Development of a CRISPR/SHERLOCK-Based Method for Rapid and Sensitive Detection of Selected Pospiviroids
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of RNA Transcripts
2.2. Plant RNA Extraction
2.3. Design of RPA Primers and crRNA
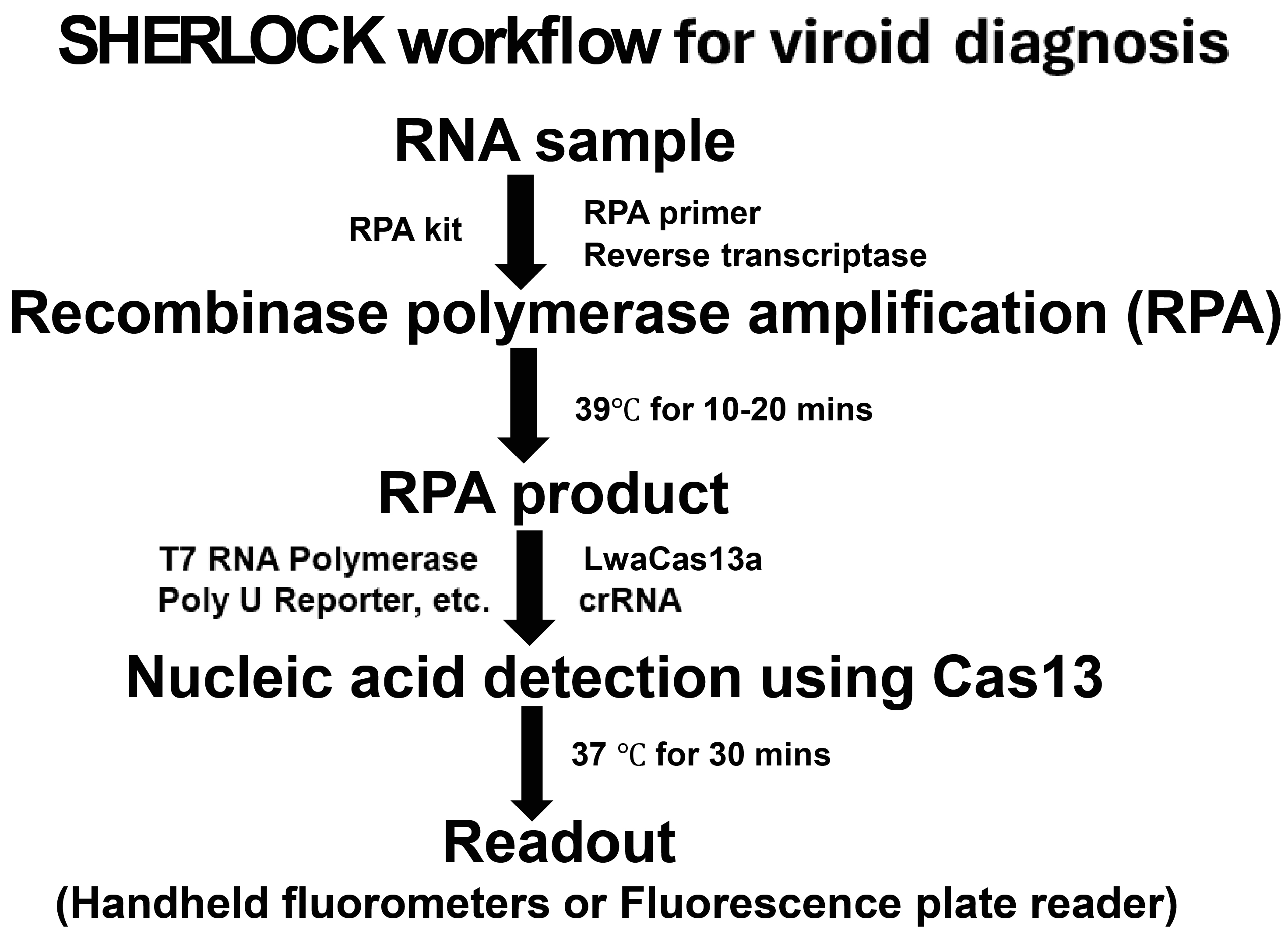
2.4. Recombinase Polymerase Amplification (RPA)
2.5. Nucleic Acid Detection Using Cas13a
2.6. Comparative Sensitivity Analysis between Real-Time RT-PCR and CRISPR-Cas13 Based-Detection
3. Results
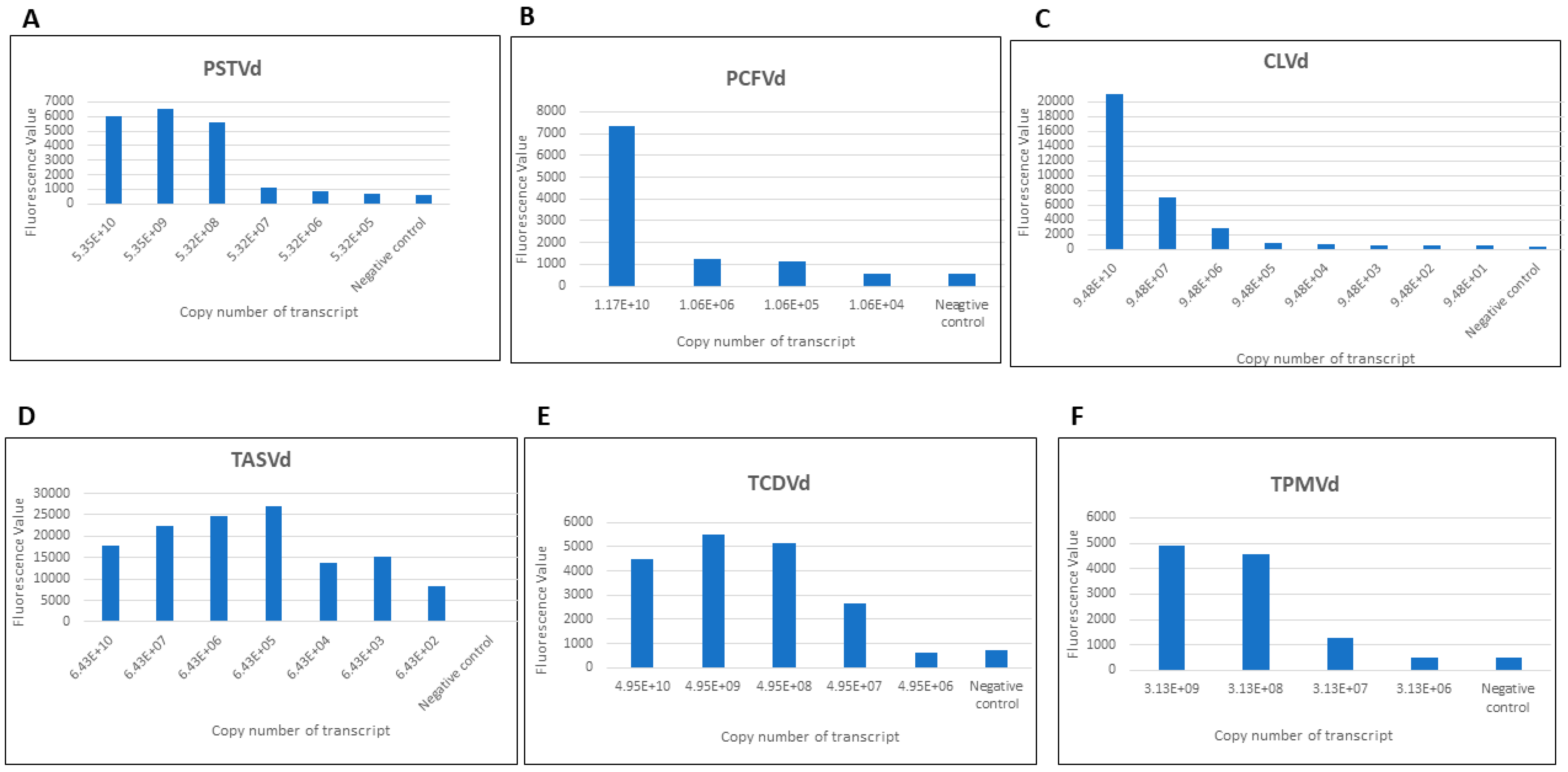
3.1. Detection of Pospiviroids with CRISPR-Cas13a
3.2. The Limits of Detection (LOD) of Monoplex and Specificity CRISPR-Cas13a Assays
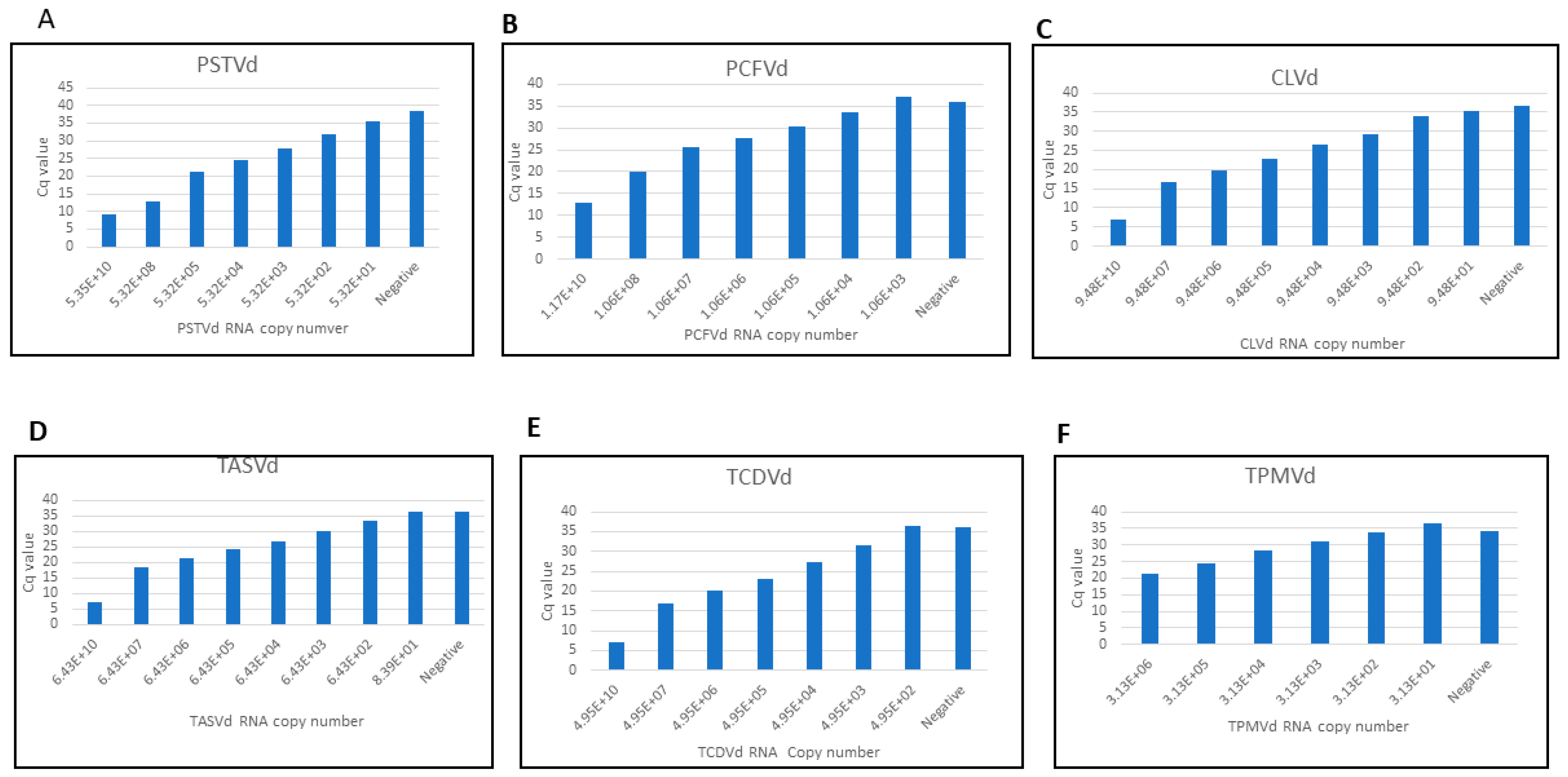
3.3. Comparative Sensitivity Analysis between Real-Time RT-PCR and CRISPR-Cas13a Based-Detection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortolá, B.; Daròs, J.A. Viroids: Non-coding circular RNAs Able to autonomously replicate and infect higher plants. Biology 2023, 12, 172. [Google Scholar] [CrossRef]
- Di Serio, F.; Flores, R.; Verhoeven, J.T.; Li, S.F.; Pallás, V.; Randles, J.W.; Sano, T.; Vidalakis, G.; Owens, R.A. Current status of viroid taxonomy. Arch. Virol. 2014, 159, 3467–3478. [Google Scholar] [CrossRef]
- Ding, B. The biology of viroid-host interactions. Annu. Rev. Phytopathol. 2009, 47, 105–131. [Google Scholar] [CrossRef]
- Flores, R.; Gas, M.E.; Molina-Serrano, D.; Nohales, M.Á.; Carbonell, A.; Gago, S.; De la Peña, M.; Daròs, J.A. Viroid replication: Rolling-circles, enzymes and ribozymes. Viruses 2009, 1, 317–334. [Google Scholar] [CrossRef]
- Palukaitis, P. What has been happening with viroids? Virus Genes 2014, 49, 175–184. [Google Scholar] [CrossRef]
- Zhang, Y.; Nie, Y.; Wang, L.; Wu, J. Viroid Replication, Movement, and the Host Factors Involved. Microorganisms 2024, 12, 565. [Google Scholar] [CrossRef]
- Verhoeven, J.T.; Roenhorst, J.W.; Owens, R.A. Mexican papita viroid and tomato planta macho viroid belong to a single species in the genus Pospiviroid. Arch. Virol. 2011, 156, 1433–1437. [Google Scholar] [CrossRef]
- Owens, R.A.; Flores, R.; Di Serio, F.; Li, S.F.; Pallas, V.; Randles, J.W.; Sano, T.; Vidalakis, G. Viroids. In Virus Taxonomy, Classification and Nomenclature of Viruses, Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: London, UK, 2012; pp. 1221–1234. [Google Scholar] [CrossRef]
- Singh, R.P.; Nie, X.; Singh, M. Tomato chlorotic dwarf viroid: An evolutionary link in the origin of pospiviroids. J. Gen. Virol. 1999, 80 Pt 11, 2823–2828. [Google Scholar] [CrossRef]
- Matsushita, Y.; Usugi, T.; Tsuda, S. Host range and properties of tomato chlorotic dwarf viroid. Eur. J. Plant Pathol. 2009, 12, 349–352. [Google Scholar] [CrossRef]
- Verhoeven, J.T.J.; Jansen, C.C.C.; Willemen, T.M.; Kox, L.F.F.; Owens, R.A.; Roenhorst, J.W. Natural infections of tomato by Citrus exocortis viroid, Columnea latent viroid, Potato spindle tuber viroid and Tomato chlorotic dwarf viroid. Eur. J. Plant Pathol. 2004, 110, 823–831. [Google Scholar] [CrossRef]
- EFSA. Scientific Opinion on the assessment of the risk of solanaceous pospiviroids for the EU territory and the identification and evaluation of risk management options. EFSA J. 2011, 9, 2330. [Google Scholar] [CrossRef]
- Verhoeven, J.T.H.J.; Huner, L.; Virscek Marn, M.; Mavric Plesko, I.; Roenhorst, J.W. Mechanical transmission of Potato spindle tuber viroid between plants of Brugmansia suaveoles, Solanumjasminoides and potatoes and tomatoes. Eur. J. Plant Pathol. 2010, 128, 417–421. [Google Scholar] [CrossRef]
- Matsushita, Y.; Tsuda, S. Seed transmission of potato spindle tuber viroid, tomato chlorotic dwarf viroid, tomato apical stunt viroid, and Columnea latent viroid in horticultural plants. Eur. J. Plant Pathol. 2016, 145, 1007–1011. [Google Scholar] [CrossRef]
- Sharma, S.K.; Gupta, O.P.; Pathaw, N.; Sharma, D.; Maibam, A.; Sharma, P.; Sanasam, J.; Karkute, S.G.; Kumar, S.; Bhattacharjee, B. CRISPR-Cas-Led revolution in diagnosis and management of emerging plant viruses: New avenues toward food and nutritional security. Front. Nutr. 2021, 8, 751512. [Google Scholar] [CrossRef]
- Zhang, W.; Jiao, Y.; Ding, C.; Shen, L.; Li, Y.; Yu, Y.; Huang, K.; Li, B.; Wang, F.; Yang, J. Rapid detection of tomato spotted wilt virus with cas13a in tomato and frankliniella occidentalis. Front. Microbiol. 2021, 12, 745173. [Google Scholar] [CrossRef]
- Duan, X.; Ma, W.; Jiao, Z.; Tian, Y.; Ismail, R.G.; Zhou, T.; Fan, Z. Reverse transcription-recombinase-aided amplification and CRISPR/Cas12a-based visual detection of maize chlorotic mottle virus. Phytopathol. Res. 2022, 4, 23. [Google Scholar] [CrossRef]
- Marqués, M.C.; Sánchez-Vicente, J.; Ruiz, R.; Montagud-Martínez, R.; Márquez-Costa, R.; Gómez, G.; Carbonell, A.; Daròs, J.A.; Rodrigo, G. Diagnostics of infections produced by the plant viruses TMV, TEV, and PVX with CRISPR-Cas12 and CRISPR-Cas13. ACS Synth. Biol. 2022, 11, 2384–2393. [Google Scholar] [CrossRef]
- Jiao, J.; Liu, Y.; Yang, M.; Zheng, J.; Liu, C.; Ye, W.; Song, S.; Bai, T.; Song, C.; Wang, M.; et al. The engineered CRISPR-Mb2Cas12a variant enables sensitive and fast nucleic acid-based pathogens diagnostics in the field. Plant Biotechnol. J. 2023, 21, 1465–1478. [Google Scholar] [CrossRef]
- Lei, R.; Kuang, R.; Peng, X.; Jiao, Z.; Zhao, Z.; Cong, H.; Fan, Z.; Zhang, Y. Portable rapid detection of maize chlorotic mottle virus using RT-RAA/CRISPR-Cas12a based lateral flow assay. Front. Plant Sci. 2023, 14, 1088544. [Google Scholar] [CrossRef]
- Wang, J.; Huang, X.; Chen, S.; Chen, J.; Liang, Z.; Chen, B.; Yang, X.; Zhou, G.; Zhang, T. On-site and visual detection of sorghum mosaic virus and rice stripe mosaic virus based on reverse transcription-recombinase-aided amplification and CRISPR/Cas12a. Front. Genome Ed. 2023, 5, 1124794. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, S.; Dong, Z.; Fan, Q.; Lei, R.; Kuang, R.; Zhang, Y. One-Stepreverse-transcription recombinase-aided amplification CRISPR/CAS12a-based lateral flow assay for fast field screening and accurate differentiation of four major tobamoviruses infecting tomato and pepper. J. Agric. Food Chem. 2023, 71, 17025–17035. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, H.J.; Cho, I.S.; Jeong, R.D. Identification of viruses infecting phalaenopsis orchids using nanopore sequencing and development of an RT-RPA-CRISPR/Cas12a for rapid visual detection of nerine latent virus. Int. J. Mol. Sci. 2024, 25, 2666. [Google Scholar] [CrossRef]
- Xu, T.; Yang, X.; Feng, X.; Luo, H.; Luo, C.; Jia, M.A.; Lei, L. Sensitive and visual detection of brassica yellows virus using reverse transcription loop-mediated isothermal amplification-coupled CRISPR-Cas12 assay. Phytopathology 2024, 114, 474–483. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Wacker, M.J.; Godard, M.P. Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J. Biomol. Tech. 2005, 16, 266–271. [Google Scholar]
- Flores, R.; Gago-Zachert, S.; Serra, P.; Sanjuán, R.; Elena, S.F. Viroids: Survivors from the RNA world? Annu. Rev. Microbiol. 2014, 68, 395–414. [Google Scholar] [CrossRef]
- Drygin, Y.; Gasanova, T.; Butenko, K. Polyclonal antibodies against potato spindle tuber viroid RNA. Front. Biosci. 2022, 14, 7. [Google Scholar] [CrossRef]
- Nie, X.Z.; Singh, R.P. Viroid detection and identification by bioassay. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: New York, NY, USA, 2017; pp. 347–356. [Google Scholar] [CrossRef]
- Gucek, T.; Trdan, S.; Jakse, J.; Javornik, B.; Matousek, J.; Radisek, S. Diagnostic techniques for viroids. Plant Pathol. 2017, 66, 339–358. [Google Scholar] [CrossRef]
- Li, Y.; Mansour, H.; Wang, T.; Poojari, S.; Li, F. Naked-Eye Detection of Grapevine Red-Blotch Viral Infection Using a Plasmonic CRISPR Cas12a Assay. Anal. Chem. 2019, 91, 11510–11513. [Google Scholar] [CrossRef]
- Aman, R.; Mahas, A.; Marsic, T.; Hassan, N.; Mahfouz, M.M. Efficient, Rapid, and Sensitive Detection of Plant RNA Viruses with One-Pot RT-RPA-CRISPR/Cas12a Assay. Front. Microbiol. 2020, 11, 610872. [Google Scholar] [CrossRef]
- Mahas, A.; Hassan, N.; Aman, R.; Marsic, T.; Wang, Q.; Ali, Z.; Mahfouz, M.M. LAMP-Coupled CRISPR-Cas12a Module for Rapid and Sensitive Detection of Plant DNA Viruses. Viruses 2021, 13, 466. [Google Scholar] [CrossRef]
- Alon, D.M.; Hak, H.; Bornstein, M.; Pines, G.; Spiegelman, Z. Differential Detection of the Tobamoviruses Tomato Mosaic Virus (ToMV) and Tomato Brown Rugose Fruit Virus (ToBRFV) Using CRISPR-Cas12a. Plants 2021, 10, 1256. [Google Scholar] [CrossRef]
- Costa, L.C.; Atha, B., 3rd; Hu, X.; Lamour, K.; Yang, Y.; O’Connell, M.; McFarland, C.; Foster, J.A.; Hurtado-Gonzales, O.P. High-throughput detection of a large set of viruses and viroids of pome and stone fruit trees by multiplex PCR-based amplicon sequencing. Front. Plant Sci. 2022, 13, 1072768. [Google Scholar] [CrossRef]
- Fernandez I Marti, A.; Parungao, M.; Hollin, J.; Selimotic, B.; Farrar, G.; Seyler, T.; Anand, A.; Ahmad, R. A novel, precise and high-throughput technology for viroid detection in cannabis (MFDetectTM). Viruses 2023, 15, 1487. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Li, Z.X.; Du, Y.J.; Li, S.F.; Zhang, Z.X. A universal probe for simultaneous detection of six pospiviroids and natural infection of potato spindle tuber viroid (PSTVd) in tomato in China. J. Integr. Agric. 2023, 22, 790–798. [Google Scholar] [CrossRef]
- Guček, T.; Jakše, J.; Radišek, S. Optimization and validation of singleplex and multiplex rt-qpcr for detection of citrus bark cracking viroid (CBCVd), hop latent viroid (HLVd), and hop stunt viroid (HSVd) in hops (Humulus lupulus). Plant Dis. 2023, 107, 3592–3601. [Google Scholar] [CrossRef]



| Combination | RPA Primer | Sequence (5′-3′) | crRNA Using LwaCas13a(5′-3′) | Target |
|---|---|---|---|---|
| PSTVd combination 1 | PSTVd-RPA-For-1 | GAAATTAATACGACTCACTATAGGGACTAAACTCGTGGTTCCTGTGGTTCACAC | GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACCCCUGAAGCGCUCCUCCGAGCCGCCUUC | PSTVd |
| PSTVd-RPA-Rev-3 | CTCCCCACCGTCCTTATTGCCAGTTCGCT | |||
| PSTVd combination 2 | PSTVd-RPA-For-2 | GAAATTAATACGACTCACTATAGGGAGTAATTCCCGCCGAAACAGGGTTTTCAC | GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACAGGGGGCGAGGGGUGGUCCUGCGGGCGC | |
| PSTVd-RPA-Rev-2 | TTCTCGGGAGCTTCAGTTGTTTCCACCGGGTA | |||
| PCFVd combination 1 | PCFVd-RPA-For-1 | GAAATTAATACGACTCACTATAGGGTAGGGAAAAGAAAGGGGAAGCAAGCATCTC | GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACCUUCUCCGCCCGGUCUGUCCAGGUUUCC | PCFVd |
| PCFVd-RPA-Rev-1 | CTGCTGGGATTACTCCTGTCAGAAGACGGT | |||
| PCFVd combination 2 | PCFVd-RPA-For-2 | GAAATTAATACGACTCACTATAGGGAAACAGGGTTTTCACCCTTCCTTTCTTCG | GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACGUGCGCGAGAAGGCCGACGCGGACCGGU | |
| PCFVd-RPA-Rev-2 | GCACCTCTGTCAGTTGTATCCACCGGGTAG | |||
| CLVd combination 1 | CLVd-RPA-For-3 | GAAATTAATACGACTCACTATAGGGCCATGCAAAAGAAAAAAGAACGGGAGGG | GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACGCUCGGUCUGAGUUGCCCCGGGGCUCCU | CLVd |
| CLVd-RPA-Rev-3 | CTCCTGTCTGAACAGGGCAACGCCCTCGAC | |||
| CLVd combination 2 | CLVd-RPA-For-3 | GAAATTAATACGACTCACTATAGGGCCATGCAAAAGAAAAAAGAACGGGAGGG | ||
| CLVd-RPA-Rev-4 | AGGAAGGGTGAAAACCCTGTTTCAGCTGGG | |||
| TASVd combination 1 | TASVd-RPA-For-2 | GAAATTAATACGACTCACTATAGGGCAGCTGAAACAGGGTTTTCACCCTTCC | GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACGGCGAGCGCCGAAGACCUUCCGGCGAGA | TASVd |
| TASVd-RPA-Rev-2 | CCGTGGAGTCGAAGCTTCAGTTGTTTCC | |||
| TASVd combination 2 | TASVd-RPA-For-2 | GAAATTAATACGACTCACTATAGGGCAGCTGAAACAGGGTTTTCACCCTTCC | ||
| TASVd-RPA-Rev-5 | AGATAGAGAAAAAGAGCCGTGGAGTCGAAGC | |||
| TCDVd combination | TCDVd-RPA-For-1 | GAAATTAATACGACTCACTATAGGGGTGGTTCCTGTGGTTCACACCTGACCTCC | GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACGUUUCCCCGGGGAUCCCUGAAGCGCUCC | TCDVd |
| TCDVd-RPA-Rev-1 | CCTGTTTCGCCTTCCACAAGCTCCCTGC | |||
| TPMVd combination | TPMVd-RPA-For-1 | GAAATTAATACGACTCACTATAGGGGTGGTTCCTGTGGTTCACACCTGACCTCC | GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACGGGAUCCCUGAAGCGCUCCUUUGGCCGC | TPMVd |
| TPMVd-RPA-Rev-1 | CAGCGGGGATTACTCCTGTCTGGGAGAC |
| PSTVd Transcript | PCFVd Transcript | CLVd Transcript | TASVd Transcript | TCDVd Transcript | TPMVd Transcript | Healthy Host | |
|---|---|---|---|---|---|---|---|
| PSTVd combination 1 | + | - | - | - | + | - | - |
| PCFVd combination 1 | - | + | - | - | - | - | - |
| CLVd combination 1 | - | - | + | + | + | + | - |
| TASVd combination 1 | - | - | + | + | + | + | - |
| TCDVd combination | + | - | - | + | + | + | - |
| TPMVd combination | + | - | - | - | + | + | - |
| Real-Time RT-PCR Primer | Sequence (5′-3′) |
|---|---|
| PSTVd-For | CTGGAGCGAACTGGCAATAA |
| PSTVd-Rev | CCGAAGAAAGGAAGGGTGAAA |
| PCFVd-For | CTTTCTTCGGGTTTCCTTCCT |
| PCFVd-Rev | AAGCACCTCTGTCAGTTGTATC |
| CLVd-For | ACCCTTCCTTTCTTCTGGTTTC |
| CLVd-Rev | CCGGAGACCAAGCTAAGATAGA |
| TASVd-For | ACCCTTCCTTTCTTCTGGTTTC |
| TASVd-Rev | GGAGTCGAAGCTTCAGTTGTT |
| TCDVd-For | TCCTTTCTTCTGCGGTTTCC |
| TCDVd-Rev | TCGGGAGCTTCAGTTGTTTC |
| TPMVd-For | CTTCCTTTCTTCGGGTTTCCT |
| TPMVd-Rev | TGGGAGCTTCAGTTGTTTCC |
| PSTVd Transcript | PCFVd Transcript | CLVd Transcript | TASVd Transcript | TCDVd Transcript | TPMVd Transcript | Healthy Host | |
|---|---|---|---|---|---|---|---|
| PSTVd real-time RT-PCR primers | + | - | + | + | + | - | - |
| PCFVd real-time RT-PCR primers | - | + | - | - | - | - | - |
| CLVd real-time RT-PCR primers | - | - | + | + | + | - | - |
| TASVd real-time RT-PCR primers | - | - | + | + | + | + | - |
| TCDVd real-time RT-PCR primers | - | - | + | + | + | - | - |
| TPMVd real-time RT-PCR primers | + | + | + | + | + | + | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, Y.; Gnanasekaran, P.; Pappu, H.R. Development of a CRISPR/SHERLOCK-Based Method for Rapid and Sensitive Detection of Selected Pospiviroids. Viruses 2024, 16, 1079. https://doi.org/10.3390/v16071079
Zhai Y, Gnanasekaran P, Pappu HR. Development of a CRISPR/SHERLOCK-Based Method for Rapid and Sensitive Detection of Selected Pospiviroids. Viruses. 2024; 16(7):1079. https://doi.org/10.3390/v16071079
Chicago/Turabian StyleZhai, Ying, Prabu Gnanasekaran, and Hanu R. Pappu. 2024. "Development of a CRISPR/SHERLOCK-Based Method for Rapid and Sensitive Detection of Selected Pospiviroids" Viruses 16, no. 7: 1079. https://doi.org/10.3390/v16071079
APA StyleZhai, Y., Gnanasekaran, P., & Pappu, H. R. (2024). Development of a CRISPR/SHERLOCK-Based Method for Rapid and Sensitive Detection of Selected Pospiviroids. Viruses, 16(7), 1079. https://doi.org/10.3390/v16071079










