Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences
Abstract
:1. Introduction
2. Materials and Methods
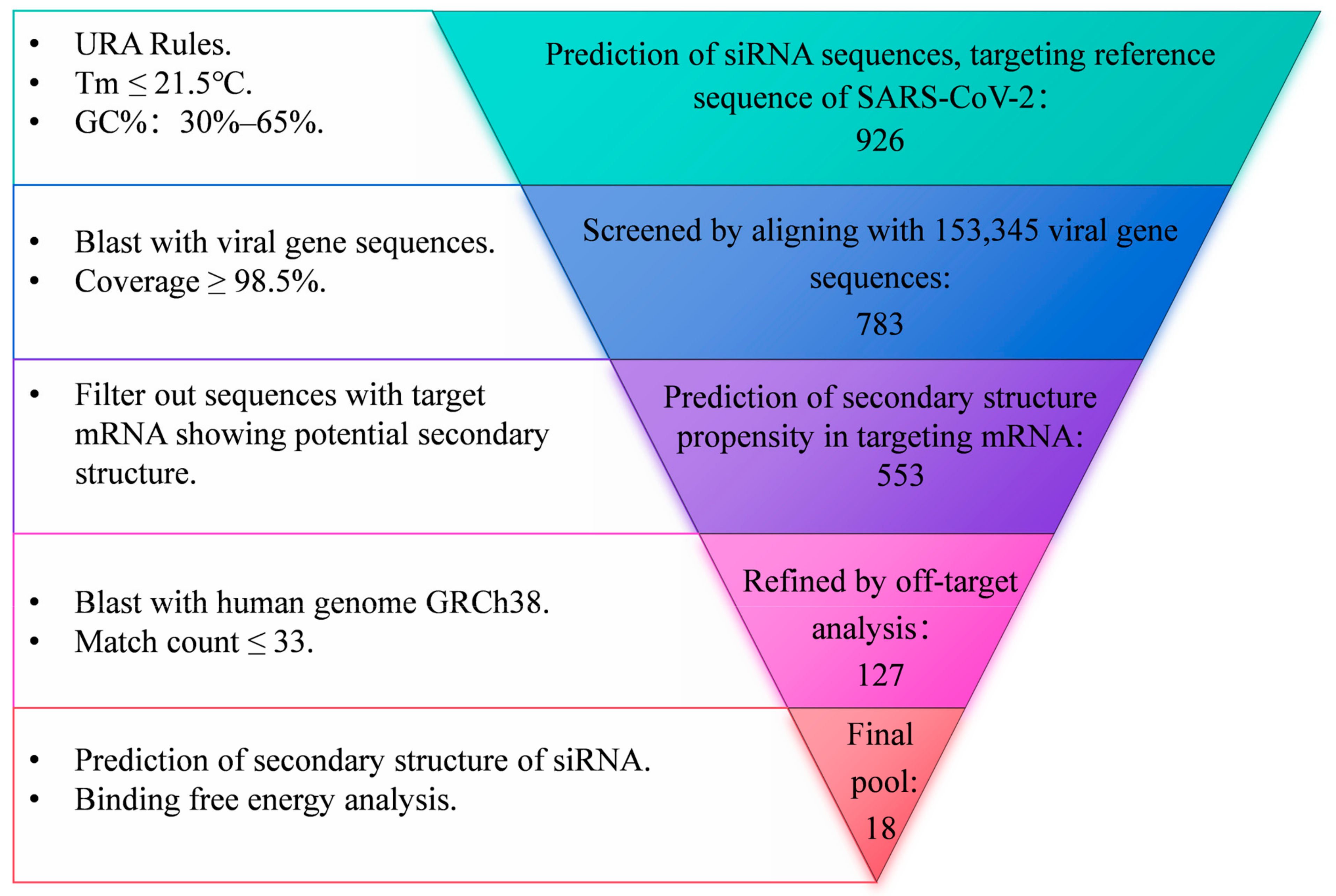
2.1. Design of siRNA Sequences
2.2. Assembly of siRNA Nanoparticles
2.3. Transmission Electron Microscopy (TEM) and Dynamic Light Scattering (DLS)
2.4. Cell Cultures
2.5. SARS-CoV-2 GFP/ΔN trVLP Production
2.6. SARS-CoV-2 GFP/ΔN trVLP Infection
2.7. Confocal Microscopy
2.8. RNA Extraction and RT-qPCR
2.9. Statistical Analysis
3. Results
3.1. Design and Verification of siRNAs Targeting Highly Conserved Regions of SARS-CoV-2
3.2. Construction and Characterization of siRNA Nanoparticles
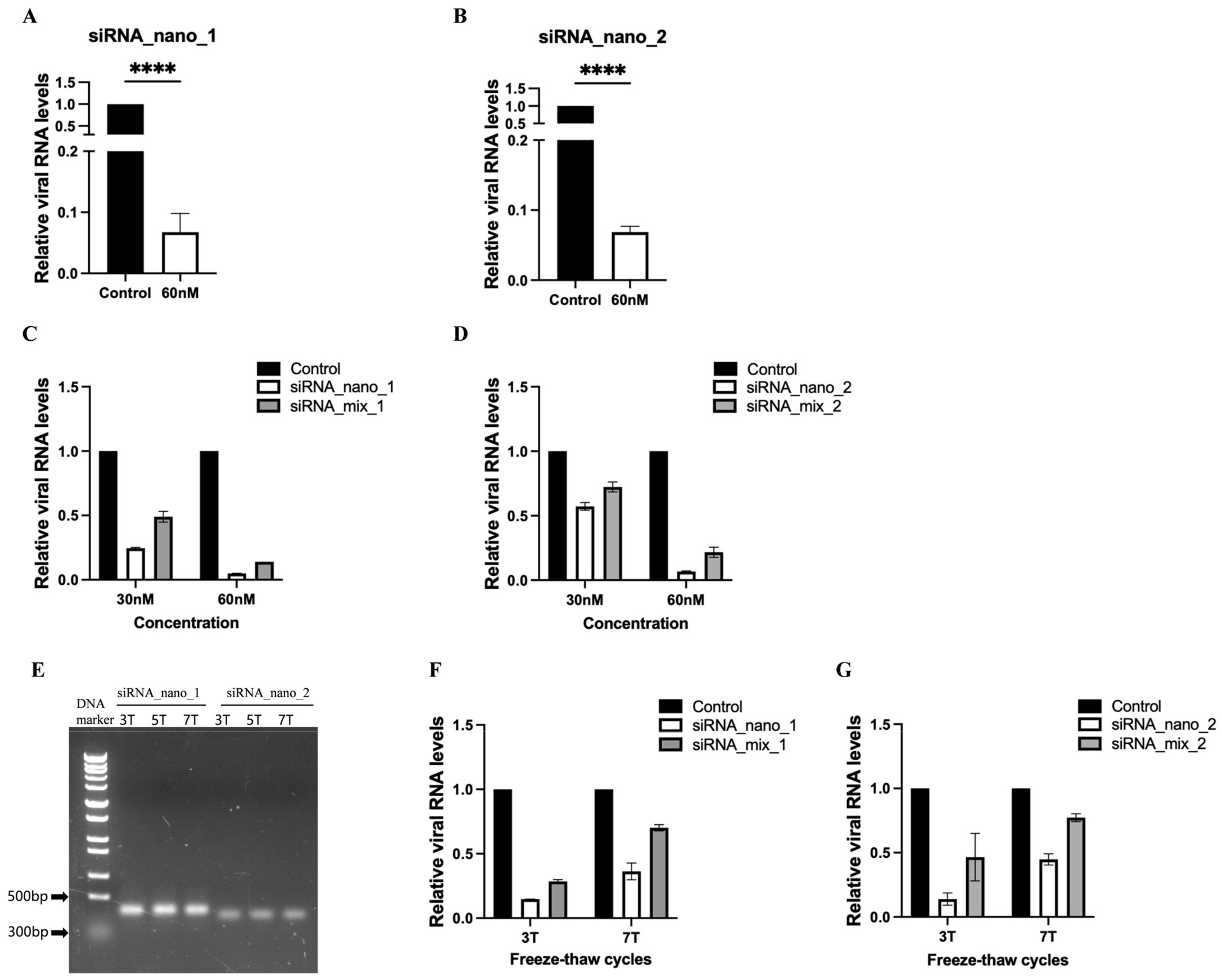
3.3. siRNA Nanoparticles Efficiently Inhibited SARS-CoV-2 Replication
3.4. siRNA Nanoparticles Enter Cells Directly through Cellular Endocytic Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chan, J.F.; Yuan, S.; Chu, H.; Sridhar, S.; Yuen, K.Y. COVID-19 drug discovery and treatment options. Nat. Rev. Microbiol. 2024, 22, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Chung, Y.S.; Jo, H.J.; Lee, N.J.; Kim, M.S.; Woo, S.H.; Park, S.; Kim, J.W.; Kim, H.M.; Han, M.G. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020, 11, 3–7. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Steiner, S.; Kratzel, A.; Barut, G.T.; Lang, R.M.; Aguiar Moreira, E.; Thomann, L.; Kelly, J.N.; Thiel, V. SARS-CoV-2 biology and host interactions. Nat. Rev. Microbiol. 2024, 22, 206–225. [Google Scholar] [CrossRef]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Aigner, A. Therapeutic siRNA: State-of-the-Art and Future Perspectives. Biodrugs 2022, 36, 549–571. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Huang, H.W.; Liu, C.Y.; Hong, C.F.; Chan, Y.L. Inhibition of SARS-CoV replication by siRNA. Antivir. Res. 2005, 65, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.; Davis, A.; Supramaniam, A.; Acharya, D.; Kelly, G.; Tayyar, Y.; West, N.; Zhang, P.; McMillan, C.L.D.; Soemardy, C.; et al. A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 2021, 29, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Tolksdorf, B.; Heinze, J.; Niemeyer, D.; Röhrs, V.; Berg, J.; Drosten, C.; Kurreck, J. Development of a highly stable, active small interfering RNA with broad activity against SARS-CoV viruses. Antivir. Res. 2024, 226, 105879. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, I.G.; Khayat, A.S.; Stransky, B.; Santos, S.; Assumpção, P.; de Souza, J.E.S. A small interfering RNA (siRNA) database for SARS-CoV-2. Sci. Rep. 2021, 11, 8849. [Google Scholar] [CrossRef] [PubMed]
- Ambike, S.; Cheng, C.C.; Feuerherd, M.; Velkov, S.; Baldassi, D.; Afridi, S.Q.; Porras-Gonzalez, D.; Wei, X.; Hagen, P.; Kneidinger, N.; et al. Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread. Nucleic Acids Res. 2021, 50, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.; Supramaniam, A.; Tayyar, Y.; Kelly, G.; McMillan, N.A.J.; Morris, K.V. An intranasally delivered ultra-conserved siRNA prophylactically represses SARS-CoV-2 infection in the lung and nasal cavity. Antivir. Res. 2024, 222, 105815. [Google Scholar] [CrossRef] [PubMed]
- Brady, D.K.; Gurijala, A.R.; Huang, L.; Hussain, A.A.; Lingan, A.L.; Pembridge, O.G.; Ratangee, B.A.; Sealy, T.T.; Vallone, K.T.; Clements, T.P. A guide to COVID-19 antiviral therapeutics: A summary and perspective of the antiviral weapons against SARS-CoV-2 infection. FEBS J. 2024, 291, 1632–1662. [Google Scholar] [CrossRef] [PubMed]
- Bowden-Reid, E.; Ledger, S.; Zhang, Y.; Di Giallonardo, F.; Aggarwal, A.; Stella, A.O.; Akerman, A.; Milogiannakis, V.; Walker, G.; Rawlinson, W.; et al. Novel siRNA therapeutics demonstrate multi-variant efficacy against SARS-CoV-2. Antivir. Res. 2023, 217, 105677. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Park, J.H.; Sailor, M.J. Rekindling RNAi Therapy: Materials Design Requirements for In Vivo siRNA Delivery. Adv. Mater. 2019, 31, e1903637. [Google Scholar] [CrossRef] [PubMed]
- Kanasty, R.; Dorkin, J.R.; Vegas, A.; Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013, 12, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Keam, S.J. Vutrisiran: First Approval. Drugs 2022, 82, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Syed, Y.Y. Nedosiran: First Approval. Drugs 2023, 83, 1729–1733. [Google Scholar] [CrossRef]
- Scott, L.J. Givosiran: First Approval. Drugs 2020, 80, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Lamb, Y.N. Inclisiran: First Approval. Drugs 2021, 81, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J.; Keam, S.J. Lumasiran: First Approval. Drugs 2021, 81, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Patisiran: First Global Approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Viard, M.; Koyfman, A.Y.; Martins, A.N.; Kasprzak, W.K.; Panigaj, M.; Desai, R.; Santhanam, A.; Grabow, W.W.; Jaeger, L.; et al. Multifunctional RNA Nanoparticles. Nano Lett. 2014, 14, 5662–5671. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Kasprzak, W.K.; Bindewald, E.; Kireeva, M.; Viard, M.; Kashlev, M.; Shapiro, B.A. In Silico Design and Enzymatic Synthesis of Functional RNA Nanoparticles. Acc. Chem. Res. 2014, 47, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Viard, M.; Kagiampakis, I.; Case, C.L.; Dobrovolskaia, M.A.; Hofmann, J.; Vrzak, A.; Kireeva, M.; Kasprzak, W.K.; KewalRamani, V.N.; et al. Triggering of RNA Interference with RNA–RNA, RNA–DNA, and DNA–RNA Nanoparticles. ACS Nano 2015, 9, 251–259. [Google Scholar] [CrossRef]
- Afonin, K.A.; Bindewald, E.; Yaghoubian, A.J.; Voss, N.; Jacovetty, E.; Shapiro, B.A.; Jaeger, L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010, 5, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, H.; Bao, G. Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano 2015, 9, 8655–8671. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Grabow, W.W.; Walker, F.M.; Bindewald, E.; Dobrovolskaia, M.A.; Shapiro, B.A.; Jaeger, L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 2011, 6, 2022–2034. [Google Scholar] [CrossRef] [PubMed]
- Ui-Tei, K.; Naito, Y.; Takahashi, F.; Haraguchi, T.; Ohki-Hamazaki, H.; Juni, A.; Ueda, R.; Saigo, K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004, 32, 936–948. [Google Scholar] [CrossRef]
- Reynolds, A.; Leake, D.; Boese, Q.; Scaringe, S.; Marshall, W.S.; Khvorova, A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004, 22, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Amarzguioui, M.; Prydz, H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004, 316, 1050–1058. [Google Scholar] [CrossRef] [PubMed]
- Rangan, R.; Zheludev, I.N.; Hagey, R.J.; Pham, E.A.; Wayment-Steele, H.K.; Glenn, J.S.; Das, R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: A first look. RNA 2020, 26, 937–959. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Akiyama, M.; Sakakibara, Y. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat. Commun. 2021, 12, 941. [Google Scholar] [CrossRef] [PubMed]
- Welcome to the DuplexFold Web Server [EB/OL]. Available online: https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/DuplexFold/DuplexFold.html (accessed on 5 January 2024).
- Kasprzak, W.; Bindewald, E.; Kim, T.J.; Jaeger, L.; Shapiro, B.A. Use of RNA structure flexibility data in nanostructure modeling. Methods 2011, 54, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Zhu, Y.; Wang, Y.; Li, J.; Zhang, J.; Gong, M.; Ren, W.; Li, S.; Zhong, J.; Zhang, L.; et al. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLOS Pathog. 2021, 17, e1009439. [Google Scholar] [CrossRef] [PubMed]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, W.; Fan, M.; Zhang, J.; Peng, Y.; Huang, F.; Wang, N.; He, L.; Zhang, L.; Holmdahl, R.; et al. Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification. Comput. Struct. Biotechnol. J. 2021, 19, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, D.X.; Tam, J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008, 369, 344–349. [Google Scholar] [CrossRef]
- Toussi, S.S.; Hammond, J.L.; Gerstenberger, B.S.; Anderson, A.S. Therapeutics for COVID-19. Nat. Microbiol. 2023, 8, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Abbasian, M.H.; Mahmanzar, M.; Rahimian, K.; Mahdavi, B.; Tokhanbigli, S.; Moradi, B.; Sisakht, M.M.; Deng, Y. Global landscape of SARS-CoV-2 mutations and conserved regions. J. Transl. Med. 2023, 21, 152. [Google Scholar] [CrossRef] [PubMed]
- Moazzam, M.; Zhang, M.; Hussain, A.; Yu, X.; Huang, J.; Huang, Y. The landscape of nanoparticle-based siRNA delivery and therapeutic development. Mol. Ther. 2024, 32, 284–312. [Google Scholar] [CrossRef] [PubMed]
- Otter, C.J.; Bracci, N.; Parenti, N.A.; Ye, C.; Asthana, A.; Blomqvist, E.K.; Tan, L.H.; Pfannenstiel, J.J.; Jackson, N.; Fehr, A.R.; et al. SARS-CoV-2 nsp15 endoribonuclease antagonizes dsRNA-induced antiviral signaling. Proc. Natl. Acad. Sci. USA 2024, 121, e2320194121. [Google Scholar] [CrossRef] [PubMed]
- Simeoni, M.; Cavinato, T.; Rodriguez, D.; Gatfield, D. I(nsp1)ecting SARS-CoV-2–ribosome interactions. Commun. Biol. 2021, 4, 715. [Google Scholar] [CrossRef] [PubMed]
- Calleja, D.J.; Lessene, G.; Komander, D. Inhibitors of SARS-CoV-2 PLpro. Front. Chem. 2022, 10, 876212. [Google Scholar] [CrossRef]
- Corona, A.; Madia, V.N.; De Santis, R.; Manelfi, C.; Emmolo, R.; Ialongo, D.; Patacchini, E.; Messore, A.; Amatore, D.; Faggioni, G.; et al. Diketo acid inhibitors of nsp13 of SARS-CoV-2 block viral replication. Antivir. Res. 2023, 217, 105697. [Google Scholar] [CrossRef]
- Afonin, K.A.; Dobrovolskaia, M.A.; Ke, W.; Grodzinski, P.; Bathe, M. Critical review of nucleic acid nanotechnology to identify gaps and inform a strategy for accelerated clinical translation. Adv. Drug Deliv. Rev. 2022, 181, 114081. [Google Scholar] [CrossRef] [PubMed]
- Kanarskaya, M.A.; Pyshnyi, D.V.; Lomzov, A.A. Diversity of Self-Assembled RNA Complexes: From Nanoarchitecture to Nanomachines. Molecules 2023, 29, 10. [Google Scholar] [CrossRef] [PubMed]
- Poppleton, E.; Urbanek, N.; Chakraborty, T.; Griffo, A.; Monari, L.; Göpfrich, K. RNA origami: Design, simulation and application. RNA Biol. 2023, 20, 510–524. [Google Scholar] [CrossRef]
- Afonin, K.A.; Dobrovolskaia, M.A.; Church, G.; Bathe, M. Opportunities, Barriers, and a Strategy for Overcoming Translational Challenges to Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2020, 14, 9221–9227. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Mishra, G.; Sharma, A.K.; Gothwal, A.; Kesharwani, P.; Gupta, U. Intranasal Drug Delivery: A Non-Invasive Approach for the Better Delivery of Neurotherapeutics. Pharm. Nanotechnol. 2017, 5, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Song, X.J.; Chen, X.; Wang, J.W.; Cui, Y.L. Advances and future perspectives of intranasal drug delivery: A scientometric review. J. Control. Release 2024, 367, 366–384. [Google Scholar] [CrossRef] [PubMed]



| ID | Strand | Sequence (5′-3′) | Secondary Structure | Coverage | Target Genes |
|---|---|---|---|---|---|
| siRNA1 | guide | UGUGUUUUCUCGUUGAAACCA | ((.(((((......))))))) (−0.8) | 99.83% | NSP1_N |
| passenger | GUUUCAACGAGAAAACACACG | (((((.....)))).)..... (−3.3) | |||
| siRNA2 | guide | UAUUACCGUUCUUACGAAGAA | ..((..(((....)))..)). (−2) | 99.69% | NSP1_C |
| passenger | CUUCGUAAGAACGGUAAUAAA | ((((....))).)........ (−2.3) | |||
| siRNA3 | guide | UAUUAUUGGGUAAACCUUGGG | (......((.....))....) (−0.5) | 99.36% | Nucleocapsid |
| passenger | CAAGGUUUACCCAAUAAUACU | ..((..(((.....)))..)) (−4) | |||
| siRNA4 | guide | UUGUAUAUGCGAAAAGUGCAU | .(((((.........))))). (−0.1) | 99.52% | RdRp |
| passenger | GCACUUUUCGCAUAUACAAAA | ((.......)).......... (−2.9) | |||
| siRNA5 | guide | AAUAAACACGCCAAGUAGGAG | ..........((.....)).. (−1.8) | 99.90% | Spike_S2 |
| passenger | CCUACUUGGCGUGUUUAUUCU | ((.....))...(......). (−2.8) | |||
| siRNA6 | guide | GAAUUCCAAGCUAUAACGCAG | .........((......)).. (−3.9) | 99.69% | Spike_RBD |
| passenger | GCGUUAUAGCUUGGAAUUCUA | ((......))........... (−2.3) | |||
| siRNA7 | guide | UUUCAACGUACACUUUGUUUC | ....((((.......)))).. (−4.7) | 99.77% | Spike_NTD |
| passenger | AACAAAGUGUACGUUGAAAUC | ............(.......) (−4.9) | |||
| siRNA8 | guide | GUAGCUUUGAGCGUUUCUGCU | ((((............)))). (−0.5) | 99.77% | NSP13_Stem |
| passenger | CAGAAACGCUCAAAGCUACUG | (((.(..(((...)))).))) (−0.8) | |||
| siRNA9 | guide | UUAGUUACACGAUAACCAGUA | (..((((.....))))....) (−2.8) | 99.93% | NSP13_1B |
| passenger | CUGGUUAUCGUGUAACUAAAA | .(((((((...)))))))... (−0.7) | |||
| siRNA10 | guide | ACAUCAUGCGUGAUAACACCC | ..((((....))))....... (−2.6) | 98.89% | NSP13_helicase |
| passenger | GUGUUAUCACGCAUGAUGUUU | (...(((((....)))))..) (−0.6) | |||
| siRNA11 | guide | UAAGAAUGGUCUACGUAUGCA | (..(.(((.....))).)..) (−2.7) | 99.46% | NSP13_ZBD |
| passenger | CAUACGUAGACCAUUCUUAUG | ((((...(((....))))))) (−0.4) | |||
| siRNA12 | guide | AGCUUUAGGGUUACCAAUGUC | .((....((....))...)). (−0.5) | 99.87% | NSP14 |
| passenger | CAUUGGUAACCCUAAAGCUAU | ...((((.........)))). (−3.3) | |||
| siRNA13 | guide | UAAACGAUAUGUUCGAAGGCA | ....(((.....)))...... (−2.5) | 99.79% | NSP15_NendoU domain |
| passenger | CCUUCGAACAUAUCGUUUAUG | (...(((.....))).....) (−1.8) | |||
| siRNA14 | guide | UCUACUUGACCAUCAACUCUA | (....((((...))))....) (−3) | 99.66% | NSP15_Middle domain |
| passenger | GAGUUGAUGGUCAAGUAGACU | .((((.((......)).)))) (−2.3) | |||
| siRNA15 | guide | AAAAUCUAGCACCAUAAUCAA | ..................... (−1.9) | 99.85% | NSP3 _Single-stranded poly(A) binding domain |
| passenger | GAUUAUGGUGCUAGAUUUUAC | (...((........))....) (−3.5) | |||
| siRNA16 | guide | ACAAACACGGUUUAAACACCG | .......((((......)))) (−2) | 99.76% | NSP3 _PLpro |
| passenger | GUGUUUAAACCGUGUUUGUAC | (((..(((((...)))))))) (−0.1) | |||
| siRNA17 | guide | AAUUACAACCGUCUACAACAU | ..................... (−2.9) | 99.84% | NSP3_C |
| passenger | GUUGUAGACGGUUGUAAUUCA | (...((.(....).))...). (−3.3) | |||
| siRNA18 | guide | GUAAACUACGUCAUCAAGCCA | ..................... (−2.6) | 99.54% | NSP5 |
| passenger | GCUUGAUGACGUAGUUUACUG | (........).(((....))) (−3.1) |
| siRNA Nanoparticles | siRNA | RNA Motif | Length | Target Gene |
|---|---|---|---|---|
| siRNA_nano_1 | siRNA1 | CUCCUACUUGGCGUGUUUAUUCCUGUCAAUCAUGGCAAGUGUGUUUUCUCGUUGAAACCA | 60 | NSP1_N |
| siRNA2 | UGGUUUCAACGAGAAAACACACUUGUCAUGUGUAUGUUGCCUUAUUACCGUUCUUACGAAGAA | 63 | NSP1_C | |
| siRNA5 | UUCUUCGUAAGAACGGUAAUAAGGCACAUACUUUGUUGAUAGGAAUAAACACGCCAAGUAGGAG | 64 | Spike_S2 | |
| siRNA_nano_2 | siRNA8 | AGCAGAGACGCUCGGAGCUGCUGUUUUGGUCUACUUGACCAUCAACUCUA | 50 | NSP15_Middle domain |
| siRNA14 | UAGAGUUGGUGGUCGAGUAGGCCUUUUGCAAAAUCUAGCACCAUAAUCAA | 50 | NSP3 _Single-stranded poly(A) binding domain | |
| siRNA15 | UUGAUUAUGGUGCUAGGUUUUGCUUUUCAGUAGCUUUGAGCGUUUCUGCU | 50 | NSP13_Stem |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Lu, S.; Xiao, J.; Xu, N.; Li, Y.; Xu, J.; Deng, M.; Xuanyuan, H.; Zhang, Y.; Wu, F.; et al. Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences. Viruses 2024, 16, 1072. https://doi.org/10.3390/v16071072
Sun J, Lu S, Xiao J, Xu N, Li Y, Xu J, Deng M, Xuanyuan H, Zhang Y, Wu F, et al. Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences. Viruses. 2024; 16(7):1072. https://doi.org/10.3390/v16071072
Chicago/Turabian StyleSun, Jianan, Siya Lu, Jizhen Xiao, Nuo Xu, Yingbin Li, Jinfeng Xu, Maohua Deng, Hanlu Xuanyuan, Yushi Zhang, Fangli Wu, and et al. 2024. "Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences" Viruses 16, no. 7: 1072. https://doi.org/10.3390/v16071072
APA StyleSun, J., Lu, S., Xiao, J., Xu, N., Li, Y., Xu, J., Deng, M., Xuanyuan, H., Zhang, Y., Wu, F., Jin, W., & Liu, K. (2024). Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences. Viruses, 16(7), 1072. https://doi.org/10.3390/v16071072







