; Kumar, S.; Li, W.; et al. Aliidiomarina halalkaliphila sp. nov., a Haloalkaliphilic Bacterium Isolated from a Soda Lake in Inner Mongolia Autonomous Region, China. Int. J. Syst. Evol. Microbiol. 2022, 72, 005263. [Google Scholar] [CrossRef] [PubMed]
- Kurata, A.; Uchimura, K.; Kobayashi, T.; Horikoshi, K. Collagenolytic Subtilisin-like Protease from the Deep-Sea Bacterium Alkalimonas Collagenimarina AC40T. Appl. Microbiol. Biotechnol. 2010, 86, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Liu, H.; Huang, Y.; Dai, X.; Zhou, Y. Aliidiomarina indica sp. nov., Isolated from Deep Seawater. Int. J. Syst. Evol. Microbiol. 2021, 71, 005122. [Google Scholar] [CrossRef] [PubMed]
- Duchaud, E.; Boussaha, M.; Loux, V.; Bernardet, J.-F.; Michel, C.; Kerouault, B.; Mondot, S.; Nicolas, P.; Bossy, R.; Caron, C.; et al. Complete Genome Sequence of the Fish Pathogen Flavobacterium Psychrophilum. Nat. Biotechnol. 2007, 25, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Bernardet, J.-F.; Bowman, J.P. The Genus Flavobacterium. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 481–531. ISBN 978-0-387-25497-5. [Google Scholar]
- Abraham, W.-R.; Lünsdorf, H.; Vancanneyt, M.; Smit, J. Cauliform Bacteria Lacking Phospholipids from an Abyssal Hydrothermal Vent: Proposal of Glycocaulis abyssi gen. nov., sp. nov., Belonging to the Family Hyphomonadaceae. Int. J. Syst. Evol. Microbiol. 2013, 63, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.; Pan, X.-C.; Mei, R.; Wang, Y.-N.; Liu, X.-Y.; Wang, X.-B.; Tang, Y.-Q.; Wu, X.-L. Glycocaulisalkaliphilus sp. nov., a Dimorphic Prosthecate Bacterium Isolated from Crude Oil. Int. J. Syst. Evol. Microbiol. 2015, 65, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Hu, Y.; Zhou, S.; Zheng, Y.; Zhang, X.-H. Glycocaulis profundi sp. nov., a Marine Bacterium Isolated from Seawater of the Mariana Trench. Int. J. Syst. Evol. Microbiol. 2020, 70, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.I.; Turova, T.; Kuznetsov, B.; Briantseva, I.; Gorlenko, V. Roseinatronobacter thiooxidans gen. nov., sp. nov., a New Alkaliphilic Aerobic Bacteriochlorophyll-Alpha-Containing Bacteria from a Soda Lake. Mikrobiologiia 2000, 69, 89–97. [Google Scholar] [PubMed]
- Boldareva, E.; Bryantseva, I.; Tsapin, A.; Nelson, K.; Sorokin, D.Y.; Tourova, T.; Boichenko, V.; Stadnichuk, I.; Gorlenko, V. The New Alkaliphilic Bacteriochlorophyll A-Containing Bacterium Roseinatronobacter monicus sp. nov. from the Hypersaline Soda Mono Lake (California, United States). Microbiology 2007, 76, 82–92. [Google Scholar] [CrossRef]
- Trutschel, L.R.; Rowe, A.R.; Sackett, J.D. Complete Genome Sequence of Roseinatronobacter sp. Strain S2, a Chemolithoheterotroph Isolated from a pH 12 Serpentinizing System. Microbiol. Resour. Announc. 2023, 12, e00288-23. [Google Scholar] [CrossRef]
- Pálmai, T.; Szabó, B.; Lengyel, E.; Kotut, K.; Krienitz, L.; Padisák, J. Growth Response of the Picoplanktic Picocystis salinarum and the Microplanktic Limnospira (Arthrospira) fusiformis Strains from Lake Nakuru (Kenya) to Rapidly Changing Environmental Conditions. Hydrobiologia 2023, 851, 1873–1889. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Clooney, A.G.; Sutton, T.D.; Ryan, F.J.; Daly, K.M.; Nolan, J.A.; McDonnell, S.A.; Khokhlova, E.V.; Draper, L.A.; Forde, A. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe 2019, 26, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.C.; Zablocki, O.; Zayed, A.A.; Howell, A.; Bolduc, B.; Sullivan, M.B. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe 2020, 28, 724–740. [Google Scholar] [CrossRef] [PubMed]
- Szafrański, S.P.; Slots, J.; Stiesch, M. The Human Oral Phageome. Periodontology 2000 2021, 86, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, Y.; Zeng, Z.; Wang, Z. Metagenomics of Wastewater Phageome Identifies an Extensively Cored Antibiotic Resistome in a Swine Feedlot Water Treatment Environment. Ecotoxicol. Environ. Saf. 2021, 222, 112552. [Google Scholar] [CrossRef] [PubMed]
- Verbanic, S.; Deacon, J.M.; Chen, I.A. The Chronic Wound Phageome: Phage Diversity and Associations with Wounds and Healing Outcomes. Microbiol. Spectr. 2022, 10, e02777-21. [Google Scholar] [CrossRef] [PubMed]
- Jurasz, H.; Pawłowski, T.; Perlejewski, K. Contamination Issue in Viral Metagenomics: Problems, Solutions, and Clinical Perspectives. Front. Microbiol. 2021, 12, 745076. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of Morphology-Based Taxa and Change to Binomial Species Names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Handley, P.; Coykendall, A.; Beighton, D.; Hardie, J.M.; Whiley, R.A. Streptococcus Crista Sp. Nov., a Viridans Streptococcus with Tufted Fibrils, Isolated from the Human Oral Cavity and Throat. Int. J. Syst. Evol. Microbiol. 1991, 41, 543–547. [Google Scholar] [CrossRef]
- Villarroel, J.; Kleinheinz, K.A.; Jurtz, V.I.; Zschach, H.; Lund, O.; Nielsen, M.; Larsen, M.V. HostPhinder: A Phage Host Prediction Tool. Viruses 2016, 8, 116. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, X.; Wang, J.; Li, S.C. DeepHost: Phage Host Prediction with Convolutional Neural Network. Brief. Bioinform. 2022, 23, bbab385. [Google Scholar]
- Zielezinski, A.; Barylski, J.; Karlowski, W.M. Taxonomy-Aware, Sequence Similarity Ranking Reliably Predicts Phage–Host Relationships. BMC Biol. 2021, 19, 223. [Google Scholar] [CrossRef]
- Tang, T.; Hou, S.; Fuhrman, J.A.; Sun, F. Phage–Bacterial Contig Association Prediction with a Convolutional Neural Network. Bioinformatics 2022, 38, i45–i52. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Sun, Y. Predicting the Hosts of Prokaryotic Viruses Using GCN-Based Semi-Supervised Learning. BMC Biol. 2021, 19, 250. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.H.; Yang, S.; Liu, Z.; Zhang, Y.; Wang, X.; Xu, Z.; Wang, J.; Li, S.C. PhageScope: A Well-Annotated Bacteriophage Database with Automatic Analyses and Visualizations. Nucleic Acids Res. 2024, 52, D756–D761. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, Q.; Li, M.; Xu, J.; Wang, C.; Zhang, J.; Xiao, M.; Bin, Y.; Xia, J. PhaGAA: An Integrated Web Server Platform for Phage Genome Annotation and Analysis. Bioinformatics 2023, 39, btad120. [Google Scholar] [CrossRef] [PubMed]
- Amgarten, D.; Iha, B.K.V.; Piroupo, C.M.; da Silva, A.M.; Setubal, J.C. vHULK, a New Tool for Bacteriophage Host Prediction Based on Annotated Genomic Features and Neural Networks. PHAGE 2022, 3, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Dhall, A.; Patiyal, S.; Choudhury, S.; Arora, A.; Raghava, G.P.S. An Ensemble Method for Prediction of Phage-Based Therapy against Bacterial Infections. Front. Microbiol. 2023, 14, 1148579. [Google Scholar] [CrossRef]
- Kochetkova, T.V.; Grabarnik, I.P.; Klyukina, A.A.; Zayulina, K.S.; Elizarov, I.M.; Shestakova, O.O.; Gavirova, L.A.; Malysheva, A.D.; Shcherbakova, P.A.; Barkhutova, D.D.; et al. Microbial Communities of Artisanal Fermented Milk Products from Russia. Microorganisms 2022, 10, 2140. [Google Scholar] [CrossRef]
- Riquelme, C.; Câmara, S.; Enes Dapkevicius, M.d.L.N.; Vinuesa, P.; da Silva, C.C.G.; Malcata, F.X.; Rego, O.A. Characterization of the Bacterial Biodiversity in Pico Cheese (an Artisanal Azorean Food). Int. J. Food Microbiol. 2015, 192, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Nwaiwu, O.; Aduba, C.C.; Igbokwe, V.C.; Sam, C.E.; Ukwuru, M.U. Traditional and Artisanal Beverages in Nigeria: Microbial Diversity and Safety Issues. Beverages 2020, 6, 53. [Google Scholar] [CrossRef]
- Bustamante, F.; Maury-Sintjago, E.; Leal, F.C.; Acuña, S.; Aguirre, J.; Troncoso, M.; Figueroa, G.; Parra-Flores, J. Presence of Listeria Monocytogenes in Ready-to-Eat Artisanal Chilean Foods. Microorganisms 2020, 8, 1669. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.C.; de Araújo, J.P.A.; Fusieger, A.; de Carvalho, A.F.; Nero, L.A. Microbiological Quality and Safety of Brazilian Artisanal Cheeses. Braz. J. Microbiol. 2021, 52, 393–409. [Google Scholar] [CrossRef]
- Ruta, S.; Murray, M.; Kampff, Z.; McDonnell, B.; Lugli, G.A.; Ventura, M.; Todaro, M.; Settanni, L.; van Sinderen, D.; Mahony, J. Microbial Ecology of Pecorino Siciliano PDO Cheese Production Systems. Fermentation 2023, 9, 620. [Google Scholar] [CrossRef]


| Sample Name | Date of Sampling | Sequenced Reads | Filtered Reads | Classified Reads | Limnospira spp. Reads (Metagenome) | as % of Total Metagenome Reads | |
|---|---|---|---|---|---|---|---|
| 2b 1 | August 2018 | 56,581 | 39,990 | Virome | 7842 | - | - |
| S2B1 (failure) | September 2021 | 191,356 | 75,833 | Virome | 7453 | ||
| S2B2 (failure) | September 2021 | 5,523,647 | 4,432,696 | Virome | 3575 | ||
| S2B3 (failure) | September 2021 | 3,018,468 | 2,809,813 | Virome | 3233 | ||
| S1B2-21-9 | September 2021 | 29,963 | 28,342 | Metagenome | 14,871 | 0 | 0.00 |
| 5,877,945 | 5,716,372 | Virome | 5292 | - | - | ||
| S1B2-oct | October 2021 | 25,520 | 24,044 | Metagenome | 14,671 | 3006 | 20.49 |
| 6,021,430 | 5,862,093 | Virome | 5970 | - | - | ||
| S1B2-1-22 | January 2022 | 22,502 | 21,011 | Metagenome | 10,981 | 641 | 5.84 |
| 3,894,117 | 3,814,643 | Virome | 3388 | - | - | ||
| S1B7-21-9 | September 2021 | 25,548 | 24,186 | Metagenome | 10,527 | 0 | 0.00 |
| 5,924,463 | 5,747,853 | Virome | 14,079 | - | - | ||
| S1B7-1-22 | January 2022 | 16,765 | 15,244 | Metagenome | 8291 | 335 | 4.04 |
| 4,232,472 | 4,123,599 | Virome | 2936 | - | - | ||
| S2B14-21-9 | September 2021 | 27,112 | 25,573 | Metagenome | 14,466 | 0 | 0.00 |
| 4,151,790 | 4,079,681 | Virome | 2740 | - | - | ||
| S2B14-11-21 | November 2021 | 22,998 | 20,743 | Metagenome | 10,880 | 300 | 2.76 |
| 2,582,342 | 2,520,442 | Virome | 1392 | - | - | ||
| S2B14-Dec-21 | December 2021 | 18,847 | 17,491 | Metagenome | 9978 | 304 | 3.05 |
| 4,741,802 | 4,628,992 | Virome | 1744 | - | - | ||
| S2B3-21-9 | September 2021 | 15,712 | 13,022 | Metagenome | 5815 | 0 | 0.00 |
| 4,064,953 | 3,983,395 | Virome | 2310 | - | - | ||
| S2B3-oct | October 2021 | 20,305 | 18,817 | Metagenome | 8911 | 1451 | 16.28 |
| 4,680,796 | 4,455,834 | Virome | 20,406 | - | - | ||
| S2B3-7-1-22 | January 2022 | 31,805 | 30,038 | Metagenome | 18,632 | 4477 | 24.03 |
| 4,141,294 | 4,035,443 | Virome | 6323 | - | - | ||
| S2B7-oct | October 2021 | 19,848 | 17,262 | Metagenome | 6650 | 573 | 8.62 |
| 5,461,758 | 5,294,359 | Virome | 5147 | - | - | ||
| S2C1-21-9 | September 2021 | 23,440 | 22,129 | Metagenome | 13,325 | 999 | 7.50 |
| 4,023,482 | 3,921,215 | Virome | 7782 | - | - | ||
| S2C1-oct | October 2021 | 23,308 | 20,753 | Metagenome | 8174 | 119 | 1.45 |
| 3,732,816 | 3,473,164 | Virome | 6965 | - | - | ||
| S2C1-1-22 | January 2022 | 17,192 | 15,584 | Metagenome | 6930 | 983 | 14.18 |
| 2,131,399 | 2,090,652 | Virome | 2461 | - | - | ||
| Sample | Accession | Length | PhaMer | PhaTYP | PhaGCN | CHERRY | Top Hit (Blastn) |
|---|---|---|---|---|---|---|---|
| 2b | NODE_36_length_16638_cov_4.583057 | 16,638 | phage | virulent | unknown | Limnospira indica | No hit |
| S1B2-21-9 | NODE_112_length_13274_cov_6.270061 | 13,274 | phage | virulent | unknown | Arthrospira platensis | Uncultured Caudovirales phage |
| S1B2-oct | NODE_531_length_5912_cov_158.66289 | 5912 | phage | virulent | unknown | Arthrospira platensis | No hit |
| S1B7-1-22 | NODE_323_length_5912_cov_106.02519 | 5912 | phage | virulent | unknown | Arthrospira platensis | No hit |
| s2b3-7-1-22 | NODE_396_length_8718_cov_28.387802 | 8718 | phage | virulent | unknown | Arthrospira platensis | No hit |
| s1b7-21-9 | NODE_370_length_7665_cov_20.110174 | 7665 | phage | virulent | unknown | Limnospira maxima | Arthrospira platensis C1 chromosome |
| S1B2-21-9 | NODE_240_length_5397_cov_2.393985 | 5397 | phage | virulent | unknown | Limnospira maxima | Arthrospira platensis C1 chromosome |
| s2b3-oct | NODE_485_length_7782_cov_19.334848 | 7782 | phage | virulent | unknown | Limnospira maxima | Arthrospira platensis C1 chromosome |
| s2b7-oct | NODE_238_length_7533_cov_3.470494 | 7533 | phage | virulent | unknown | Limnospira maxima | Arthrospira platensis C1 chromosome |
| S1B2-oct | NODE_439_length_7299_cov_6.697591 | 7299 | phage | temperate | unknown | Limnospira maxima | Arthrospira sp. TJSD092 chromosome |
| s1b7-21-9 | NODE_382_length_7496_cov_22.413668 | 7496 | phage | virulent | unknown | Limnospira maxima | Arthrospira platensis C1 chromosome |
| s2b3-oct | NODE_517_length_7292_cov_23.274012 | 7292 | phage | temperate | unknown | Limnospira maxima | Arthrospira platensis C1 chromosome |
| s2b3-7-1-22 | NODE_469_length_7495_cov_5.71420 | 7495 | phage | virulent | unknown | Limnospira maxima | Arthrospira sp. TJSD092 chromosome |
| s1b7-21-9 | NODE_261_length_11129_cov_16.54904 | 11,129 | phage | temperate | unknown | Arthrospira platensis | Limnospira fusiformis SAG 85.79 chromosome |
| s2b3-oct | NODE_379_length_10119_cov_16.70065 | 10,119 | phage | temperate | unknown | Arthrospira platensis | Limnospira fusiformis SAG 85.79 chromosome |
| s1b7-21-9 | NODE_461_length_6172_cov_15.65381 | 6172 | phage | virulent | unknown | Limnospira maxima | Arthrospira sp. TJSD092 chromosome |
| s2b3-7-1-22 | NODE_281_length_11272_cov_4.703082 | 11,272 | phage | virulent | unknown | Limnospira maxima | Limnospira fusiformis KN01 |
| s2b3-oct | NODE_321_length_12009_cov_19.52816 | 12,009 | phage | temperate | unknown | Limnospira maxima | Arthrospira platensis C1 chromosome |
| s2b3-oct | NODE_463_length_8258_cov_20.730962 | 8258 | phage | temperate | unknown | Limnospira maxima | Arthrospira sp. TJSD092 chromosome |
| s2b14-21-9 | NODE_28_length_38639_cov_12.619237 | 38,639 | phage | virulent | unknown | Arthrospira platensis | No hit |
| Web Server | Host Prediction Tool | Host Assignment |
|---|---|---|
| PhaBOX [43] | CHERRY [45] | Arthrospira platensis |
| PhageScope [78] | DeepHost [73] | Salmonella enterica |
| PhaGAA [79] | vHULK [80] | Escherichia coli |
| PhageTB [81] | Custom; BLAST [77] | Mycolicibacterium smegmatis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDonnell, B.; Parlindungan, E.; Vasiliauskaite, E.; Bottacini, F.; Coughlan, K.; Krishnaswami, L.P.; Sassen, T.; Lugli, G.A.; Ventura, M.; Mastroleo, F.; et al. Viromic and Metagenomic Analyses of Commercial Spirulina Fermentations Reveal Remarkable Microbial Diversity. Viruses 2024, 16, 1039. https://doi.org/10.3390/v16071039
McDonnell B, Parlindungan E, Vasiliauskaite E, Bottacini F, Coughlan K, Krishnaswami LP, Sassen T, Lugli GA, Ventura M, Mastroleo F, et al. Viromic and Metagenomic Analyses of Commercial Spirulina Fermentations Reveal Remarkable Microbial Diversity. Viruses. 2024; 16(7):1039. https://doi.org/10.3390/v16071039
Chicago/Turabian StyleMcDonnell, Brian, Elvina Parlindungan, Erika Vasiliauskaite, Francesca Bottacini, Keith Coughlan, Lakshmi Priyadarshini Krishnaswami, Tom Sassen, Gabriele Andrea Lugli, Marco Ventura, Felice Mastroleo, and et al. 2024. "Viromic and Metagenomic Analyses of Commercial Spirulina Fermentations Reveal Remarkable Microbial Diversity" Viruses 16, no. 7: 1039. https://doi.org/10.3390/v16071039






